Abstract
Phosphoinositide 3-kinases (PI 3-kinases) are regulated by a diverse range of upstream activators, including receptor tyrosine kinases (RTKs), G-protein-coupled receptors (GPCRs), and small GTPases from the Ras, Rho and Rab families. For the Class IA PI 3-kinase PI3Kβ, two mechanisms for GPCR-mediated regulation have been described: direct binding of Gβγ subunits to the C2-helical domain linker of p110β, and Dock180/Elmo1-mediated activation of Rac1, which binds to the Ras-Binding Domain of p110β. We now show that the integration of these dual pathways is unexpectedly complex. In breast cancer cells, expression of constitutively activated Rac1 (CA-Rac1) along with either GPCR stimulation or expression of Gβγ led to an additive PI3Kβ-dependent activation of Akt. Whereas CA-Rac1-mediated activation of Akt was blocked in cells expressing a mutated PI3Kβ that cannot bind Gβγ, Gβγ and GPCR-mediated activation of Akt was preserved when Rac1 binding to PI3Kβ was blocked. Surprisingly, PI3Kβ-dependent CA-Rac1 signaling to Akt was still seen in cells expressing a mutant p110β that cannot bind Rac1. Instead of directly binding to PI3Kβ, CA-Rac1 acts by enhancing Gβγ coupling to PI3Kβ, as CA-Rac1-mediated Akt activation was blocked by inhibitors of Gβγ. Cells expressing CA-Rac1 exhibited a robust induction of macropinocytosis, and inhibitors of macropinocytosis blocked the activation of Akt by CA-Rac1 or lysophosphatidic acid. Our data suggest that Rac1 can potentiate the activation of PI3Kβ by GPCRs through an indirect mechanism, by driving the formation of macropinosomes that serve as signaling platforms for Gβγ coupling to PI3Kβ.
Introduction
Class I PI 3-kinases comprise four distinct catalytic and seven distinct regulatory subunits. They mediate a wide range of non-redundant signaling events in distinct tissues and cell types [1]. PI3Kβ, composed of either p85α or p85β regulatory subunits and the p110β catalytic subunit, has been implicated in thrombosis, spermatogenesis, and tumorigenesis in tumors lacking the PTEN tumor suppressor [2–5]. At the cellular level, PI3Kβ plays a role in vesicular trafficking, macroautophagy, and integrin signaling [3,4,6–8].
While all class I PI 3-kinases integrate activating and inhibitory inputs from multiple upstream regulators, the control of PI3Kβ activity is particularly complex. PI3Kβ is predicted to be strongly activated by receptor tyrosine kinases (RTKs) through SH2 domains in p85 [9]. PI3Kβ is also activated by G-protein-coupled receptors (GPCRs), as Gβγ subunits from trimeric G-proteins bind directly to a surface loop in p110β [10]. Gβγ and tyrosine phosphorylated peptides show strong synergistic activation of PI3Kβ in vitro [11]. However, several studies have shown that RTK activation of PI3Kβ is weak relative to PI3Kα, even in cells that contain similar levels of both isoforms [12,13]. The reason for this is not yet clear, but could reflect the specific targeting of PI3Kβ to cellular regions that preclude its binding to RTKs. In contrast, studies in leukocytes show that PI3Kβ is selectively responsive to a combined RTK-GPCR stimulus [14]. Finally, GPCRs can also activate PI3Kβ through the Dock180/Elmo1-mediated activation of the small Rho GTPase Rac1, which binds to the Ras-Binding Domain (RBD) of p110β [15]. The ability of Rac1 (and Cdc42) to stimulate PI3Kβ activity suggests that the activation of these GTPases downstream from RTKs might also activate PI3Kβ, although this has not yet been tested.
Given the dual pathways by which GPCRs activate PI3Kβ, we sought to examine the integration of these inputs in intact cells. Unlike previous studies in MEFs [15,16], we do not see a requirement for Rac1 binding to p110β during GPCR-mediated activation of PI3Kβ. However, we have uncovered a novel mechanism for the effects of Rac1 on PI3Kβ. Our data suggest that activated Rac1 drives the formation of macropinosomes, which enhance the coupling of Gβγ to PI3Kβ. These studies highlight the increasingly complex biology of PI3Kβ regulation in mammalian cells.
Experimental procedures
Antibodies and reagents
Mouse myc, rabbit pT308-Akt, and rabbit Akt antibodies were purchased from Cell Signaling Technology. Mouse FLAG, mouse GRK2, and mouse α-tubulin antibodies were purchased from Sigma. Rabbit GFP antibody was a gift from Dr Erik Snapp, Janelia Research Campus, HHMI; in some figures GFP antibody was from Cell Signaling Technology. Lysophosphatidic acid (LPA) and epidermal growth factor (EGF) were purchased from Sigma–Aldrich and Millipore, respectively. Rhodamine phalloidin and 70 kDa Rhodamine Dextran were purchased from Invitrogen. 5-(n-Ethyl-N-isopropyl) amiloride (EIPA) and Latrunculin B were from Sigma–Aldrich and CalBiochem, respectively. The expression construct for GRKct-PM was a gift from Dr Philip Wedegaertner, Thomas Jefferson University. The mammalian expression constructs for GFP-CA-Rac1 and GFP-DN-Rac1 were assembled by cloning eGFP and Rac1(Q61L) or Rac1(T17N), respectively, into pcDNA3.1 vectors.
Cell lines and transfections
HEK293T cells and the human breast cancer cell line MDA-MB-231 were obtained from American Type Culture Collection (ATCC) and maintained in DMEM containing 10% FBS and supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. MDA-MB-231 knockdown cells, and cells in which endogenous p110β is replaced by physiological levels of murine wild-type p110β or a GPCR-uncoupled mutant (p110βKK-DD), have been previously described [17]. When indicated, stable p110β knockdown cells were transiently transfected with wild-type or mutant p110β, p110βKK-DD, or a Rac binding-deficient mutant (S205D/K224A; p110βRBD), using Lipofectamine 3000 (Thermo-Fisher) according to manufacturer instructions.
Akt activation
Cells were transiently transfected with GFP-CA-Rac1 or a bicistronic vector encoding Gβγ using Lipofectamine 3000 as per manufacturer instructions. Alternatively, cells were starved for 18 h in serum-free medium supplemented with 0.1% BSA and then stimulated with 10 μM LPA for 5 min at 37°C. Cells were lysed in 2× Laemmli sample buffer with phosphatase inhibitor cocktails (Sigma–Aldrich and Calbiochem) and boiled for 5 min, and lysates were separated by SDS–PAGE and blotted as described. Blots were visualized using ECL (Thermo-Fisher) and quantitated by densitometry using ImageJ.
Inhibitor treatments
When indicated, cells were transfected with GFP-DN-Rac1 or GRKct-PM 48 h prior to experiments. When indicated, cells were treated with TGX-221 (100 nM for 30 min prior to assay), pertussis toxin (200 ng/ml for 1 h prior to assay), EIPA (100 nM for 3 h prior to assay), or Latrunculin B (0.5 μM for 15 min prior to assay). Control cells in the TGX-221, EIPA, and Latrunculin B experiments were treated with 0.1% DMSO.
EGF stimulation and phalloidin staining
MDA-MB-231 cells were plated on glass coverslips and transfected with the indicated plasmids. After 48 h, cells were starved for 18 h in serum-free medium supplemented with 0.1% BSA and stimulated with 5 nM EGF for 5 min at 37°C. Cells were then fixed with 4% paraformaldehyde, stained with rhodamine phalloidin, and imaged using a 1.4 N.A. 60× objective and a Nikon Eclipse fluorescence microscope.
Protein purification
GST-CA-Rac1Q61L in pGEX6p-1 was transformed into BL21 Escherichia coli and protein expression was induced with 0.4 mM isopropyl β-D-thiogalactoside overnight at 18°C. Bacterial cells were harvested by centrifugation and pellets resuspended in 1× PBS containing 4 mM DTT, 2 mM EDTA, 2 mM PMSF, 2.5 units/ml nuclease (Thermo Scientific), and protease inhibitor tablets (Roche Diagnostics). The cells were sonicated and TritonX-100 was added to a final concentration of 1%. Lysates were rotated at 4°C for 20 min and centrifuged at 27 000 × g for 15 min at 4°C. Cleared lysates were incubated with glutathione-agarose beads (Thermo Scientific) on a rotating wheel at 4°C for 2 h. The beads were washed three times in 50 mM Tris pH 8.0, 150 mM NaCl and stored in 50% glycerol at −20°C. For use in kinase assays, CA-Rac1 was cleaved overnight at 4°C with PreScission Protease (GE Healthcare) in 50 mM Tris pH 8.0, 150 mM NaCl, 5 mM MgCl2, and 1 mM DTT. The cleaved material was eluted, brought to 10% glycerol, and stored at −80°C. Purification of heterologously expressed Gβ1γ2 in Sf9 cells following infection with recombinant baculovirus has been recently described [18]. Proteins were stored at −80°C until use.
In vitro binding assay
HEK293T cells were transfected with murine or human myc-p110βWT or myc-p110βKK-DD. After 48 h, cells were lysed in 20 mM Tris pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1% TritonX-100, and protease inhibitors. Glutathione beads containing GST or GST-CA-Rac1 were loaded with 2 mM GTPγS for 20 min at 37°C in loading buffer (20 mM Tris pH X.X, 5 mM EDTA, 25 mM EDTA, and 1 mM DTT). MgCl2 was then added at a final concentration of 10 mM, and the beads were placed on ice for 10 min. GST or GST-CA-Rac1 beads (~25 μg of protein per sample) were added to cell lysates, and the samples were incubated on a rotating wheel at 4°C for 2 h. The beads were washed three times in lysis buffer, boiled in 2× Laemmli sample buffer, and analyzed on a 7.5% SDS–PAGE followed by blotting with a myc antibody.
Lipid kinase assay
Myc-tagged human p110β and HA-tagged bovine p85 were co-expressed in HEK293T cells and immunopurified with myc antibodies and Protein G beads. The beads were washed with 40 mM HEPES pH 7.4, 0.1% BSA, 1 mM EGTA, 7 mM MgCl2, 120 mM NaCl, 1 mM β-glycerophosphate, 1 mM DTT, and resuspended in 40 μl of the same buffer. The beads were incubated in the presence of 45 μg sonicated phosphatidylinositol and 100 μM ATP containing 10 μCi [32P]-ATP, without or with 10 μM Rac1, with shaking for 10 min. Reactions were stopped by the addition of 80 μl of 3.2 M HCl and 160 μl 1 : 1 chloroform:methanol, vortexed, and centrifuged at 13 000×g for 5 min. Lipids in the lower phase were separated by the thin layer chromatgraphy in chloroform:methanol:water:ammonium hydroxide 60 : 47 : 11.3 : 2, and quantitated using a Molecular Dynamics Phosphorimager.
Macropinocytosis assay
Acid washed 18 mm coverslips were rinsed twice with PBS and placed in 12-well plates. p110β knockdown cells transfected with wild-type p110β, p110βKK-DD, or p110βRBD, along with GFP-CA-Rac1 or GFP. After 48 h, cells were seeded at 75 × 103 cells per coverslip in complete medium. Cells were pre-incubated with 0.1% DMSO or 25 μM EIPA for 90 min, washed, and then incubated with 1 mg/ml 70 KDa rhodamine-Dextran for 30 min at 37°C. Cells were transferred to ice, washed five times with ice-cold PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were then washed twice with 1× PBS, mounted on glass slides using DAPI Fluoromount-G® (SouthernBiotech), sealed with nail polish and imaged immediately. Image acquisition was performed using a Nikon Eclipse fluorescence microscope equipped with phase contrast and rhodamine, DAPI and FITC filters. Images were acquired with a 40×, 0.075 N.A. objective.
Analysis of macropinosomes
Macropinosomes were analyzed by a modification of the method of Commisso et al. [19]. Briefly, images were analyzed in ImageJ. For a given experiment, a phase contrast image and corresponding fluorescence image with bright discrete macropinocytotic punctae were used to define analysis settings. The Set Scale function used the following parameters: distance in pixels: 620; known distance: 0.1; unit of length: mm. Background subtraction was performed using a rolling ball radius of 10 pixels. Images were adjusted using the Image:Adjust:Threshold commands, selecting Dark Background and Auto. The resulting thresholding was compared with a duplicate of the original image to make sure that all macropinosomes were selected, and manual adjustments were made to the threshold value, if necessary; the Apply function was then chosen. The outline of the cell was traced from the phase contrast image using the Polygon selections function, and this outline was superimposed on the thresholded fluorescent image. Finally, the number of macropinosomes was counted using the Analyze Particles function, using a Size setting of 0.00000044178-Infinity to select particles greater than 0.79 μm. Thresholding settings from the first image were recorded and used for the analysis of subsequent images from the same data set.
Statistical analysis
Statistical analyses were done using one-way ANOVA. A P-value less than 0.05 was considered statistically significant.
Results
To study the integration of signaling from Gβγ and Rac1 by PI3Kβ, we used stable MDA-MB-231 cells in which endogenous p110β is knocked down and replaced by physiological levels of wild-type p110β or a mutant (p110βKK-DD) that is defective for binding to Gβγ [17]. In vitro, this mutation has no effect on basal PI3Kβ activity or activation by tyrosine phosphopeptides, but abolishes activation by Gβγ [10]. MDA-MB-231 cells expressing mutant PI3Kβ show normal EGF-stimulated Akt activation but are defective for responses to GPCR ligands [17].
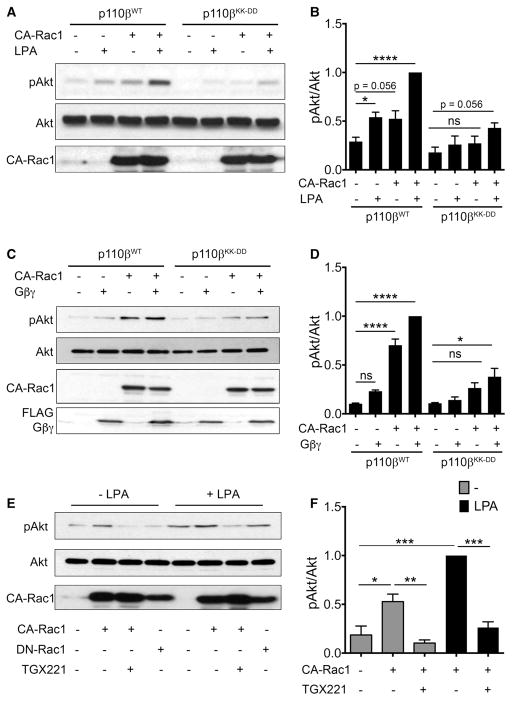
To produce comparable levels of Rac activation in cells expressing wild-type versus mutant p110β, we transfected the cells with constitutively active Rac1 (Rac1Q61L; CA-Rac1). Phosphorylation of Akt at T308 was used as readout for PI3Kβ activation. In cells expressing wild-type PI3Kβ, treatment with LPA or transfection of CA-Rac1 led to a two-fold increase in Akt phosphorylation, and Akt phosphorylation increased nearly four-fold when cells were stimulated with both CA-Rac1 and LPA (Figure 1A, B). LPA-stimulated Akt phosphorylation was markedly reduced in cells expressing GPCR-uncoupled PI3Kβ, consistent with our previous work [17]. Surprisingly, stimulation of Akt phosphorylation by CA-Rac1 in cells expressing PI3KβKK-DD was also reduced, with no significant difference from untreated cells. The additive activation of Akt by LPA plus CA-Rac1 was reduced by ~60% in cells expressing GPCR-uncoupled p110β. Similar data were obtained when we transfected cells with recombinant Gβγ rather than stimulating with LPA. In cells expressing PI3KβKK-DD, activation of Akt by Gβγ and CA-Rac1, as well as the additive activation in cells expressing both Gβγ and CA-Rac1, were both inhibited (Figure 1C, D). To demonstrate that the activation of Akt by CA-Rac1 is a valid readout for activation of PI3Kβ, we treated cells with the PI3Kβ-specific inhibitor TGX-221. Akt activation by CA-Rac1 and additive activation by CA-Rac1 and LPA were both abolished by treatment with TGX-221 (Figure 1E, F).
Figure 1. CA-Rac1 does not activate GPCR-uncoupled p110β.
(A) MDA-MB-231 cells stably expressing wild-type or GPCR uncoupled p110β (p110βKK-DD) were transiently transfected with control plasmid or GFP-CA-Rac1, and stimulated with or without 10 μM LPA for 5 min. Representative immunoblots show pT308-Akt, total Akt, and GFP-CA-Rac1. (B) Quantitation of pAkt/Akt ratios from immunoblots in (A). pAkt/Akt ratios in each experiment were normalized to that seen in LPA-stimulated cells expressing wild-type p110β and GFP-CA-Rac1. The data represent the mean ± SEM from four independent experiments. (C) MDA-MB-231 cells stably expressing wild-type p110β or p110βKK-DD were transiently transfected with control plasmid, GFP-CA-Rac1, and/or Flag-Gβγ plasmids. Representative immunoblots show pT308-Akt, total Akt, GFP-CA-Rac1, and Flag-Gβγ. (D) Quantitation of pAkt/Akt ratios from immunoblots in (C). pAkt/Akt ratios in each experiment were normalized to that seen in cells simultaneously expressing wild-type p110β, GFP-CA-Rac1, and Gβγ. The data represent the mean ± SD from two independent experiments. (E) Parental MDA-MB-231 cells were transiently transfected with GFP-CA-Rac1 or GFP-DN-Rac1, and stimulated with LPA and treated with TGX-221 as indicated. Representative immunoblots show pT308-Akt, total Akt, and GFP-CA-Rac1. (F) Quantitation of pAkt/Akt ratios from (E) pAkt/Akt ratios in each experiment were normalized to that seen in cells expressing GFP-CA-Rac1 without TGX-221 treatment. Quantitation of the DN-Rac1 data in (E) is shown in Figure 3A. *P < 0.05; **P < 0.01; ****P < 0.0001; ns: not significant.
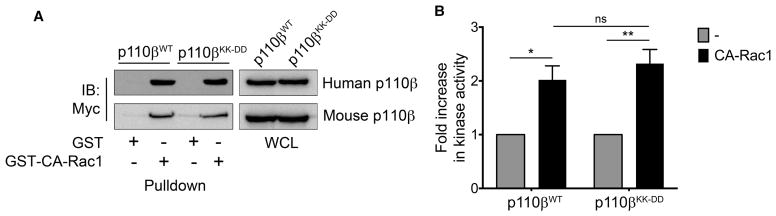
Based on the structure of p110β [9], the binding sites for Gβγ (in the C2-helical linker) and Rac1 (in the RBD) reside in distinct domains of the protein, suggesting that a point mutation at one site should not affect binding at the other site. To confirm that mutation of the Gβγ-binding site did not affect Rac1 binding to the RBD, we measured the binding of recombinant PI3Kβ to immobilized GST-CA-Rac1. Similar levels of wild-type and mutant PI3Kβ bound to GST-CA-Rac1, but not GST beads (Figure 2A). We also measured the activation of recombinant PI3Kβ by CA-Rac1 in vitro and observed a two-fold activation of both wild-type and mutant PI3Kβ (Figure 2B). Therefore, the requirement for an intact Gβγ binding in p110β for Rac1 activation of PI3Kβ is observed in cells but not in vitro.
Figure 2. GPCR-uncoupled p110β binds and is activated by CA-Rac1 in vitro.
(A) Representative immunoblots showing binding of human (upper panel) and mouse (lower panel) myc-tagged wild-type p110β or p110βKK-DD to GST or GST-CA-Rac1 beads. (B) In vitro lipid kinase activity of human wild-type p110β or p110βKK-DD with or without 10 μM recombinant GST-CA-Rac1. Values were normalized to those obtained for each protein in the absence of GFP-CA-Rac1. The data represent the mean ± SEM from three independent experiments. *P < 0.05; **P < 0.01; ns: not significant.
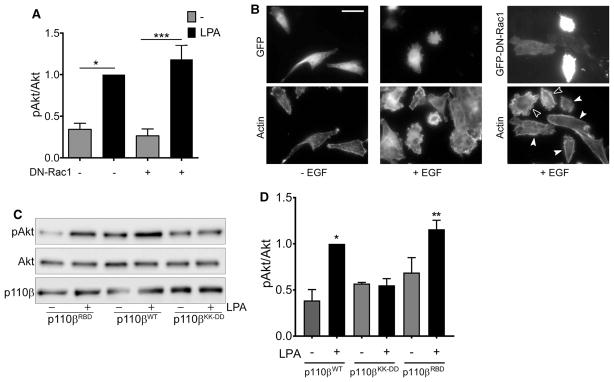
The relationship between Gβγ and Rac during the activation of PI3Kβ was not reciprocal, as LPA stimulation of Akt phosphorylation was unaffected by overexpression of dominant negative Rac1 (DN-Rac1) (Figure 3A). To confirm that the expression of DN-Rac1 was interfering with Rac1 signaling, we expressed GFP or GFP-DN-Rac1 in parental MDA-MB-231 cells and stimulated with EGF. Cells expressing GFP (Figure 3B; middle panel) and non-transfected cells (Figure 3B; right panel, open arrowheads) showed extensive membrane ruffiing in response to EGF (compare to unstimulated cells in Figure 3B, left panel). In contrast, EGF-stimulated membrane ruffiing was inhibited in cells expressing GFP-DN-Rac1 (Figure 3B; right panel, solid arrowheads).
Figure 3. LPA-stimulated activation of PI3Kβ does not require Rac1 binding.
(A) Parental MDA-MB-231 cells were transfected with GFP or GFP-DN-Rac1 and stimulated with or without 10 μM LPA for 5 min. pAkt/Akt ratios in each experiment were normalized to that seen in LPA-stimulated cells without DN-Rac1. The data represent the mean ± SEM from three independent experiments. Western blot data from one of the three experiments is shown in Figure 1E. (B) Representative micrographs of parental MDA-MB-231 cells transfected with GFP or GFP-DN-Rac1 (upper panels) and stimulated without or with EGF. Cells were stained with rhodamine phalloidin to visualize F-actin (lower panels). Open arrowheads indicate untransfected cells that show membrane ruffiing. Closed arrowheads show transfected cells with reduced ruffiing. Scale bar = 50 μm. (C) Stable p110β knockdown cells were transiently transfected with wild-type p110β, p110βKK-DD, or p110βRBD. Forty-eight hours after transfection, cells were starved overnight and then stimulated with 10 μM LPA for 5 min. Cells were lysed and blotted for p110β, Akt, and pT308-Akt. (D) Quantitation of pAkt/Akt immunoblots in Figure 3D. pAkt/Akt ratios in each experiment were normalized to that seen in cells expressing wild-type p110β and stimulated with LPA. The data represent the mean ± SEM from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant.
We also measured LPA-stimulated Akt phosphorylation in MDA-MB-231 p110β knockdown cells transiently transfected with wild-type p110β, p110βKK-DD, or a mutant p110β that is defective for binding to Rac1 (S205D/K224A [15]; p110βRBD;). LPA stimulation of pT308-Akt was observed in cells expressing wild-type p110β, as well as in cells expressing the Rac1 binding-deficient p110βRBD (Figure 3C, D). In contrast, the expression of GPCR-uncoupled p110βKK-DD did not support Akt phosphorylation. These data show that in breast cancer cells, LPA stimulation of Akt does not require PI3Kβ binding to Rac1.
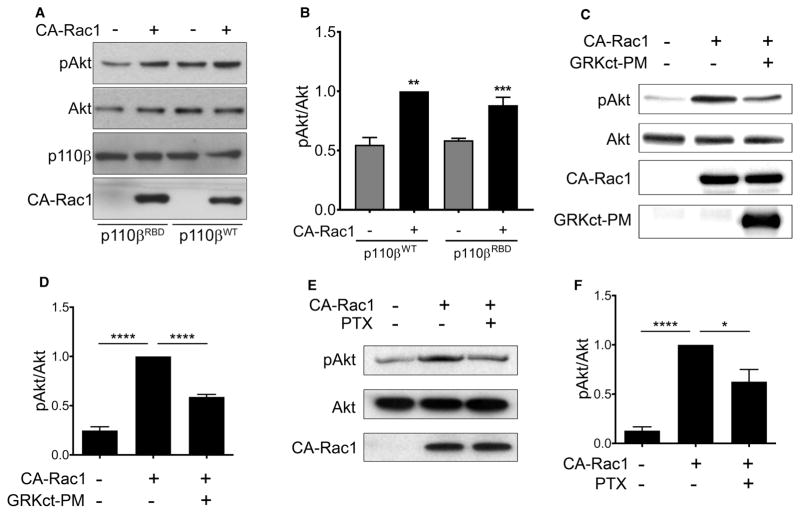
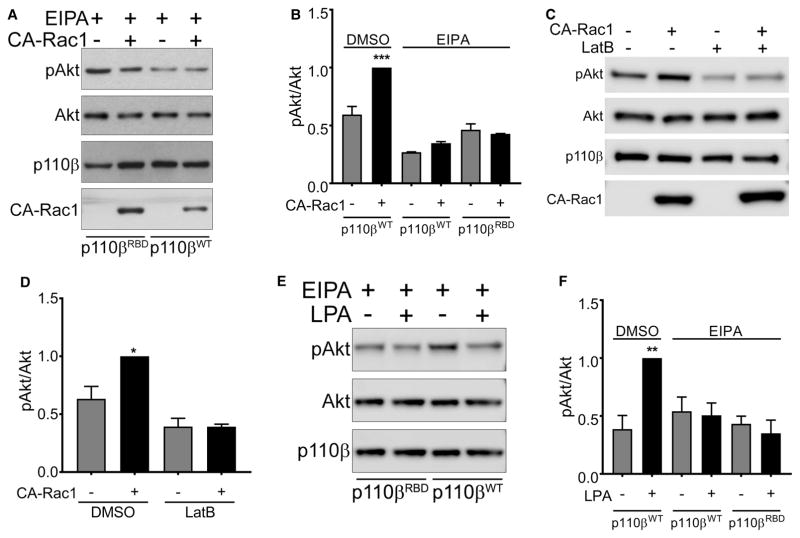
We next tested whether loss of p110β binding to Rac1 would affect the activation of Akt by CA-Rac1. Surprisingly, despite the fact that CA-Rac1 activation of Akt was PI3Kβ dependent (Figure 1F), we observed similar activation of Akt in p110β knockdown cells transiently expressing wild-type p110β versus p110βRBD (Figure 4A, B). These data suggest that CA-Rac1 activates PI3Kβ indirectly, by enhancing Gβγ coupling to PI3Kβ. To test this model, we measured Akt phosphorylation in cells transfected with CA-Rac1 alone or with a membrane-targeted C-terminal fragment of the Gβγ effector GRK (GRKct-PM), which sequesters Gβγ subunits [20]. Activation of Akt by CA-Rac1 in serum-starved cells was inhibited by ~50% in cells expressing GRKct-PM (Figure 4C, D). In an alternative approach, we measured CA-Rac1-mediated activation of Akt in serum-starved cells treated with pertussis toxin (PTX), which inhibits Gαi-coupled GPCRs [21]. PTX caused a 40% decrease in Akt phosphorylation in cells expressing CA-Rac1 (Figure 4E, F). These two independent methods demonstrate that the activation of PI3Kβ by CA-Rac1 requires Gβγ signaling.
Figure 4. CA-Rac1-mediated activation of PI3Kβ requires Gβγ.
(A) Stable p110β knockdown cells were transiently transfected with wild-type p110β, p110βKK-DD or p110βRBD, with or without GFP-CA-Rac1. After 48 h, cells were lysed and blotted for p110β, Akt, and pT308-Akt. (B) Quantitation of pAkt/Akt immunoblots in (A). pAkt/Akt ratios in each experiment were normalized to that seen in cells expressing wild-type p110β and GFP-CA-Rac1. The data represent the mean ± SEM from three independent experiments. (C) Parental MDA-MB-231 cells were transfected with GFP-CA-Rac1 with or without co-transfection of GRKct-PM. Representative immunoblots show pT308-Akt, total Akt, GFP-CA-Rac1, and GRKct-PM. (D) Quantitation of pAkt/Akt levels from immunoblots in (C). pAkt/Akt ratios in each experiment were normalized to that seen in cells expressing GFP-CA-Rac1 without GRKct-PM. The data represent the mean ± SEM from five independent experiments. (E) MDA-MB-231 cells were transfected with GFP-CA-Rac1 with or without PTX treatment. Representative immunoblots show pT308-Akt, total Akt, and GFP-CA-Rac1. (F) Quantitation of pAkt/Akt levels from immunoblots in (E). pAkt/Akt ratios in each experiment were normalized to that seen in cells transfected with GFP-CA-Rac1 without PTX treatment. The data represent the mean ± SEM from three independent experiments. *P < 0.05; ****P < 0.0001.
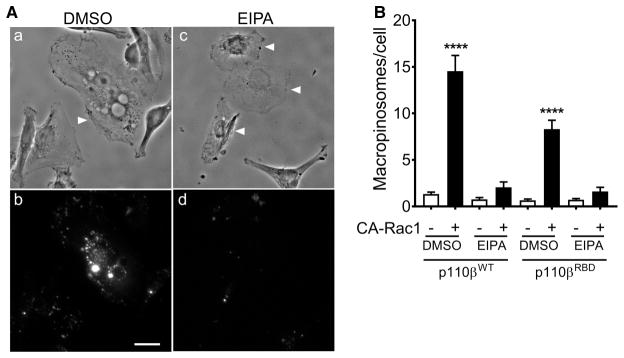
These data show that the expression of CA-Rac activates PI3Kβ by a pathway that is independent of direct PI3Kβ-Rac1 binding, but requires Gβγ, and PI3Kβ binding to Gβγ. Interestingly, Swanson and colleagues have recently shown that macropinosomes can serve as signaling scaffolds for the propagation of GPCR signaling to PI3K [22]. Given that the expression of activated Rac1 stimulated the formation of macropinosomes in Raw 264.7 macrophages [23], we measured macropinocytosis in MDA-MB-231 cells transfected with CA-Rac1. Transfection of CA-Rac1 caused an increase in phase-bright cytosolic vesicles in cells expressing wild-type p110β (Figure 5A, upper panels), and CA-Rac1 caused a seven-fold increase in the uptake of 70 kDa rhodamine-dextran (Figure 5A, lower panels and Figure 5B); this uptake was blocked by the macropinocytosis inhibitor EIPA. CA-Rac1 stimulation of dextran uptake was somewhat reduced in cells expressing the Rac1-binding defective mutant of p110β, but still increased by three-fold (Figure 5B). Notably, activation of Akt by the expression of CA-Rac1 was blocked by the treatment of cells with EIPA (Figure 6A, B) or another the macropinocytosis inhibitor, Latrunculin B (Figure 6C, D). Similarly, LPA stimulation of Akt was blocked by EIPA (Figure 6E, F). Thus, both GPCR- and CA-Rac1-mediated activation of Akt requires macropinocytosis, which acts by enhancing Gβγ coupling to PI3Kβ.
Figure 5. CA-Rac1 stimulates macropinocytosis.
(A) Stable p110β knockdown cells were transfected with wild-type p110β and GFP-CA-Rac1. After 48 h, cells were pretreated with 0.1% DMSO or 25 μM EIPA for 90 min, incubated with 1 mg/ml 70 kDa rhodamine-Dextran for 30 min at 37°C, fixed and imaged. (B) Stable p110β knockdown cells were transfected with wild-type p110β or p110βRBD, with or without GFP-CA-Rac1. The number of macropinosomes per cell (defined as vesicles greater than 0.75 μm in diameter) in GFP-positive cells were counted as described in the Experimental Procedures. Scale bar = 20 μm.
Figure 6. Stimulation of PI3Kβ by CA-Rac1 and LPA requires macropinocytosis.
(A) Stable p110β knockdown cells were transiently transfected with wild-type p110β, p110βKK-DD or p110βRBD, without or with GFP-CA-Rac1. After 48 h, cells were treated with 25 μM EIPA for 3 h, lysed and blotted for p110β, Akt and pT308-Akt. (B) Quantitation of pAkt/Akt immunoblots in Figure 6A. The data represent the mean ± SEM from three independent experiments. pAkt/Akt ratios in each experiment were normalized to that seen in cells expressing wild-type p110β and transfected with GFP-CA-Rac-1, but treated with 0.1% DMSO. (C) Stable p110β knockdown cells were transiently transfected with wild-type p110β without or with GFP-CA-Rac1. After 48 h, cells were treated for 15 min with 0.5 μM Latrunculin B, lysed and blotted for p110β, Akt and pT308-Akt. (D) Quantitation of pAkt/Akt immunoblots in Figure 6C. pAkt/Akt ratios in each experiment were normalized to that seen in cells expressing wild-type p110β and GFP-CA-Rac1. The data represent the mean ± SD from two independent experiments. (E) Stable p110β knockdown cells were transiently transfect with wild-type p110β, p110βKK-DD or p110βRBD. After 48 h, cells were starved overnight, treated with 25 μM EIPA for 3 h, and stimulated for 5 min with 10 μM LPA. Cells were lysed and blotted for p110β, Akt and pT308-Akt. (F) Quantitation of pAkt/Akt immunoblots in Figure 6E. pAkt/Akt ratios in each experiment were normalized to that seen in cells expressing wild-type p110β and stimulated with LPA but treated with DMSO. The data represent the mean ± SEM from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant. Note: The western blot data in Figures 4A and 6A are taken from the same set of experiments, as are the data in Figures 3C and 6E. In each case, cells were treated with 0.1% DMSO or 25 μM EIPA. The EIPA arms of these experiments are presented separately in Figure 6, for clarity of the narrative. Because of this, blots for the controls (cells expressing wild-type p110β and treated with DMSO) are shown in Figures 4 and 3, and are not reproduced in Figure 6. We have included the quantitation of the controls (labeled DMSO) from Figures 3D and 4B in the bar graphs shown in Figure 6B,F, to facilitate a comparison of DMSO- versus EIPA-treated cells.
Discussion
The regulation of the PI3Kβ isoform of Class IA PI 3-kinase is unusually complex, given its ability to directly bind small GTPases (Rac1, Cdc42, Rab5, though the RBD and helical domains [15,24], Gβγ (through the C2-helical linker [10]) and tyrosine phosphorylated proteins (through its p85 regulatory subunit). Previous studies in MEFs have focused on the coupling between Rac- and Gβγ-mediated activation at the level of p110β itself, and have suggested the existence of a feed–forward loop involving Rac1-mediated activation of PI3Kβ- and PI3Kβ-mediated activation of Rac1 [25], as well as the enhancement of GPCR signaling to PI3Kβ by Rac1-mediated targeting of PI3Kβ to lipid rafts [16]. The present study identifies another layer of regulation, in which Rac1-stimulated macropinosomes enhance Gβγ coupling to PI3Kβ independently of Rac1 binding to p110β.
We have previously shown that GPCR signaling in PI3Kβ in MDA-MB-231 cells is required for tumor cell invasion and metastasis, but not for tumor cell growth [17]. To better understand the roles of Rac1 and Gβγ in the regulation of PI3Kβ in breast cancer cells, we used T308 phosphorylation of Akt as a readout for PI3Kβ activity in p110β knockdown cells expressing physiological levels of wild-type or mutant p110β. We examined activation by both a GPCR ligand (LPA) and expression of constitutively active Rac1 (CA-Rac1). This latter approach was used to avoid potential effects of p110β mutants on the activation of endogenous Rac1. Importantly, experiments with TGX-221 showed that in MDA-MB-231 cells, activation of Akt by LPA or CA-Rac1 is dependent on PI3Kβ, validating our use of pT308-Akt as a reporter. While an appropriate assay in breast cancer cells, this approach may not be useful in cells of hematopoietic origin (e.g. macrophages and neutrophils), which also express high levels of PI3Kγ. Indeed, GPCR stimulation of PIP3 in macrophages is largely PI3Kγ dependent [14]. Although MDA-MB-231 cells do express PI3Kγ [26], this isoform does not appear to contribute significantly to GPCR signaling to Akt in these cells.
Stimulation of MDA-MB-231 cells with CA-Rac1 and LPA, or overexpression of Gβγ, led to additive stimulation of Akt. However, the effect of mutations in the Gβγ- and Rac1-binding sites in p110β was not reciprocal. Mutation of the Gβγ-binding site caused a loss of Gβγ- and LPA-stimulated Akt activation, as well as a loss of Rac1-mediated activation. In contrast, mutation of the Rac1-binding site in p110β, or expression of dominant negative Rac1, had no effect on Gβγ-mediated Akt activation. Even more surprising, mutation of the Rac1-binding domain in p110β had no effect on the activation of Akt by either LPA or CA-Rac.
Studies in MEFs have revealed dual pathways for PI3Kβ activation by GPCRs, involving either direct binding of Gβγ to PI3Kβ [10] or Gβγ-stimulation of the ELMO/DOCK/Rac1 pathway, which leads to the binding of activated Rac1 to PI3Kβ [15] and targeting of PI3Kβ to lipid rafts [16]. For unclear reasons, we cannot detect a requirement for Rac1 binding during LPA-stimulated activation of PI3Kβ in MDA-MB-231 cells. This could be due either to differences in the expression/activity of ELMO/DOCK, or other differences between MEFs and breast cancer cells. Lipid raft-mediated signaling has been documented in MDA-MB-231 cells [27], but our data suggest that Rac1-mediated targeting of PI3Kβ to lipid rafts is not critical for Gβγ- or GPCR-mediated PI3Kβ activation in MDA-MB-231 cells.
The finding that the activation of PI3Kβ by CA-Rac1 does not require an intact Rac1-binding site in p110β clearly indicates an indirect mechanism for Rac1 activation of PI3Kβ. However, this mechanism is specific for GPCR signaling to PI3Kβ, as Rac1-mediated activation is blocked by the mutation of the Gβγ-binding site in p110β, treatment of cells with PTX, or expression of GRKct-PM, which sequesters endogenous Gβγ subunits.
How might Rac1 activation enhance Gβγ coupling to PI3Kβ? Rac could work by inhibiting a GRK or other Gαi GAP so as to prolong GPCR-stimulated Gβγ signaling, although this would be unlikely to affect signaling by overexpressed Gβγ. Alternatively, studies by Swanson and colleagues have proposed that macropinocytotic cups can serve as signaling platforms that enhance GPCR signaling. In bone marrow-derived macrophages, inhibitors of macropinocytosis block stimulation of Akt by the CXCR4 receptor, and both PIP3 and phosphory-lated Akt can be detected in macropinocytotic cups that form in CXCL12-stimulated bone marrow-derived macrophages [22]. In MDA-MB-231 cells expressing wild-type p110β, the expression of CA-Rac1 causes a large increase in the production of phase-bright vesicles, and a seven-fold stimulation of macropinocytosis. Moreover, the activation of Akt by CA-Rac1 is blocked by treatment with the macropinocytosis inhibitor EIPA, or disruption of the actin cytoskeleton with Latrunculin B, which also blocks macropinocytosis. EIPA also blocks the activation of Akt by LPA, demonstrating a requirement for macropinocytosis during physiological regulation of GPCR signaling to PI3Kβ. Of note, while the effects of amiloride (the parent compound of EIPA) on EGF-stimulated macropinocytosis in A431 cells involve the inhibition of Rac activation [28], this would not be affect signaling in cells expressing constitutively active Rac1.
In the present study, we focused on GPCR signaling to PI3Kβ. It has been suggested that signaling by RTKs like the CSF-1R can also be enhanced by macropinosomes formation, but only at low levels of ligand stimulation [22]. However, PI3Kβ does not appear to couple effectively to RTKs [12–14], so we have not pursued the role of macropinosomes in RTK/PI3Kβ signaling. In contrast, studies in neutrophils and macrophages have shown that PI3Kβ can serve as a coincidence detector from simultaneous RTK-GPCR stimuli [14]. A potential role for macropinosomes in responses to a combined RTK-GPCR stimulus in breast cancer cells remains an interesting question.
The role of macropinosomes (or macropinocytotic cups) in enhancing Gβγ signaling to PI3Kβ that we observe in breast cancer cells has some parallels to the requirement for lipid raft targeting of PI3Kβ described in MEFs [16]. In both cases, a membrane subdomain enhances the interaction between PI3Kβ and Gβγ. The need for a membrane based amplification system may be inherent in the p110β–Gβγ interaction, which is relatively weak compared with that of PI3Kγ and Gβγ, and only occurs on membranes [10]. A major difference in the two systems is that in MEFs, targeting of PI3Kβ to rafts requires a direct Rac1–PI3Kβ interaction, whereas in breast cancer cells, Rac1 is required to produce the macropinocytotic cup, but its direct binding to PI3Kβ is not required. The precise mechanisms that target Gβγ, PI3Kβ or both to macropinocytotic cups will be an important avenue for future investigation.
Acknowledgments
Funding
This manuscript was supported by NIH grants GM112524 and CA100324, and by the Albert Einstein College of Medicine Cancer Center [P30 CA013330], and by the Deutsche Forschungsgemeinschaft (DFG).
We thank Dr Philip Wedegaertner, Thomas Jefferson University, for the GRKct-PM construct, and Drs Joel Swanon, University of Michigan, and Sergio Grinstein, Hosptial for Sick Kids, Toronto, for helpful discussions and suggestions.
Abbreviations
- ATCC
American Type Culture Collection
- EGF
epidermal growth factor
- EIPA
5-(n-ethyl-N-isopropyl) amiloride
- GPCRs
G-protein-coupled receptors
- LPA
lysophosphatidic acid
- MEFs
mouse embryonic fibroblasts
- RBD
Ras-Binding Domain
- RTKs
receptor tyrosine kinases
Footnotes
Author Contribution
Z.E. and B.D.K. helped design the study and prepare the figures, contributed to the manuscript, and performed and analyzed experiments. G.S. performed and analyzed the macropinocytosis experiments. Y.Y. performed the experiments shown in Figure 2B. JLP purified the recombinant CA-Rac1. A.S. and B.N. purified the recombinant Gβγ. A.R.B. and J.M.B. conceived and co-ordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Competing Interests
J.M.B. is on the Scientific Advisory board of Karus Therapeutics.
References
- 1.Backer JM. The regulation of class IA PI 3-kinases by inter-subunit interactions. Curr Top Microbiol Immunol. 2010;346:87–114. doi: 10.1007/82_2010_52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciraolo E, Morello F, Hobbs RM, Wolf F, Marone R, Iezzi M, et al. Essential role of the p110β subunit of phosphoinositide 3-OH kinase in male fertility. Mol Biol Cell. 2010;21:704–711. doi: 10.1091/mbc.E09-08-0744. https://doi.org/10.1091/mbc.E09-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CE, et al. PI 3-kinase p110β: a new target for antithrombotic therapy. Nat Med. 2005;11:507–514. doi: 10.1038/nm1232. https://doi.org/10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 4.Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, et al. Essential roles of PI(3)K-p110β in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. https://doi.org/10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciraolo E, Iezzi M, Marone R, Marengo S, Curcio C, Costa C, et al. Phosphoinositide 3-kinase p110β activity: key role in metabolism and mammary gland cancer but not development. Sci Signal. 2008;1:ra3. doi: 10.1126/scisignal.1161577. https://doi.org/10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gratacap MP, Guillermet-Guibert J, Martin V, Chicanne G, Tronchère H, Gaits-Iacovoni F, et al. Regulation and roles of PI3Kβ, a major actor in platelet signaling and functions. Adv Enzyme Regul. 2011;51:106–116. doi: 10.1016/j.advenzreg.2010.09.011. https://doi.org/10.1016/j.advenzreg.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Dou Z, Pan JA, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, et al. Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell. 2013;50:29–42. doi: 10.1016/j.molcel.2013.01.022. https://doi.org/10.1016/j.molcel.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, Hawkins PT, et al. Structure of lipid kinase p110β/p85β elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. https://doi.org/10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dbouk HA, Vadas O, Shymanets A, Burke JE, Salamon RS, Khalil BD, et al. G protein-coupled receptor-mediated activation of p110β by Gβγ is required for cellular transformation and invasiveness. Sci Signal. 2012;5:ra89. doi: 10.1126/scisignal.2003264. https://doi.org/10.1126/scisignal.2003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier U, Babich A, Nürnberg B. Roles of non-catalytic subunits in Gβγ-induced activation of class I phosphoinositide 3-kinase isoforms β and γ. J Biol Chem. 1999;274:29311–29317. doi: 10.1074/jbc.274.41.29311. https://doi.org/10.1074/jbc.274.41.29311. [DOI] [PubMed] [Google Scholar]
- 12.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, et al. A pharmacological map of the PI3-K family defines a role for p110α in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. https://doi.org/10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, et al. The p110β isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110γ. Proc Natl Acad Sci U S A. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. https://doi.org/10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houslay DM, Anderson KE, Chessa T, Kulkarni S, Fritsch R, Downward J, et al. Coincident signals from GPCRs and receptor tyrosine kinases are uniquely transduced by PI3Kβ in myeloid cells. Sci Signal. 2016;9:ra82. doi: 10.1126/scisignal.aae0453. https://doi.org/10.1126/scisignal.aae0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, et al. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153:1050–1063. doi: 10.1016/j.cell.2013.04.031. https://doi.org/10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cizmecioglu O, Ni J, Xie S, Zhao JJ, Roberts TM. Rac1-mediated membrane raft localization of PI3K/p110β is required for its activation by GPCRs or PTEN loss. eLife. 2016;5:776. doi: 10.7554/eLife.17635. https://doi.org/10.7554/eLife.17635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalil BD, Hsueh C, Cao Y, Abi Saab WF, Wang Y, Condeelis JS, et al. GPCR signaling mediates tumor metastasis via PI3Kβ. Cancer Res. 2016;76:2944–2953. doi: 10.1158/0008-5472.CAN-15-1675. https://doi.org/10.1158/0008-5472.CAN-15-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shymanets A, Prajwal, Vadas O, Czupalla C, LoPiccolo J, Brenowitz M, et al. Different inhibition of Gβγ-stimulated class IB phosphoinositide 3-kinase (PI3K) variants by a monoclonal antibody. Specific function of p101 as a Gβγ-dependent regulator of PI3Kγ enzymatic activity. Biochem J. 2015;469:59–69. doi: 10.1042/BJ20150099. https://doi.org/10.1042/BJ20150099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Commisso C, Flinn RJ, Bar-Sagi D. Determining the macropinocytic index of cells through a quantitative image-based assay. Nat Protoc. 2014;9:182–192. doi: 10.1038/nprot.2014.004. https://doi.org/10.1038/nprot.2014.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irannejad R, Wedegaertner PB. Regulation of constitutive cargo transport from the trans-Golgi network to plasma membrane by Golgi-localized G protein βγ subunits. J Biol Chem. 2010;285:32393–32404. doi: 10.1074/jbc.M110.154963. https://doi.org/10.1074/jbc.M110.154963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smrcka AV. G protein βγ subunits: central mediators of G protein-coupled receptor signaling. Cell Mol Life Sci. 2008;65:2191–2214. doi: 10.1007/s00018-008-8006-5. https://doi.org/10.1007/s00018-008-8006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacitto R, Gaeta I, Swanson JA, Yoshida S. CXCL12-induced macropinocytosis modulates two distinct pathways to activate mTORC1 in macrophages. J Leukoc Biol. 2017;101:683–692. doi: 10.1189/jlb.2A0316-141RR. https://doi.org/10.1189/jlb.2A0316-141RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii M, Kawai K, Egami Y, Araki N. Dissecting the roles of Rac1 activation and deactivation in macropinocytosis using microscopic photo-manipulation. Sci Rep. 2013;3:2385. doi: 10.1038/srep02385. https://doi.org/10.1038/srep02385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salamon RS, Dbouk HA, Collado D, Lopiccolo J, Bresnick AR, Backer JM. Identification of the Rab5 binding site in p110β: assays for PI3Kβ binding to Rab5. Methods Mol Biol. 2015;1298:271–281. doi: 10.1007/978-1-4939-2569-8_23. https://doi.org/10.1007/978-1-4939-2569-8_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuzugullu H, Baitsch L, Von T, Steiner A, Tong H, Ni J, et al. A PI3K p110β–Rac signalling loop mediates Pten-loss-induced perturbation of haematopoiesis and leukaemogenesis. Nat Commun. 2015;6:8501. doi: 10.1038/ncomms9501. https://doi.org/10.1038/ncomms9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brazzatti JA, Klingler-Hoffmann M, Haylock-Jacobs S, Harata-Lee Y, Niu M, Higgins MD, et al. Differential roles for the p101 and p84 regulatory subunits of PI3Kγ in tumor growth and metastasis. Oncogene. 2012;31:2350–2361. doi: 10.1038/onc.2011.414. https://doi.org/10.1038/onc.2011.414. [DOI] [PubMed] [Google Scholar]
- 27.Badana A, Chintala M, Varikuti G, Pudi N, Kumari S, Kappala VR, et al. Lipid raft integrity is required for survival of triple negative breast cancer cells. J Breast Cancer. 2016;19:372–384. doi: 10.4048/jbc.2016.19.4.372. https://doi.org/10.4048/jbc.2016.19.4.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koivusalo M, Welch C, Hayashi H, Scott CC, Kim M, Alexander T, et al. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J Cell Biol. 2010;188:547–563. doi: 10.1083/jcb.200908086. https://doi.org/10.1083/jcb.200908086. [DOI] [PMC free article] [PubMed] [Google Scholar]