Key Points
AML blasts express both activating and inhibitory NKRLs at diagnosis.
An overall activating NK ligand phenotype on blasts is associated with improved survival and reduced relapse after induction chemotherapy.
Abstract
Natural killer (NKs) cells provide rapid responses to viral-infected and malignant cells, including acute myeloid leukemia (AML) blasts. The balance among inhibitory and activating signals, delivered by multiple interactions between ligands on target cells and NK receptors, determines the posture of the NK cell response to either one of target cell elimination or tolerance. The aim of this work was to study the influence of the differential expression of activating and inhibitory NK receptor ligands (NKRLs) by leukemic blasts on clinical outcome in newly diagnosed AML patients. Leukemic cells and clinical data from 66 patients undergoing induction chemotherapy were obtained from the Australasian Leukemia Lymphoma Group tissue bank. Expression of 6 activating (MICA, MICAB, CD155, CD112, ULBP1, and ULBP2/5/6) and 3 inhibitory (HLA class I, PD-L1, and PD-L2) NKRLs was analyzed by flow cytometry. AML blasts displayed heterogeneous expression of NKRLs. MICA, CD112, and ULBP1 were most frequently expressed. ULBP1 expression was significantly associated with improved 2-year overall survival (51.4% vs 11.4%), relapse-free survival (42.5% vs 10.0%), and reduced relapse (44.1% vs 78.6%). We calculated a net score of activating minus inhibitory ligands and demonstrated that the expression of an overall activating NK ligand phenotype was associated with superior 2-year overall survival (59.6% vs 24.4%) and reduced relapse (31.5% vs 68.2%). Our study provides clinical evidence for the role of NK cell–mediated immunoediting against AML, mediated by the expression of NKRLs on blasts, and supports investigation into strategies to enhance NK cell function to improve outcomes in patients with AML.
Visual Abstract
Introduction
Natural killer (NK) cells are innate lymphoid cells with cytolytic and cytokine-secreting functions and have an important role in cancer immunoediting, including against acute myeloid leukemia (AML).1,2 The ability of NK cells to eliminate AML has been demonstrated in vitro and also through clinical observations in haploidentical allogeneic hematopoietic stem cell transplantation (alloHSCT), where the presence of killer immunoglobulin receptor (KIR) ligand mismatch in the graft-versus-host direction confers enhanced protection against AML relapse.3-6
NK cell activity is modulated by a number of germ-line encoded inhibitory and activating receptors. The interaction between HLA class I molecules on target cells and KIR or the lectin heterodimer CD94/natural killer group 2 member A delivers an inhibitory signal to NK cells, and the corollary of this is observed where the lack of HLA class I expression (ie, “missing self”) on tumor cells allows NK cells to detect and kill malignant transformed cells.7 In addition to this primary NK and target cell interaction, engagement of the NK cell receptors, including NKG2D, NKp30, NKp44, and NKp46, with ligands on target cells promotes NK cell activation.8 Furthermore, a single ligand can bind different receptors with opposite immunological functions, as in the case of CD155 and its receptors CD226 (which promotes an activating signaling) and TIGIT or CD96 (that conversely act as inhibitory receptors). As a result, the final balance among inhibitory and activating signals delivered by multiple ligand/receptor interactions determines the eventual posture of the NK cell response to one of target cell elimination or tolerance.9
Several studies have demonstrated that host-intrinsic NK-cell responsiveness contributes to prognosis in patients with AML. High frequency of circulating NK cells,10 NK cell cytolytic activity, and NK cell interferon-γ secretion potency have been described to correlate with a favorable outcome,11 whereas functional defects in NK cells have been associated with failure to achieve remission, disease progression, and short survival duration.12-14
In addition to host-intrinsic determinants of NK cell reactivity, the expression of NK cell receptor ligands (NKRLs) by AML blasts may influence NK cell–mediated immune surveillance and therefore prognosis. Expression of NKG2D receptor ligands by AML blasts enhances NK cell recognition and cytolysis in vitro.3 However, a detailed analysis of the prognostic impact of NKRL expression by AML, and the combinatorial effect of simultaneous expression of inhibitory and activating ligands on AML blasts, has not been well described. The aim of this work was to investigate the impact of the differential expression of activating and inhibitory NKRLs by AML blasts on clinical outcome in newly diagnosed patients undergoing induction chemotherapy. We hypothesized that patients with blasts expressing NKRLs that facilitated NK cell activation (namely with a low-inhibitory and high-activating ligand expression pattern) would have improved immune surveillance and reduced disease relapse.
Methods
Bone marrow samples
Cryopreserved bone marrow aspirate samples from 66 patients with AML were obtained from the Australasian Leukemia and Lymphoma Group tissue bank. Prior to sample collection, written informed consent had been provided by patients for future research use. All patients underwent induction chemotherapy with curative intent. Patients with acute promyelocytic leukemia and those receiving palliative chemotherapy only were excluded from the cohort. Deidentified clinical data, including diagnostic information, cytogenetics, treatment outcome, and survival, were also provided by the Australasian Leukemia and Lymphoma Group. The experimental plan was approved by the institutional human research ethics committee.
Mutation status
Patients were stratified by mutation status using European Leukemia Net criteria (ELN).15 Sequence analysis of targeted regions within 26 genes involved in myeloid malignancies was performed in duplicate using Access Array methodology (Fluidigm) to prepare amplicon-based, indexed libraries that were sequenced to a depth of ∼1000 reads per amplicon on a MiSeq instrument using v2 chemistry (Illumina). Alignment, variant calling, and annotation were performed using an in-house amplicon pipeline. Variants were evaluated using multiple functional and quality filters to identify likely pathogenic variants. FLT3-ITDs and the ASXL1 NM_015338.5:c.1934dup;p.Gly646Trpfs*12 were assessed using capillary electrophoresis.
Mutations in CEBPA were assessed by conventional Sanger sequencing.
Flow cytometric acquisition and analysis of NKRLs on bone marrow blasts
Cryopreserved bone marrow samples were thawed and stained with the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Life Technologies) and fluorescein isothiocyanate–, phycoerythrin-, phycoerythrin-Cychrome 7–, PerCP Cychrome5.5–, allophycocyanin–, and allophycocyanin Cychrome 7–conjugated antibodies directed to human CD45, CD33, CD34, MICA, MICB, CD155, CD112, ULBP1, ULBP2/5/6, HLA class I, PD-L1, and PD-L2 (BD Biosciences) to analyze the expression of NKRLs on AML blasts. Samples were fixed in 2% paraformaldehyde and analyzed using an LSRFortessa flow cytometer (BD Biosciences) and Flow Jo software (Tree Star Inc). Surface expression of NKRLs was measured by the relative fluorescence intensity (RFI) of the ligands on AML blasts compared with unstained controls.
Statistical analysis
Overall survival (OS) and relapse-free survival (RFS) were calculated from the time of AML diagnosis using the Kaplan-Meier method. For patients who achieved complete remission (CR), cumulative incidence of relapse (CIR) was calculated from the time of diagnosis with nonrelapse mortality considered as a competing risk. Patient outcomes were censored at the time of alloHSCT. Cox regression was used to investigate relationships between NKRL expression and OS and RFS. Gray’s test was used to evaluate associations with CIR. NKRL scores were combined with cytogenetic risk and ELN groups in bivariable models for associations with OS, RFS, and CIR. Differences in baseline characteristics between patient subgroups were assessed using Fisher’s exact test. Significance level was set at α < 0.05. Statistical analyses were performed using R v3.1.0 (Comprehensive R Archive Network Project).
Results
Baseline patient characteristics
Patient characteristics are summarized in Table 1. Median age at diagnosis was 56 years (range 16-78 years). Seven patients had favorable cytogenetics (10.6%); 46 patients had intermediate cytogenetics (69.7%), and 13 patients unfavorable cytogenetics (19.7%). The molecular mutation pattern was determined to further stratify patients according to the last ELN criteria. The most common mutations were NPM1 (33%), FLT3-internal tandem duplication (30%), and DNMT3A (18%). Twelve out of 66 patients simultaneously harbored NPM1 and FLT3-ITD mutations, and 4 of them were also mutated for DNMT3A. All other mutations (among which RUNX1, ASXL1, TP53) were less frequent (<20% of positive patients). Interestingly, the mutational status could “upgrade” 13 of the 46 patients classified as having an intermediate cytogenetic risk to a more favorable ELN risk and “downgrade” 12 out of 46 patients to an adverse ELN risk. Overall, 20 patients (33.3%) had favorable molecular risk according to the ELN classification; 15 (25%) patients had intermediate risk, and 25 (41.7%) patients had adverse risk. Forty-five patients (68.2%) achieved CR after induction chemotherapy; 11 (16.7%) did not respond to induction, and 10 (15.2%) either experienced treatment-related mortality or were not available for response assessment. Sixteen patients (24%) received alloHSCT as consolidation therapy. Median follow-up after diagnosis was 41.2 months (95% confidence interval 19.6-57.6 months).
Table 1.
Baseline patient characteristics
| All | Group 0 | Group 1 | P | |
|---|---|---|---|---|
| Patients, n (%) | 66 (100) | 32 (48) | 34 (52) | |
| Age at diagnosis, median (range), y | 56 (16-78) | 48 (16-78) | 60 (26-77) | .3 (>60 vs <60) |
| Sex, M/F | 37/29 | 17/15 | 20/14 | |
| % BM blasts at diagnosis, median (range) | 78 (20-97) | 81 (21-94) | 70 (20-97) | |
| Cytogenetic risk, n (%) | .7 (favorable vs intermediate/adverse) | |||
| Favorable | 7 (10.6) | 4 (12.5) | 3 (8.8) | |
| Intermediate | 46 (69.7) | 20 (62.5) | 26 (76.5) | |
| Adverse | 13 (19.7) | 8 (25) | 5 (14.7) | |
| ELN, evaluable, n (%) | 60 (91) | 30 (93.8) | 30 (88.2) | .08 (favorable vs intermediate/adverse) |
| Favorable | 20 (33.3) | 7 (23.3) | 13 (43.3) | |
| Intermediate | 15 (25) | 7 (23.3) | 8 (26.7) | |
| Adverse | 25 (41.7) | 16 (53.3) | 9 (30) | |
| Response to iCT, n (%) | ||||
| CR | 45 (68.2) | 22 (68.8) | 23 (67.6) | |
| NR | 11 (16.7) | 6 (18.8) | 5 (14.7) | |
| n.e./death | 10 (15.2) | 4 (12.5) | 6 (17.6) | |
| AlloHSCT, n (%) | ||||
| No | 46 (69.7) | 20 (62.5) | 26 (76.5) | |
| Yes | 16 (24.2) | 11 (34.4) | 5 (14.7) | |
| n.a. | 4 (6) | 1 (3.1) | 3 (8.8) |
All patients (All) and patients stratified according to their flow cytometry NKRL group (Group 0, NK receptor ligand inhibitory pattern; and Group 1, NKRL activating pattern) are shown separately.
BM, bone marrow; CR, complete remission; F, female; iCT, induction chemotherapy; M, male; n.a., not available; n.e., not evaluable; NR, nonresponse.
AML cells express variable levels of NKRLs
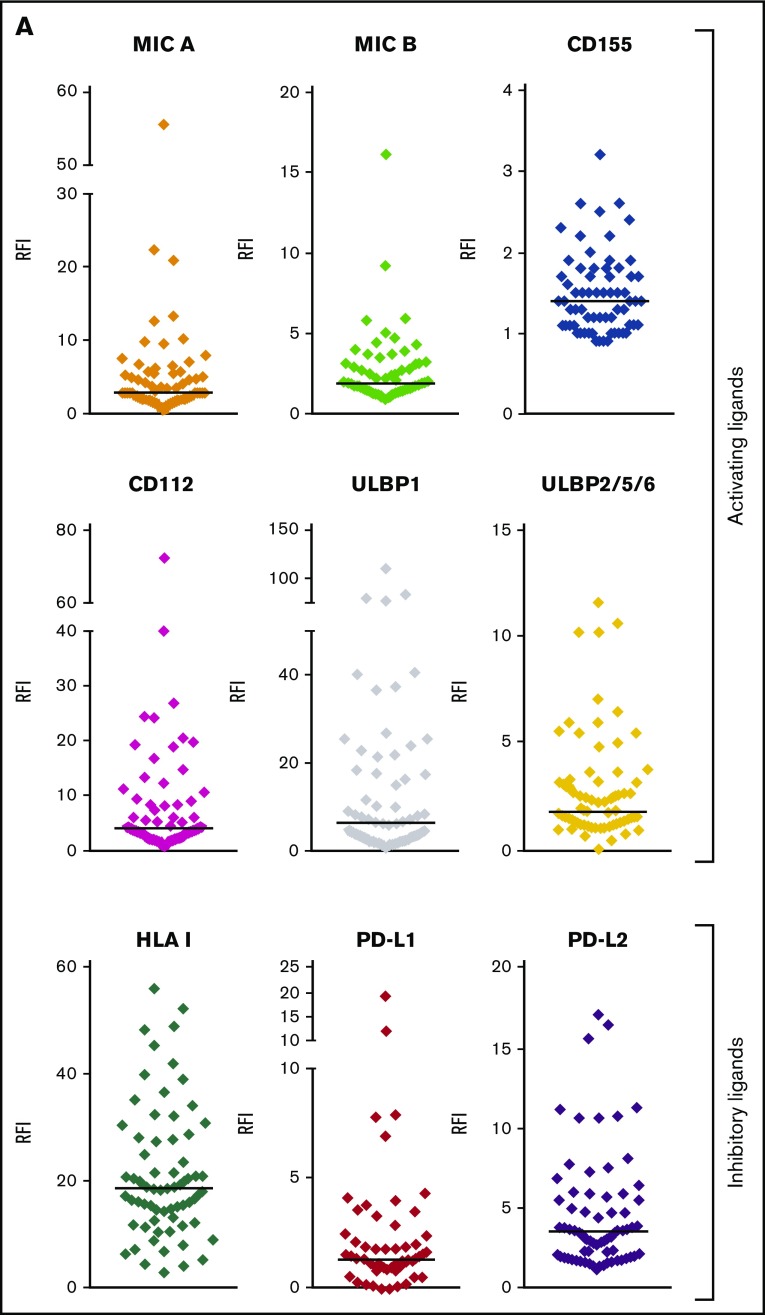
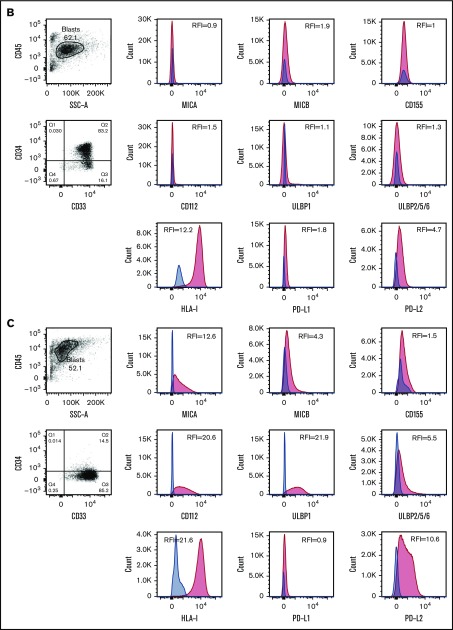
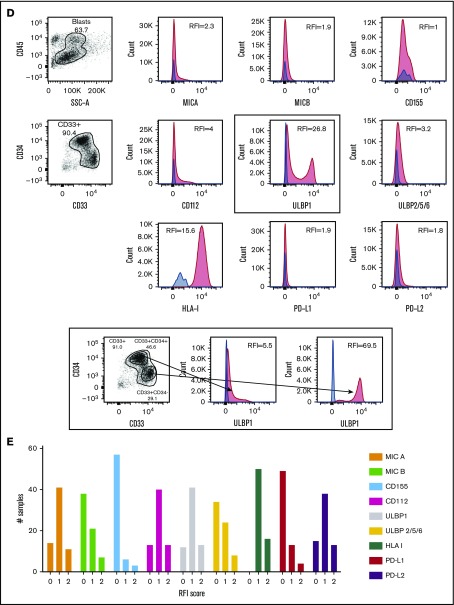
Among the NKRLs examined, MICA, MICB, CD155, CD112, ULBP1, and ULBP2/5/6 are known to confer an activating signal to NK cells, and HLA class I, PD-L1, and PD-L2 are known to inhibit NK cell function. We observed that these NKRLs were variably expressed on AML blasts (Figure 1A). Among activating ligands, MICA, CD112, and ULBP1 were frequently expressed by AML blasts (78.8%, 80.3%, and 81.8% of patients, respectively) and also displayed the highest intensity of expression (median RFI of 3, 4.2, and 6.5, respectively). All patients expressed HLA class I molecules at intermediate or high levels at diagnosis (median RFI 18.6), whereas PD-L2 expression on blasts was more frequent (77.3% patients PD-L2 positive vs 25.8% PD-L1 positive) and expressed to a higher intensity compared with PD-L1 (median PD-L2 RFI 3.6 vs PD-L1 RFI 1.3). There was also significant variation in the number of NKRLs expressed by AML blasts from each patient, with some patients demonstrating an absence of most NKRLs (representative example in Figure 1B) and others showing a broad ligand expression profile (representative example in Figure 1C). Interestingly, in some cases where distinct immunophenotypic subclones of blasts coexisted in the same patient (ie, CD33posCD34pos and CD33posCD34dim), there were differences in the NKRL expression in the separate blast subpopulations (representative flow cytometry plots in Figure 1D).
Figure 1.
NKRLs are variably expressed by AML blasts. The expression of 6 activating (MICA, MICB, CD155, CD112, ULBP1, and ULBP2/5/6) and 3 inhibitory (PD-L1, PD-L2, and HLA class I) NKRLs was detected by flow cytometry on cryopreserved bone marrow blasts harvested from 66 patients with a new diagnosis of AML. Side scatter (SSC-A) and CD45 were used to gate the blast population. CD33 and CD34 were used as additional markers of disease. The RFI was calculated for each ligand as follows: mean fluorescence intensity (MFI) of the ligand/MFI of the unstained control. (A) NKRL RFIs calculated and plotted for all 66 patients. Each replicate is shown; black horizontal line corresponds to median values. (B) Representative plots of a patient expressing only inhibitory ligands (HLA-I and PD-L2). (C) Representative plots of a patient with high expression of both activating and inhibitory NKRLs. (D) Representative plots of a patient with a double blast population (CD33posCD34pos and CD33posCD34dim), where the expression of ULBP1 was strikingly different: the CD33posCD34pos blasts expressed low levels of ULBP1 (RFI = 5.5), whereas the activating ligand was highly expressed on CD33posCD34dim cells (RFI = 69.5). Blue histograms correspond to unstained control MFI; red histograms correspond to NKRL MFI. (E) An RFI score ranging from 0 to 2 was calculated for each ligand as follows: (1) samples with RFI <2 (ie, less than twofold increase of median fluorescence above background) were considered negative and assigned score 0; (2) Samples with an RFI value between 2 and the III quartile were assigned score 1; (3) Samples with RFI equal to or above the III quartile were assigned score 2. Panel E represents the distribution of patients according to the RFI score for each NKRL.
In order to categorize single RFI values, an RFI score ranging from 0 to 2 was calculated for each ligand as follows: samples with RFI < 2 (ie, less than twofold increase of median fluorescence above background) were considered negative and assigned score 0; samples with an RFI value between 2 and the third quartile were assigned score 1; samples with RFI equal to or above the third quartile were assigned score 2. Figure 1E represents the distribution of patients according to the RFI score for each NKRL and summarizes the aforementioned differences in the individual ligand expression profile.
The intensity of ULBP1 expression significantly correlates with patient outcome
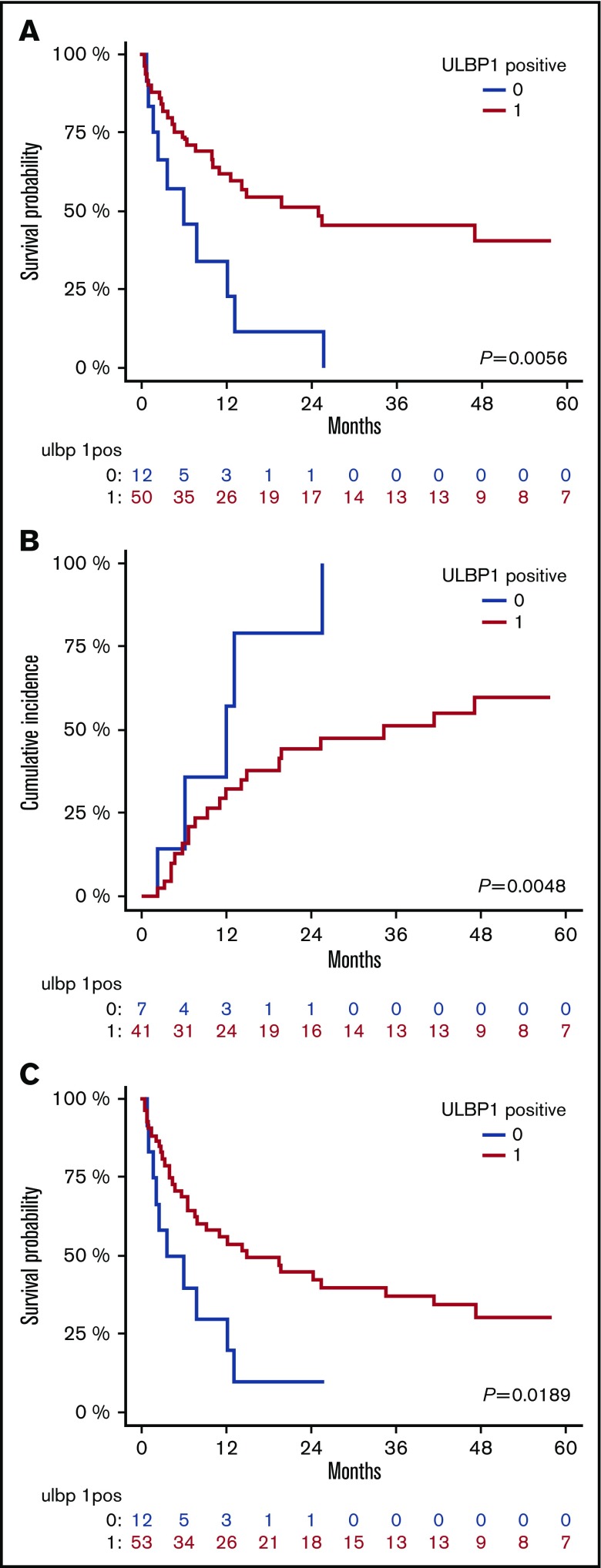
As already mentioned above, the NKG2D activating ligand ULBP1 was frequently positive on AML blasts, with 41 out of 66 patients expressing it at intermediate levels (mean RFI 6.2) and 13 patients showing a high-intensity ligand expression (mean RFI 37.2). Furthermore, the expression of ULBP1 was significantly associated with improved outcome (Figure 2). When compared with patients with AML that did not express ULBP1, patients with ULBP1-positive blasts had better 2-year OS (51.4% vs 11.4%; P < .05), RFS (42.5% vs 10.0%; P < .05), and reduced 2-year CIR (44.1% vs 78.6%; P < .005) (Figure 2). In addition, the intensity of ULBP1 expression was significantly associated with better OS, RFS, and CIR in a stepwise fashion: patients with the highest ULBP1 expression had a reduced hazard ratio of death (HR, 0.22; P < .05) or relapse (HR, 0.18; P < .05) compared with nonexpressing patients (supplemental Figure 1). ULBP1 positivity remained significantly associated with reduced relapse incidence when taking into account cytogenetic group (HR, 0.38; P < .05). There was a trend toward reduced relapse risk in patients with ULBP1-positive blasts when taking into account ELN classification; however, this was not statistically significant. All other individually analyzed ligands were not significantly associated with patients’ outcome with the exception of PD-L1, whose expression on leukemic blasts was unexpectedly found to be directly correlated with a lower CIR (HR, 0.39; P = .04), although without significant impact on OS or RFS (supplemental Table 1). The reduced incidence of relapse in PD-L1 expressing patients may be accounted for by lower incidence of adverse cytogenetic and molecular profiles in this subgroup (adverse cytogenetics 0% in PD-L1–positive patients vs 16% in PD-L1–negative patients; adverse ELN group 20% in PD-L1–positive patients vs 42% in PD-L1–negative patients).
Figure 2.
ULBP1 positivity at diagnosis correlates with better outcome. Kaplan-Meier estimates of OS (A), CIR (B), and RFS (C), according to ULBP1 expression: patients with blasts expressing and nonexpressing ULBP1 at diagnosis are shown by red and blue lines, respectively. P values are shown.
Flow cytometry net score enables patient stratification
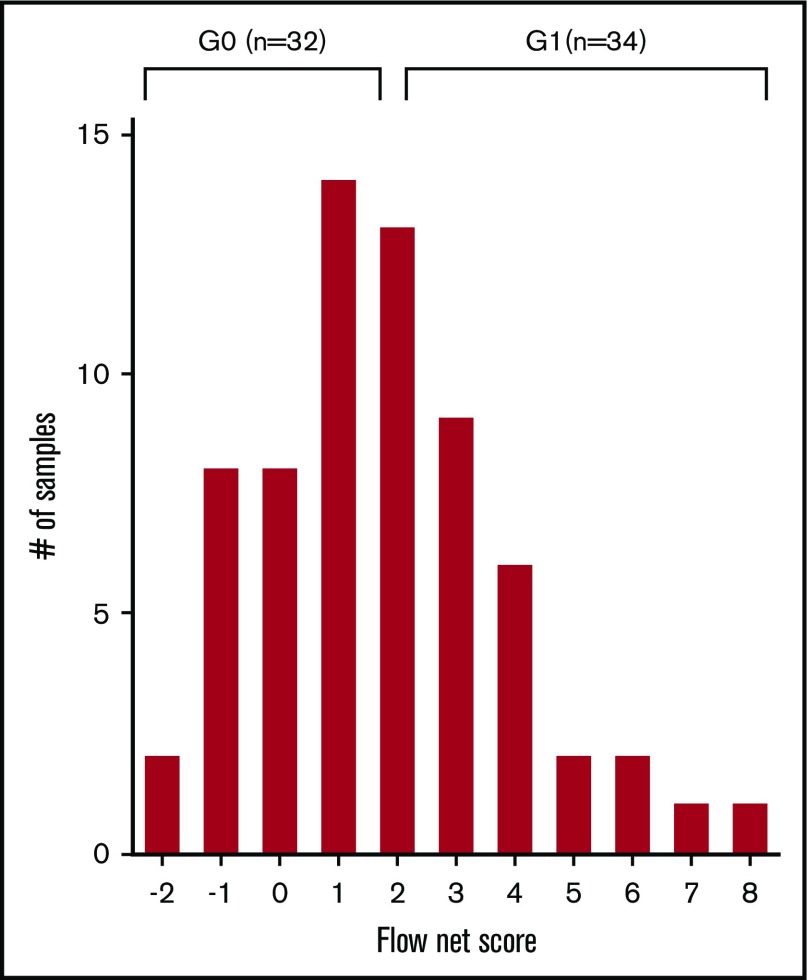
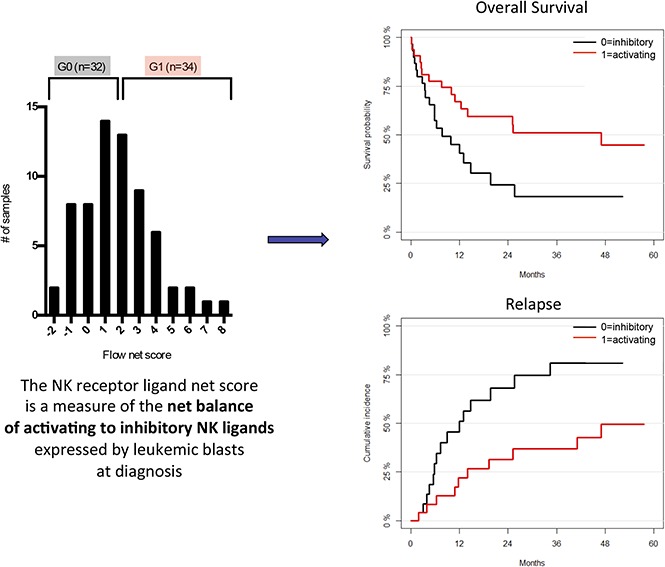
Once each patient was assigned an RFI score according to the levels of expression of a given NKRL, a flow cytometry net score including all 9 analyzed ligands was calculated for each patient as the sum of each activating ligand RFI score minus the sum of each inhibitory ligand RFI score (Figure 3). The flow net scores for the whole cohort followed a Gaussian distribution within a range from −2 (ie, highly inhibitory expression pattern) to 8 (ie, highly activating expression pattern) and centered on a flow net score of 1. The entire cohort was therefore divided into 2 groups: (1) Group 0 (G0), including subjects with score from −2 to 1, and (2) Group 1 (G1) comprising patients with scores from 2 to 8. Patients in G0 and G1 were well matched for age at diagnosis, cytogenetics, and molecular risk profile (Table 1).
Figure 3.
NKTL net score and group stratification. A flow cytometry net score ranging from −2 to 8 was calculated for each patient as follows: (sum of each activating ligand RFI score) − (sum of each inhibitory ligand RFI score). The histogram represents the distribution of patients according to their net score. A further stratification was performed in order to correlate NKRL expression to clinical outcome: patients were split into 2 homogeneous groups according to their flow cytometry net score: group 0 (G0) with score −2 to 1 and group 1 (G1) with score 2 to 8.
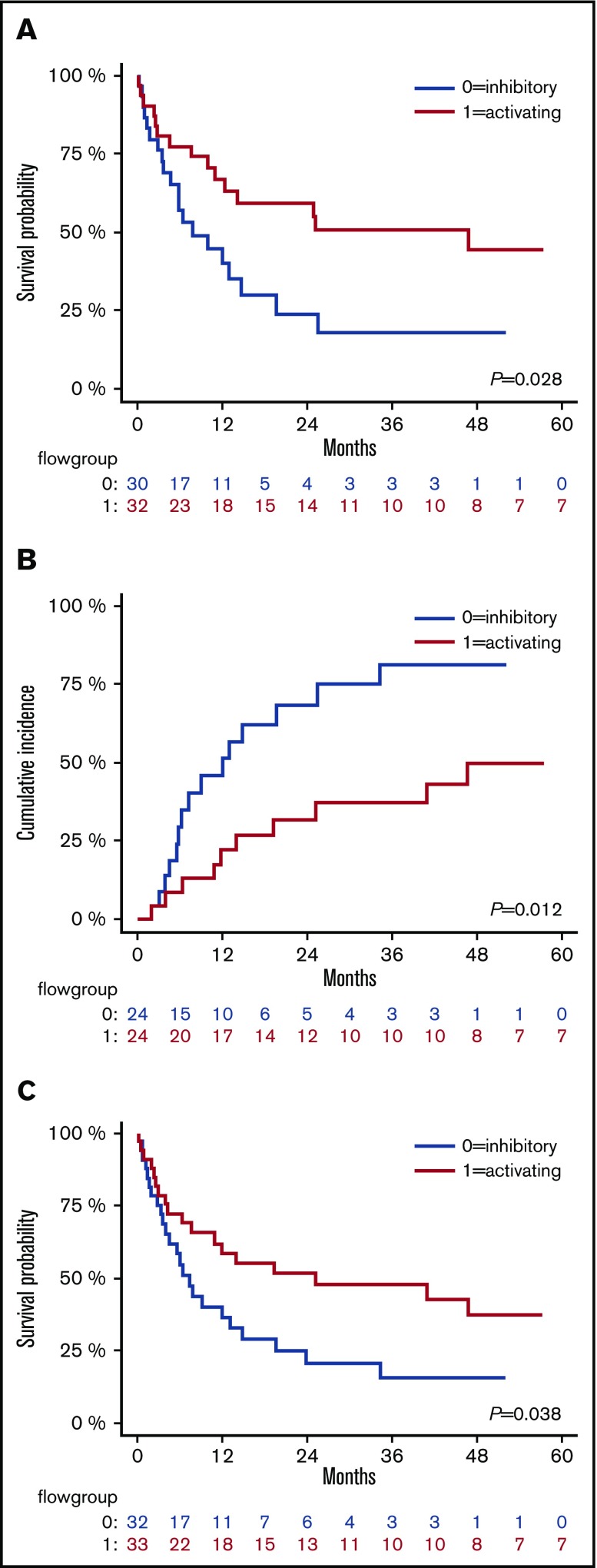
Activating NKRLs are associated with improved survival and reduced AML relapse
We assessed the impact of NKRL overall score (G0 vs G1 as described above) on patient survival and relapse. The presence of the more activating overall pattern of NKRL expression (G1) was associated with improved OS (2-year OS 59.6% vs 24.4%; HR, 0.46; log rank P < .05) and RFS (2-year RFS 51.5% vs 20.8%; HR, 0.52; P < .05) and reduced CIR (2-year CIR 31.5% vs 68.2%; HR, 0.36; P < .05) compared with patients with the more inhibitory pattern of NKRL expression (G0) (Figure 4). On multivariate analysis, the activating G1 pattern remained significantly associated with lower relapse rate when taking into account cytogenetic risk group (P = .007). There was no correlation between NKRL expression and time to AML relapse.
Figure 4.
The activating pattern of NKRLs correlates with better outcome. Kaplan-Meier estimates of OS (A), CIR (B), and RFS (C), according to flow cytometry group stratification: patients belonging to group 0 (NKRL inhibitory pattern) and 1 (NK receptor ligand activating pattern) are represented by blue and red lines, respectively. P values are shown.
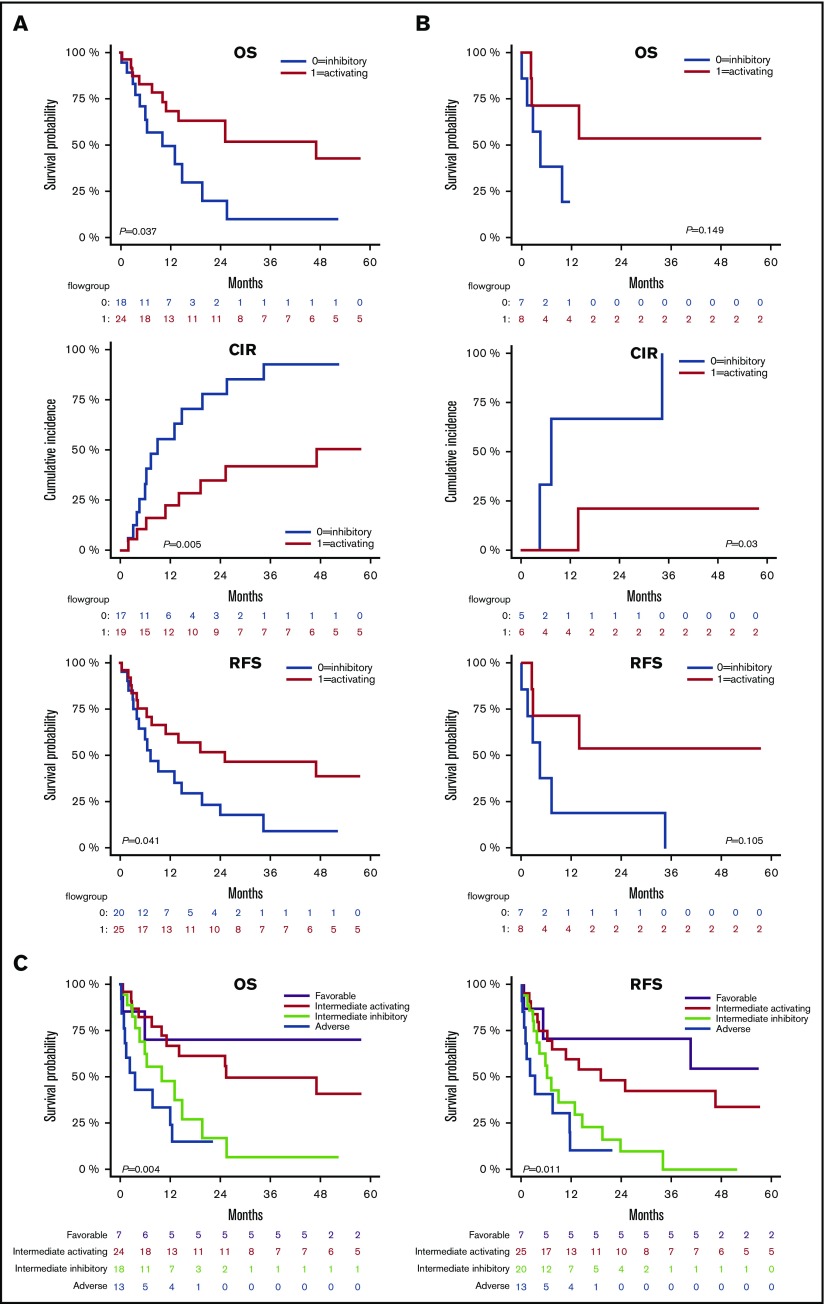
NKRL groups stratify patients with intermediate prognosis
Current cytogenetic and molecular risk classification systems inform clinicians and patients of prognosis and also facilitate decision making regarding the need for postremission consolidation therapy, including alloHSCT (supplemental Figure 2). We investigated if the NKRL overall score (G0 and G1) could add power to existing established cytogenetic and molecular risk systems to enhance their prognostic potential. In the intermediate cytogenetic risk cohort (Figure 5A), patients with a NK activating G1 score had better OS (2-year OS 62.8% vs 19.8%; HR, 0.41; P < .05) and RFS (2-year RFS 51.7% vs 17.5%; HR, 0.46; P < .05) than patients with the more inhibitory G0 score. Relapse rate was also significantly lower in G1 compared with G0, with a 2-year CIR of 34.9% vs 77.7% (HR, 0.27; P < .005). When considering patients with an intermediate ELN molecular risk (Figure 5B), there was a similar trend in favor of G1, although results were statistically significant only for CIR (2-year CIR 20.8% vs 66.7%; HR, 0.12; P < .05), likely because of the small size of this patient subgroup. No differences in OS, CIR, and RFS could be observed between G0 and G1 when individually considering favorable or adverse cytogenetic or molecular risks. Altogether, these data suggest that the NKRL expression pattern on AML blasts at diagnosis can further substratify patients with intermediate cytogenetics. Patients with intermediate cytogenetics, but an activating NKRL score (G1), had prognosis that approximated that of the cytogenetically favorable subgroup, and conversely, patients with intermediate cytogenetics but an inhibitory NKRL score (G0) had prognosis similar to patients with adverse risk cytogenetics (Figure 5C). This prognostic model incorporating cytogenetics and NKRL score therefore subdivides patients into 4 risk groups: favorable risk, intermediate-1 (intermediate cytogenetics plus NKRL score G1), intermediate-2 (intermediate cytogenetics plus NKRL score G0), and adverse risk, with significant differences in OS (P < .005) and RFS (P < .05) among groups.
Figure 5.
NKRL groups stratify patients with intermediate prognosis. Only patients belonging to the intermediate cytogenetic (A) and intermediate molecular risk according to ELN criteria32 (B) were analyzed for OS, CIR, and RFS. Patients belonging to group 0 and 1 are represented by blue and red lines, respectively. (C) Patients were stratified by cytogenetic prognosis, and subjects with intermediate cytogenetic risk were further classified according to their NKRL group: purple line corresponds to favorable cytogenetics; red line corresponds to intermediate cytogenetics and NKRL group 1 (activating pattern); light green line defines intermediate cytogenetics and NKRL group 0 (inhibitory pattern); blue line corresponds to adverse cytogenetics. Kaplan-Meier estimates for OS (left panel) and RFS (right panel) are shown. P values are shown.
Discussion
In our study, we described a heterogeneous expression of both activating and inhibitory NKRLs on AML blasts at diagnosis. Of the activating ligands, ULBP1, MICA, and CD112 were the most frequently expressed, whereas few samples were found to be positive (and with a low intensity of expression) for CD155, MICB, or ULBP2/5/6. In this cohort of patients who all received induction-remission chemotherapy with curative intent, we identified that the expression of ULBP1 was significantly associated with OS, RFS, and CIR. Furthermore, when taking into account all examined NKRLs, we showed that the balance of activating and inhibitory NKRLs expressed by AML blasts at diagnosis impacted upon survival and relapse after induction chemotherapy. Furthermore, the expression of NKRLs by AML blasts was able to stratify patients with intermediate-risk cytogenetics into 2 subgroups, with prognosis that either approximated favorable or adverse risk groups.
Several reports have previously described the expression of various NKRLs by AML blasts. We observed that leukemic blasts universally expressed HLA class I molecules at diagnosis, consistent with previous reports.16,17 In 1 series, loss of HLA class I antigens was noted to be more prevalent at the time of disease relapse, suggesting phenotypic evolution as a result of immunological selection pressure.18 This selection pressure is perhaps most evident in the setting of AML relapse following haploidentical alloSCT, whereby HLA loss relapse is a well-described mechanism of evasion from the graft-versus-leukemia effect against mismatched HLA haplotypes.19,20 Of note, in this work, the expression of HLA class I molecules by leukemic blasts was exclusively interpreted according to its inhibitory effect on NK cells, whereas the overall influence of HLA expression on AML surveillance also encompasses its positive impact on CD8+ cytotoxic T cells.
The expression of the immune checkpoint ligands PD-L1 and PD-L2 by AML blasts was more variable. We observed that 77% of patients had blasts expressing PD-L2 at diagnosis and 26% had blasts expressing PD-L1. Overall, 79% of patients had blasts expressing either PD-L1 or PD-L2, whereas 11% simultaneously expressed PD-L1 and PD-L2. These findings are consistent with previous observations that the PD-1 axis is frequently upregulated in AML and may contribute to immune evasion.21 In 2 independent cohorts, PD-L1–expressing blasts were found in 18% to 25% of patients, and 33% of patients had blasts expressing PD-L2.22,23 The expression of PD-L1 by leukemic blasts was independent of patient age, cytogenetics, and molecular mutation profile. In our cohort, we did not observe any significant correlation between survival outcomes and expression of PD-L1 or PD-L2 by AML blasts. This is consistent with disappointing clinical responses to single-agent PD-1 inhibition in AML.24 When considering the PD-1 axis, it is pertinent to note that the focus of preclinical and clinical investigation has centered upon its role in inhibiting CD8+ T-cell antitumor responses. Although a subset of mature CD56dim NK cells that express the PD-1 receptor is functionally impaired, and its antitumor activity may be partially restored upon inhibition of PD-1/PD-L1, it is likely that the inhibitory effect of PD-1 signaling on NK cells is outweighed by other activating or inhibitory signals.25
Of the activating NKRLs, ULBP1 expression was identified on AML blasts in 82% of patients, including 20% of patients with very high levels of expression. ULBPs are a family of major histocompatibility complex class I–related molecules expressed on the cell surface by a glycosylphosphatidylinositol anchor. ULBPs bind to the activating NKG2D receptor and deliver a positive signal for NK cell activation and cytotoxicity.26 The ULBPs were first investigated in the context of cytomegalovirus evasion from NK cell–mediated attack, whereby virus shedding of soluble UL16 binds to ULBPs and inhibits delivery of an activating signal to NK cells.26 ULBPs (in particular, ULBP1 and ULBP2) are expressed by colon, cervical and ovarian carcinoma, and lymphoma cell lines and contribute to endogenous NK cell–mediated immunoediting through interaction with the NKG2D receptor.27 In addition, the strength of the cytotoxic signal delivered via the NKG2D receptor is dependent on the density of ligand (eg, ULBP) expression on target cells in in vitro cytolytic assays.28 In our cohort, the expression of ULBP1 by AML blasts conferred a significant survival advantage due to a decreased incidence of disease relapse following induction chemotherapy. Furthermore, there was a stepwise improvement in survival and reduction in relapse with increasing intensity of ULBP1 expression, consistent with the preclinical observation that the strength of the cytotoxic signal through ULBP/NKG2D interactions is dose dependent. To our knowledge, the association between ULBP1 expression by AML blasts and clinical outcome following induction chemotherapy has not been reported. Salih et al reported that of 15 patients with AML 20% had blasts that expressed ULBP1.29 In a separate cohort, Nowbakht and coworkers reported that 7 out of 30 (23%) patients with AML had increased ULBP1 expression compared with progenitor cells of healthy donors.30 Increased expression of ULBP1 was associated with an increase in the susceptibility of AML blasts to NK cell–mediated cytolysis in vitro.30 Therefore, our findings are consistent with and contribute to preclinical and in vitro data that ULBP1/NKG2D interactions deliver a potent NK cell activation and cytotoxic signal that can impact the prognosis of patients with AML. Evidence for this mechanism may be further explored by in vitro functional experiments, gene expression evaluation, and ligand-receptor engagement disruption assays.
Tumor cell targets may simultaneously express both activating and inhibitory NKRLs, and the integration and relative potency of these signals determine the eventual posture of NK cells toward either elimination or tolerance.31 For example, cell lines with low expression of ligands for the activating receptor NKp30 may still undergo NK-mediated cytolysis if they express MICA or ULBP, which interact with NKG2D.32 Furthermore, the NKG2D-mediated NK cell activation is abrogated by inhibitory KIR signaling. In an ovarian carcinoma cell line that expressed NKG2D ligands, clones with high expression of HLA-Bw4 were protected from NK cell–mediated cytolysis, and blockade of HLA-Bw4/KIR interaction restored susceptibility to NK cell–mediated attack.32 In our study, in order to reflect the combinatorial effect of multiple NKRLs, we summed the expression of all ligands examined into an overall score to reflect either an overall activating or an inhibitory NKRL phenotype on AML blasts. The presence of a more activating pattern of NKRLs was associated with a reduced rate of AML relapse following induction chemotherapy, resulting in improved OS and RFS. This result is consistent with the aforementioned findings integrating multiple NK cell signals in vitro, but has not previously been reported in clinical cohorts.
Our observations may have 2 main clinical applications. First, the expression of NKRLs may represent a novel prognostic tool for patients with newly diagnosed AML, in particular with patients with intermediate-risk cytogenetics. Patients with intermediate cytogenetics AML demonstrating an activating pattern of NKRLs had a significantly better survival compared with patients with an inhibitory NKRL pattern, whose prognosis approached that of the group with adverse cytogenetic risk. We did not observe any difference in survival outcomes according to NKRL phenotype in patients with either favorable or adverse risk cytogenetics, possibly due to the smaller sample size in these subgroups or alternatively because the impact of karyotype may have significantly outweighed NKRL phenotype. Our observation requires confirmation in an independent, larger cohort prior to practical application.
The second implication of our study is that it suggests that maintenance strategies to enhance NK cell activity following achievement of remission may reduce AML relapse. Indeed, the graft-versus-leukemia effect following alloHSCT is in part mediated by alloreactive NK cells, as evidenced by the benefit of KIR mismatch in haploidentical transplants first reported by the Perugia group and also more recently by demonstration that a higher number of infused donor NK cells is associated with improved RFS after T cell–depleted alloHSCT.5,33 However, a significant proportion of AML patients are unable to proceed to alloHSCT following remission induction therapy due to either comorbidity or lack of a suitable donor. In these patients, alternative strategies to enhance endogenous NK cell–mediated immune surveillance are required. A phase 3 clinical trial using low-dose interleukin-2 combined with histamine dihydrochloride as maintenance therapy for AML demonstrated improved leukemia-free survival compared with no maintenance, an effect that was associated with peripheral blood NK cell expansion and upregulation of the activating natural cytotoxicity receptors NKp30 and NKp46, suggesting that NK cell activation against AML likely contributed to the observed survival benefit.34,35 Our results suggest that an alternative approach to prevent AML relapse may be to therapeutically enhance the expression of activating NKRLs on residual AML blasts or to reduce inhibitory NKRL signaling, thereby swaying the balance of NK cell signaling toward a more activating pattern. Rohner and coworkers reported that azacitidine upregulated ULBP1 expression on AML H60 blasts when used in combination with interferon-γ, resulting in improved sensitivity to NK-mediated killing via NKG2D signaling.36 Other strategies purported to enhance NKG2D ligand expression on tumor cells include bromodomain and extraterminal protein inhibition, heat shock protein-90 inhibitors, and histone deacetylase inhibitors.3,37,38 In addition, the use of these agents while simultaneously counteracting inhibitory NK cell signaling (eg, blockade of PD-1/PD-L1, anti-KIR, targeting of CD96/TIGIT to allow a preferential signaling through the activating receptor CD226) forms a logical combination strategy to further augment NK cell–mediated eradication of residual AML.
In conclusion, our study provides clinical evidence for the role of NK cell–mediated immunoediting against AML, determined by the differential expression of NKRLs on AML blasts at diagnosis. We have identified that the presence and intensity of ULBP1 expression are associated with AML prognosis. Furthermore, an overall activating phenotype of NKRLs is associated with improved survival following induction chemotherapy. Combination strategies to enhance NK cell activation warrant further investigation in the ongoing battle against AML.
Supplementary Material
The full-text version of this article contains a data supplement.
Authorship
Contribution: S.M. and E.W. designed the study, analyzed and interpreted data, and wrote the manuscript; T.P. participated in the design of the study; J.R. performed flow cytometry analysis; P.B. performed next-generation sequencing analysis; M.J.S. participated in data discussion and interpretation of results; R.K. participated in data discussion and interpretation of results; and D.R. designed the study, analyzed and interpreted data, supervised the project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Ritchie, Department of Clinical Hematology and Bone Marrow Transplant Unit, Royal Melbourne Hospital, Grattan St, Melbourne, VIC 3050, Australia; e-mail: david.ritchie@mh.org.au.
References
- 1.Austin R, Smyth MJ, Lane SW. Harnessing the immune system in acute myeloid leukaemia. Crit Rev Oncol Hematol. 2016;103:62-77. [DOI] [PubMed] [Google Scholar]
- 2.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235-271. [DOI] [PubMed] [Google Scholar]
- 3.Diermayr S, Himmelreich H, Durovic B, et al. . NKG2D ligand expression in AML increases in response to HDAC inhibitor valproic acid and contributes to allorecognition by NK-cell lines with single KIR-HLA class I specificities. Blood. 2008;111(3):1428-1436. [DOI] [PubMed] [Google Scholar]
- 4.Schlegel P, Ditthard K, Lang P, et al. . NKG2D signaling leads to NK cell mediated lysis of childhood AML. J Immunol Res. 2015;2015:473175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruggeri L, Capanni M, Urbani E, et al. . Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097-2100. [DOI] [PubMed] [Google Scholar]
- 6.Ruggeri L, Mancusi A, Capanni M, et al. . Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110(1):433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ljunggren HG, Kärre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11(7):237-244. [DOI] [PubMed] [Google Scholar]
- 8.Dulphy N, Chrétien AS, Khaznadar Z, et al. . Underground adaptation to a hostile environment: acute myeloid leukemia vs. natural killer cells. Front Immunol. 2016;7:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blake SJ, Dougall WC, Miles JJ, Teng MW, Smyth MJ. Molecular pathways: targeting CD96 and TIGIT for cancer immunotherapy. Clin Cancer Res. 2016;22(21):5183-5188. [DOI] [PubMed] [Google Scholar]
- 10.Boeck CL, Amberger DC, Doraneh-Gard F, et al. . Significance of frequencies, compositions, and/or antileukemic activity of (DC-stimulated) invariant NKT, NK and CIK cells on the outcome of patients with AML, ALL and CLL. J Immunother. 2017;40(6):224-248. [DOI] [PubMed] [Google Scholar]
- 11.Lion E, Willemen Y, Berneman ZN, Van Tendeloo VF, Smits EL. Natural killer cell immune escape in acute myeloid leukemia. Leukemia. 2012;26(9):2019-2026. [DOI] [PubMed] [Google Scholar]
- 12.Khaznadar Z, Boissel N, Agaugué S, et al. . Defective NK cells in acute myeloid leukemia patients at diagnosis are associated with blast transcriptional signatures of immune evasion. J Immunol. 2015;195(6):2580-2590. [DOI] [PubMed] [Google Scholar]
- 13.Stringaris K, Sekine T, Khoder A, et al. . Leukemia-induced phenotypic and functional defects in natural killer cells predict failure to achieve remission in acute myeloid leukemia. Haematologica. 2014;99(5):836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shemesh A, Brusilovsky M, Hadad U, et al. . Survival in acute myeloid leukemia is associated with NKp44 splice variants. Oncotarget. 2016;7(22):32933-32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Döhner H, Estey E, Grimwade D, et al. . Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wetzler M, Baer MR, Stewart SJ, et al. . HLA class I antigen cell surface expression is preserved on acute myeloid leukemia blasts at diagnosis and at relapse. Leukemia. 2001;15(1):128-133. [DOI] [PubMed] [Google Scholar]
- 17.Brouwer RE, van der Heiden P, Schreuder GM, et al. . Loss or downregulation of HLA class I expression at the allelic level in acute leukemia is infrequent but functionally relevant, and can be restored by interferon. Hum Immunol. 2002;63(3):200-210. [DOI] [PubMed] [Google Scholar]
- 18.Masuda K, Hiraki A, Fujii N, et al. . Loss or down-regulation of HLA class I expression at the allelic level in freshly isolated leukemic blasts. Cancer Sci. 2007;98(1):102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crucitti L, Crocchiolo R, Toffalori C, et al. . Incidence, risk factors and clinical outcome of leukemia relapses with loss of the mismatched HLA after partially incompatible hematopoietic stem cell transplantation. Leukemia. 2015;29(5):1143-1152. [DOI] [PubMed] [Google Scholar]
- 20.Vago L, Perna SK, Zanussi M, et al. . Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med. 2009;361(5):478-488. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Gajewski TF, Kline J. PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood. 2009;114(8):1545-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berthon C, Driss V, Liu J, et al. . In acute myeloid leukemia, B7-H1 (PD-L1) protection of blasts from cytotoxic T cells is induced by TLR ligands and interferon-gamma and can be reversed using MEK inhibitors. Cancer Immunol Immunother. 2010;59(12):1839-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Bueso-Ramos C, DiNardo C, et al. . Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia. 2014;28(6):1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boddu P, Kantarjian H, Garcia-Manero G, Allison J, Sharma P, Daver N. The emerging role of immune checkpoint based approaches in AML and MDS. Leuk Lymphoma. 2017:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Della Chiesa M, Pesce S, Muccio L, et al. . Features of memory-like and PD-1(+) human NK cell subsets. Front Immunol. 2016;7:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosman D, Müllberg J, Sutherland CL, et al. . ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14(2):123-133. [DOI] [PubMed] [Google Scholar]
- 27.Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exp Med. 2005;202(5):583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pende D, Rivera P, Marcenaro S, et al. . Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62(21):6178-6186. [PubMed] [Google Scholar]
- 29.Salih HR, Antropius H, Gieseke F, et al. . Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood. 2003;102(4):1389-1396. [DOI] [PubMed] [Google Scholar]
- 30.Nowbakht P, Ionescu MC, Rohner A, et al. . Ligands for natural killer cell-activating receptors are expressed upon the maturation of normal myelomonocytic cells but at low levels in acute myeloid leukemias. Blood. 2005;105(9):3615-3622. [DOI] [PubMed] [Google Scholar]
- 31.Farag SS, Fehniger TA, Ruggeri L, Velardi A, Caligiuri MA. Natural killer cell receptors: new biology and insights into the graft-versus-leukemia effect. Blood. 2002;100(6):1935-1947. [DOI] [PubMed] [Google Scholar]
- 32.Pende D, Cantoni C, Rivera P, et al. . Role of NKG2D in tumor cell lysis mediated by human NK cells: cooperation with natural cytotoxicity receptors and capability of recognizing tumors of nonepithelial origin. Eur J Immunol. 2001;31(4):1076-1086. [DOI] [PubMed] [Google Scholar]
- 33.Maggs L, Kinsella F, Chan YLT, et al. . The number of CD56dim NK cells in the graft has a major impact on risk of disease relapse following allo-HSCT. Blood Adv. 2017;1(19):1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brune M, Castaigne S, Catalano J, et al. . Improved leukemia-free survival after postconsolidation immunotherapy with histamine dihydrochloride and interleukin-2 in acute myeloid leukemia: results of a randomized phase 3 trial. Blood. 2006;108(1):88-96. [DOI] [PubMed] [Google Scholar]
- 35.Martner A, Rydström A, Riise RE, et al. . Role of natural killer cell subsets and natural cytotoxicity receptors for the outcome of immunotherapy in acute myeloid leukemia. OncoImmunology. 2015;5(1):e1041701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohner A, Langenkamp U, Siegler U, Kalberer CP, Wodnar-Filipowicz A. Differentiation-promoting drugs up-regulate NKG2D ligand expression and enhance the susceptibility of acute myeloid leukemia cells to natural killer cell-mediated lysis. Leuk Res. 2007;31(10):1393-1402. [DOI] [PubMed] [Google Scholar]
- 37.Abruzzese MP, Bilotta MT, Fionda C, et al. . Inhibition of bromodomain and extra-terminal (BET) proteins increases NKG2D ligand MICA expression and sensitivity to NK cell-mediated cytotoxicity in multiple myeloma cells: role of cMYC-IRF4-miR-125b interplay. J Hematol Oncol. 2016;9(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fionda C, Soriani A, Malgarini G, Iannitto ML, Santoni A, Cippitelli M. Heat shock protein-90 inhibitors increase MHC class I-related chain A and B ligand expression on multiple myeloma cells and their ability to trigger NK cell degranulation. J Immunol. 2009;183(7):4385-4394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.