Abstract
Background: Growing data suggest that DNA damage repair and detoxification pathways play crucial roles in radiation-induced toxicities. To determine whether common functional single-nucleotide polymorphisms (SNPs) in candidate genes from these pathways can be used as predictors of radiation pneumonitis (RP), we conducted a prospective study to evaluate the associations between functional SNPs and risk of RP.
Methods: We recruited a total of 149 lung cancer patients who had received intensity modulated radiation therapy (IMRT). GSTP1 and XRCC1 were genotyped using the SurPlexTM-xTAG method in all patients. RP events were prospectively scored using the National Cancer Institute Common Terminology Criteria for Adverse Events (CTCAE), version 4.0. Kaplan-Meier analysis was used to determine the cumulative probability of RP of grade ≥ 2. Cox proportional hazard regression was performed to identify clinical variables and SNPs associated with risk of RP grade ≥ 2, using univariate and multivariate analysis, respectively.
Results: With a median follow-up of 9 months, the incidence of RP of grade ≥ 2 was 38.3%. A predicting role in RP was observed for the GSTP1 SNP (adjusted hazard ratio 3.543; 95% CI 1.770-7.092; adjusted P< 0.001 for the Ile/Val and Val/Val genotypes versus Ile/Ile genotype). Whereas, we found that patients with XRCC1 399Arg/Gln and Gln/Gln genotypes had a lower risk of RP compares with those carrying Arg/Arg genotype (adjusted HR 0.653; 95% CI 0.342-1.245), but with no statistical significance observed (adjusted P = 0.195).
Conclusions: Our results suggested a novel association between GSTP1 SNP 105Ile/Val and risk of RP development, which suggests the potential use of this genetic polymorphism as a predictor of RP. In addition, genetic polymorphisms of XRCC1 399Arg/Gln may also be associated with RP.
Keywords: Radiation pneumonitis, Genetic polymorphism, GSTP1, XRCC1, Lung cancer
Introduction
Lung cancer is one of the most prevalent cancers and the leading cause of cancer-related deaths in China and worldwide 1-2. Radiation therapy, a key cancer treatment modality, plays a remarkable role in lung cancer management. Radiation pneumonitis (RP) is one of the most critical dose-limiting toxicities of thoracic radiation, with 5%-36% of patients experiencing symptomatic (grade 2-5) RP 3-4. The mechanisms underlying RP currently remain to be understood. Numerous factors have been associated with RP and used to estimate the risk of RP. These factors include patient-related factors such as age, sex, smoking status, and pulmonary function, and treatment-related factors, such as radiation dose, irradiated lung volume, surgery, and chemotherapy 5-7. However, despite these associations, it still is difficult to predict the occurrence of RP for individual patients. One possible explanation is genetic components, which has been associated with the development of adverse radiation responses 8. As for RP, several studies have revealed its close association with genetic polymorphisms 4, 9-12. However, these results are not conclusive, and the specific molecular mechanisms underlying RP remain unclear.
Increasing evidence suggests that glutathione S-transferase (GST) polymorphism and expression level can regulate the oxidative stress level and outcome of lung injuries 13-16. The GSTs identified so far can be categorized into four major classes, α, μ, θ and π, based on biochemical characteristics 17. GSTs in the human α, μ and θ families are encoded by different genes, while the GSTπ enzymes are encoded by a single gene, GSTP1. GSTP1 is expressed in epithelial tissues in human, and is the dominant GST enzyme in the lungs 18. Several GSTP1 variants have been described, including one bearing a single nucleotide polymorphism in exon 5 (A/G, Ile105Val, rs1695). However, not much is known about the roles the GSTP1 polymorphism plays in RP. X-ray repair cross-complementing group 1 (XRCC1) is the first gene reported to modulate cell sensitivity to ionizing radiation. A series of recent advances showed that single nucleotide polymorphisms (SNPs) of XRCC1 were related to a host of radiation-related side effects. Among these SNPs, XRCC1 Arg399Gln was the most common polymorphism and has been extensively investigated. However, some results were contradictory, and the conclusions are still in debate 19-24. In this study, we investigated the association between the risk of radiation pneumonitis in Chinese lung cancer patients and two common SNPs, GSTP1 Ile105Val and XRCC1 Arg399Gln.
Materials and Methods
Patients
From April 2014 to March 2016, 149 lung cancer patients treated with radiotherapy in Chinese PLA General Hospital (Beijing, China) were recruited with the following inclusion criteria. Lung cancer, including non-small cell lung and small cell lung cancer, has been histologically or cytologically confirmed. There was no previous or other on-going thoracic radiotherapy. There was no severe radiotherapy contraindication for the patient. The Karnofsky performance status (KPS) score was not lower than 60. Pulmonary function tests before radiotherapy were not mandatory but strongly recommended. Symptom evaluation for each patient was performed in advance. Patients' characteristics and their outcomes were unknown to the investigators performing genetic analysis. The results of genotyping were disclosed to clinical investigators after data analysis. This research protocol was approved by the Internal Review Board of Chinese PLA General Hospital (Beijing, China), and was conducted in agreement with the Helsinki Declaration. Written informed consent was obtained from each patient before entry into the study and sample collection.
Radiation treatment
All patients received image-guided IMRT with 6-MV photo beam. Patients underwent computed tomography (CT)-based treatment simulation in the supine position while immobilized in an upper body. Then, the CT images were transmitted to the Pinnacle planning system (version 9.2, Philips), where the target volume and critical normal organs were delineated. The total lung volume was then calculated as lung volume minus the tumor volume. Afterwards, the IMRT plan was generated and optimized using the Pinnacle planning system, and the doses were calculated using the collapsed cone convolution dose algorithm. Plans for each patient were optimized by direct parameter optimization, based on target volume and locations of the organs at risk. Volume of lung areas receiving radiation of > 20 Gy (V20) was limited to 25-35% of the total lung volume. The image-guided IMRT was delivered using the two predefined linear accelerators (Elekta Synergy and Varian Clinac iX). A total dose of 30-72 Gy, at a rate of 1.8-3 Gy per fraction, was given once per day, five per week.
Pulmonary toxicity assessment and follow-up
All patients were evaluated weekly during radiation and 4-6 weeks after completion of treatment. Afterwards, patients were visited every 3 months for the first 2 years and then every 6 months. Additional extra visits were required if symptoms occurred. CT examinations were performed upon each follow-up visit after completion of radiotherapy. RP was diagnosed by clinical manifestations (e.g. dyspnoea, cough, pain and low-grade fever) and radiological findings. Once diagnosed, RP was further graded by at least two radiation oncologists following the Common Toxicity Criteria for Adverse Events (CTCAE) version 4 25. Criteria for each grade were as follows. Grade 0, no change. Grade 1, RP was asymptomatic and can only be observed in radiographic findings. No intervention was indicated. Grade 2, patient manifested symptoms that limit activities of daily living (ADL). Medical intervention has been indicated. Grade 3, patient manifested severe symptoms limiting self-care ADL. Oxygen has been indicated. Grade 4, patient manifested life-threatening respiratory compromise with urgent intervention indicated (e.g. tracheotomy and intubation). Grade 5, RP has caused lethality. If the symptoms were present at baseline, worsening of symptoms of at least one grade was considered as RP. In addition, if there was pulmonary infection or thoracic disease progression, RP was excluded from the diagnosis. The diagnosis of RP of grade ≥ 2 was defined as the primary end point. The time to endpoint was calculated from the beginning of radiation. Patients were censored at the time of the last follow-up visit or death.
DNA extraction and genotyping
The SNPs selected and their dbSNP ID (rs) were as follows: GSTP1-Ile105Val (rs1695), and XRCC1- Arg399Gln (rs25487). Blood sample was collected from each patient before radiotherapy. Total genomic DNA from peripheral blood leukocytes was extracted with the Maxwell system (Promega, Madison, WI, USA). The SNP status was analyzed with the SurPlexTM-xTAG platform (Surexam, Guangzhou, China) as described by Zhu et al 26. Briefly, target genes were amplified with PCR and cleaned up with Exonuclease I and shrimp alkaline phosphatase (EXO-SAP) to remove excess nucleotides and primers. Then, the target genes were amplified again with 70 allele-specific primers linked to 70 universal tags. Using these tags, the amplified alleles were hybridized onto beads and analyzed with Luminex. The median fluorescence intensity was measured and analyzed. For quality control, samples were randomly selected and sent to independent services for DNA sequencing analysis.
Multiplex PCR was performed in a 50 ul volume containing: 1x Ex Taq polymerase buffer, 2.5 mM MgCl2, 0.2 mM dNTPs, 0.2 mM each of the primers, 1.0 U Ex Taq HS DNA polymerase (TaKaRa) and 10 ul DNA. Thermo cycling was performed using 30 cycles of 95℃ for 30 s, 56℃ for 30 s, and 72℃ for 30 s. The reaction was concluded with a final extension step of 72℃ for 10 min and the product was kept at 4℃ until use. A similar approach was used to enrich DNA fragments of GSTP1 and XRCC1 genes using 3 mM MgCl2. Exonuclease I and shrimp alkaline phosphatase (EXO-SAP) reaction was performed in a 25 μl volume containing: 1x SAP buffer, 1 U shrimp alkaline phosphatase (TaKaRa), 10 U exonuclease I (degrading any remaining PCR primers) (New England Biolabs, Ipswich, USA) and 7.5 ul PCR product. Samples were then incubated at 37℃ for 30 min, followed by 20 min incubation at 80℃ to inactivate the enzymes. Multiplex ASPE was carried out using 5 μl of treated PCR product in a final volume of 20 μl. Each reaction consisted of SAP buffer, 1.25 mM MgCl2, 5 mM biotin-dCTP, 5 mM each of dATP, dTTP, dGTP; 0.75 U of Platinum Tsp; and 25 nM ASPE primer pool. The ASPE reactions were incubated at 96℃ for 2 min and then subject to 40 cycles at 94℃ for 30 s, 52℃ for 1 min, and 74℃ for 2 min.
Statistical analysis
Patients were grouped based on their genotypes. Cox proportional hazard models were performed to identify clinical variables and genotypes associated with risk of RP, using univariate and multivariate analysis, respectively. Kaplan-Meier analysis was performed to assess the effects of different genotypes on the cumulative probability of RP. All statistical tests were two-sided, and P values < 0.05 were considered statistically significance. Statistical analyses were performed using the IBM SPSS Statistics platform (Version 20, SPSS Science, IL, USA).
Results
Characteristics of patients and clinical outcome
The demographic and clinical information of the 149 recruited lung cancer patients were summarized in Table 1. The median age was 60 years old (age ranges from 24 to 84 years old). There were 127 men (85.2%) and 22 women (14.8%). In terms of histology, 30.2% of the patients were diagnosed with squamous cell carcinoma (45 cases), 18.8% with adenocarcinoma (28 cases), and 38.5% with small cell cancer (57 cases). Over half of patients (65.2%) had stage III lung cancer, 28.2% with stage IV, and 10 patients with stage I-II. Of all patients, 71.1% were current or former smokers. Twenty-five patients (16.8%) were also diagnosed with chronic obstructive pulmonary disease (COPD). Notably, 13 patients (9.4%) had undergone surgery before radiotherapy. Most patients (89.9%) were treated with a combination of radiotherapy, chemotherapy, and target therapy. In the 121 patients who participated in pulmonary function tests, the median force expiratory volume in 1 second (FEV1) and diffusion capacity for carbon monoxide of the lung (DLCO) was 2.58 L (range, 0.83-3.63) and 18.24 mL/min/mm Hg (5.82-33.01), respectively. Most patients (84.6%) received radiation dose above 60 Gy (median, 60 Gy; range 30-72 Gy). The median V20 was 22% (5.0-35.5), median V5 58% (15.0-92.7), and median mean lung dose (MLD) was 13.8 Gy (range, 4.2-34.9 Gy). The median follow-up time was 9 months (range 3-24 months) after the initiation of radiotherapy. Fifty-seven patients (38.3%) developed RP of grade ≥ 2, and the median interval before RP diagnosis was 2.6 months (range 1-6 months). In addition, 90 patients experienced RP of grade ≥ 1, while 10 patients suffering RP grade ≥ 3, and four patients died from severe RP.
Table 1.
Patient demographics and clinical characteristics (N=149)
| Parameter (Variable) |
All Patients (n=149) |
Grade0-1RP (n=92) |
Grade≥2RP (n=57) |
|---|---|---|---|
| Sex | |||
| Male | 127(85.2%) | 82(89.1%) | 45(78.9%) |
| Female | 22(14.8%) | 10(10.9%) | 12(21.1%) |
| KPS | |||
| <80 | 24 (16.1 %) | 13(14.1%) | 11(19.3%) |
| ≥80 | 125 (83.9%) | 79(85.9%) | 46(80.7%) |
| Smoking Status | |||
| No | 43(28.9%) | 26(28.3%) | 17(29.8%) |
| Yes | 106(71.1%) | 66(71.7%) | 40(70.2%) |
| COPD | |||
| No | 124 (83.2%) | 80(87.0%) | 44(77.2%) |
| Yes | 25 (16.6%) | 12(13.0%) | 13(12.8%) |
| Tumor characteristics | |||
| Histology | |||
| Squamous cell carcinoma | 45(30.2%) | 27(29.3%) | 18(31.6%) |
| Adenocarcinoma | 28(18.8%) | 20(21.7%) | 8(14.0%) |
| Small cell lung cancer | 57(38.2%) | 33(35.9%) | 24(42.1%) |
| Others | 19(12.8%) | 12(13.1%) | 7(12.3%) |
| Stage | |||
| I- II | 10(6.7%) | 6(6.5%) | 4(7.0%) |
| III | 97(65.1%) | 59(64.1%) | 38(66.7%) |
| IV | 42(28.2 %) | 27(29.4%) | 15(26.3%) |
| Surgery27 | |||
| NO | 136(90.6%) | 85(92.4%) | 51(89.5%) |
| YES | 13(9.4%) | 7(7.6%) | 6(10.5%) |
| Chemotherapy | |||
| No | 15(10.1%) | 7(7.6%) | 8(14.0%) |
| Yes | 134(89.9%) | 85(92.4%) | 49(86.0%) |
| Median age (y) | 60(24-84) | 59(24-84) | 62(35-80) |
| Median Radiation dose(range)(Gy) | 60(30-72Gy) | 60(32-70) | 61.6(30-72) |
| Pre-RT pulmonary function | |||
| Median FEV1(range)(L) | 2.52(0.83-3.63; n=121) | 2.50(1.01-4.63; n=75) | 2.53(0.83-3.42; n=46) |
| Median FVC(range)(L) | 3.36(1.54-5.25;n=121) | 3.46(1.54-5.25; n=75) | 3.34(1.61-4.78; n=46) |
| Median DLCO(range) (mL/min/mmHg) | 18.24(5.82-33.01; n=121) | 18.04((5.82-33.01; n=75) | 18.51(9.64-26.43;n=46) |
| Bilateral Lung dose-volumehistogram | |||
| Median V5 (range) | 58.0%(15.0-92.7%) | 58.0%(15.0-92.7%) | 58.5%(29.0-91.9%) |
| Median V20(range) | 22.0%(5.0-35.5%) | 21.0%(5.0-29.6%) | 23.5%(6.0-35.5%) |
| Median MLD (range) | 13.8Gy(4.2-34.9Gy) | 13.6Gy(5.3-22.4Gy) | 14.2Gy(4.2-34.9Gy) |
RP: radiation pneumonitis; KPS: Karnofsky performance status; COPD: chronic obstructive pulmonary disease; V5: volume of normal lung receiving 5Gy or more radiation; V20: volume of normal lung receiving 20 Gy or more radiation; MLD: mean lung dose; FEV1: forced expiratory volume in 1 second; FVC: forced vital capacity; DLCO: diffusion capacity for carbon monoxide of the lung.
Genotype frequency
The genotype frequencies and distributions of the two SNPs are shown in Table 2. For GSTP1, 95 cases (63.8%) were of the genotype Ile/Ile, 49 (32.8%) were Ile/Val, and 5 (3.4%) were Val/Val. The frequency of the Ile allele and Val allele were 239 (80.2%) and 61 (19.8%), respectively. For XRCC1, 87 patients (58.4%) were Arg/Arg carriers, 52 (34.9%) were Arg/Gln, and 10 (6.7%) were Gln/Gln genotype carriers. The frequency of the Arg allele and Gln allele were 226 (75.2%) and 72 (24.2%), respectively. The allele frequencies of two SNPs observed in this study were all similar to those previously reported in Chinese population 17, 26-28.
Table 2.
Genotypic and allelic frequencies of gene polymorphisms in this study
| Gene | Exon | NCBI ds SNP ref: ID |
Amino acid no. | Genotype | Number(%) 149(%) |
Allele number(%) 298(%) |
|---|---|---|---|---|---|---|
| GSTP1 | 5 | rs1695 | 105 | A/A (Ile/Ile) | 95(63.8) | A(Ile) 239(80.2) |
| A/G (Ile/Val) | 49(32.8) | G(Val) 61(19.8) | ||||
| G/G (Val/Val) | 5(3.4) | |||||
| XRCC1 | 10 | rs25487 | 399 | G/G (Arg/Arg) | 87(58.4) | G(Arg) 226(75.8) |
| G/A (Arg/Gln) | 52(34.9) | A(Gln) 72(24.2) | ||||
| A/A (Gln/Gln) | 10(6.7) |
GTSP1: Glutathione S-transferase P1; XRCC1: X-ray repair cross-complementing group 1; dbSNP ref no.: National Center for Biotechnology Information Single Nucleotide Polymorphism database reference number (Ref SNP Cluster Report information available online at (http://www.ncbi.nlm. nih.gov)); Ile: isoleucine; Val: valine; Arg: arginine; Gln: glutamine.
Clinical factors and risk of RP
We assessed the associations between patient-, tumor-, and therapy-related factors and RP grade ≥ 2 by univariate and multivariate Cox regress analyses (Table 3). These factors include age, sex, race, KPS, tumor histology, disease stage, smoking status, COPD history, surgery, chemotherapy, radiation dose, V20, V5, MLD, FEV1, forced vital capacity (FVC) and DLCO. Age and V20 were significantly associated with RP risk (grade ≥ 2), while surgery had a borderline significant effect (P = 0.062) on RP development. Patients older than 60 years old had a higher risk of RP compared with those younger than 60 years, after adjustment for other covariates (HR 2.517, 95% CI 1.167-5.430, P = 0.019). In addition, V20 also showed strong association with the risk of RP. No other analyzed clinical factors were observed to be associated with RP risk in this study population.
Table 3.
Association between patient-, tumor-, and treatment-related characteristics and grade ≥2RP
| Parameter (Variable) |
Patients | Number of RPs(n=57) | Percent of Patients RPs | Univariate Analysis | Multivariate Analysis† | ||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | p | AHR | 95%CI | p | ||||
| Sex | |||||||||
| Male | 127 | 45 | 35.4% | 1.0 | 1.0 | ||||
| Female | 22 | 12 | 54.5% | 1.370 | 0.725-2.590 | 0.333 | 1.888 | 0.605-5.891 | 0.274 |
| Age(years) | |||||||||
| <60 | 71 | 24 | 33.8% | 1.0 | 1.0 | ||||
| ≥60 | 78 | 33 | 42.3% | 1.339 | 0.791-2.266 | 0.277 | 2.517 | 1.167-5.430 | 0.019 |
| KPS | |||||||||
| <80 | 24 | 11 | 45.8% | 1.0 | 1.0 | ||||
| ≥80 | 125 | 46 | 36.8% | 0.622 | 0.343-1.279 | 0.220 | 0.935 | 0.393-2.222 | 0.879 |
| Smoking Status | |||||||||
| NO | 43 | 17 | 39.5% | 1.0 | 1.0 | ||||
| YES | 106 | 40 | 37.7% | 1.092 | 0.619-1.927 | 0.761 | 1.690 | 0.622-4.590 | 0.304 |
| COPD | |||||||||
| No | 124 | 44 | 35.5% | 1.0 | 1.0 | ||||
| Yes | 25 | 13 | 52.0% | 1.621 | 0.872-3.013 | 0.127 | 1.043 | 0.400-2.724 | 0.931 |
| Tumor characteristics | |||||||||
| Histology | |||||||||
| Squamous cell carcinoma | 45 | 18 | 40.0% | 1.0 | 1.0 | ||||
| Adenocarcinoma | 28 | 8 | 28.6% | 0.643 | 0.279-1.478 | 0.298 | 0.800 | 0.264-2.420 | 0.800 |
| Small cell lung cancer | 57 | 24 | 42.1% | 0.978 | 0.531-1.803 | 0.943 | 1.244 | 0.560-2.765 | 0.591 |
| Others | 19 | 7 | 36.8% | 0.898 | 0.375-2.151 | 0.810 | 1.368 | 0.465-4.028 | 0.569 |
| Stage | |||||||||
| I-II | 10 | 4 | 40.0% | 1.0 | 1.0 | ||||
| III | 97 | 38 | 39.2% | 1.101 | 0.393-3.087 | 0.854 | 0.909 | 0.232-3.558 | 0.891 |
| IV | 42 | 15 | 35.7% | 0.941 | 0.312-2.838 | 0.914 | 0.967 | 0.233-4.008 | 0.963 |
| Surgery | |||||||||
| NO | 136 | 51 | 37.5% | 1.0 | 1.0 | ||||
| YES | 13 | 6 | 46.2% | 1.193 | 0.512-2.781 | 0.683 | 3.048 | 0.959-9.686 | 0.059 |
| Chemotherapy | |||||||||
| NO | 15 | 8 | 53.3% | 1.0 | 1.0 | ||||
| YES | 134 | 49 | 36.6% | 0.729 | 0.345-1.541 | 0.408 | 0.549 | 0.204-1.475 | 0.234 |
| Radiation dose | |||||||||
| <60 | 23 | 7 | 30.4% | 1.0 | 1.0 | ||||
| ≥60 | 126 | 50 | 39.7% | 1.435 | 0.650-3.186 | 0.372 | 1.080 | 0.283-4.114 | 0.910 |
| Bilateral Lung dose-volume histogram | |||||||||
| V5<58% | 75 | 28 | 37.3% | 1.0 | 1.0 | ||||
| V5≥58% | 74 | 29 | 39.2% | 1.161 | 0.691-1.952 | 0.573 | 1.133 | 0.403-3.186 | 0.813 |
| V20<20% | 56 | 14 | 25.0% | 1.0 | 1.0 | ||||
| V20≥20%<25% | 53 | 24 | 45.3% | 2.043 | 1.057-3.950 | 0.034 | 2.971 | 1.085-8.133 | 0.034 |
| V20≥25% | 40 | 19 | 47.5% | 2.182 | 1.094-4.354 | 0.027 | 5.810 | 1.391-24.27 | 0.016 |
| MLD<13.8Gy | 74 | 24 | 32.4% | 1.0 | 1.0 | ||||
| MLD≥13.8Gy | 75 | 33 | 44.0% | 1.372 | 0.811-2.321 | 0.239 | 1.605 | 0.666-3.872 | 0.292 |
| FEV1(L),n=121 | |||||||||
| <2.52 | 61 | 23 | 37.3% | 1.0 | 1.0 | ||||
| ≥2.52 | 60 | 23 | 38.7% | 1.219 | 0.683-2.174 | 0.503 | 1.449 | 0.569-3.691 | 0.437 |
| FVC(L),n=121 | |||||||||
| <3.36 | 60 | 24 | 40.0% | 1.0 | 1.0 | ||||
| ≥3.36 | 61 | 22 | 36.1% | 0.954 | 0.535-1.703 | 0.874 | 1.149 | 0.463-2.855 | 0.764 |
| DLCO (mL/min/mmHg), n=121 | |||||||||
| <18.24 | 61 | 21 | 34.4% | 1.0 | 1.0 | ||||
| ≥18.24 | 60 | 25 | 41.7% | 1.315 | 0.735-2.351 | 0.357 | 1.121 | 0.515-2.441 | 0.774 |
†Multivariate analyses were adjusted for all factors listed in Table.
RP: radiation pneumonitis; HR: hazard ratio; KPS: Karnofsky performance status; COPD: chronic obstructive pulmonary disease; V5: volume of normal lung receiving 5Gy or more radiation;V20: volume of normal lung receiving 20 Gy or more radiation; MLD: mean lung dose; FEV1: forced expiratory volume in 1 second; FVC: forced vital capacity; DLCO: diffusion capacity for carbon monoxide of the lung.
Association between GSTP1 and XRCC1 and RP risk
Correlation between GSTP1 and XRCC1 SNPs and RP of grade ≥ 2 were analyzed using the Cox proportional hazards regression models. The results of the univariate and multivariate analyses are summarized in Table 4. Of note, of the five patients harboring GSTP1 the 105Val/Val genotype, four patients (80%) experienced grade ≥ 2 RP. Further statistical analysis revealed a significant associated between GSTP1 105Ile/Val or Val/Val polymorphisms and development of RP of grade ≥ 2. Multivariate model analysis showed compared to the GSTP1 105Ile/Ile genotype, carriers of the 105Ile/Val or 105Val/Val genotypes had a 3.141-fold (95% CI, 1.536-6.424; adjusted P = 0.002) or 9.850-fold (95% CI, 2.411-40.242; adjusted P = 0.001) increased risk for developing RP, respectively. When the GSTP1 Ile/Val and Val/Val variants were grouped for analysis, the hazard ratio (HR) was 3.543 (95% CI 1.770-7.092; adjusted P = 0.000). In contrast, patients carrying XRCC1 399Arg/Gln or Gln/Gln variants did not show significantly decreased risk of RP (HR 0.652; 95% CI 0.329-1.291; adjusted P = 0.220) or (HR 0.748, 95% CI 0.148-3.778; adjusted P = 0.726) respectively. Similarly, combination of the XRCC1 399Arg/Gln and Gln/Gln genotypes, a reduced risk was observed compared with Arg/Arg carriers, albeit without achieving statistical significance (HR 0.653, 95% CI 0.342-1.245; adjusted P = 0.195).
Table 4.
Association between GSTP1 and XRCC1 and grade≥2RP
| Parameter (Variable) | Patients | Number of RPs(n=57) | Percent of Patients RPs | Univariate Analysis | Multivariate Analysis† | ||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | p | AHR | 95%CI | p | ||||
| GSTP1Ile105Val‡ | |||||||||
| Ile/Ile | 95 | 28 | 29.5% | 1.0 | 1.0 | ||||
| Ile/Val | 49 | 25 | 51.0% | 2.286 | 1.329-3.931 | 0.003 | 3.141 | 1.536-6.424 | 0.002 |
| Val/Val | 5 | 4 | 80.0% | 3.787 | 1.322-10.845 | 0.013 | 9.850 | 2.411-40.242 | 0.001 |
| Ile/Val+Val/Val | 54 | 29 | 53.7% | 2.416 | 1.433-4.075 | 0.001 | 3.543 | 1.770-7.092 | 0.000 |
| XRCC1Agr399Gln‡ | |||||||||
| Arg/Arg | 87 | 36 | 41.4% | 1.0 | 1.0 | ||||
| Arg/Gln | 52 | 18 | 34.6% | 0.732 | 0.416-1.290 | 0.281 | 0.652 | 0.329-1.291 | 0.220 |
| Gln/Gln | 10 | 3 | 30.0% | 0.653 | 0.201-2.122 | 0.479 | 0.748 | 0.148-3.778 | 0.726 |
| Arg/Gln+Gln/Gln | 62 | 21 | 33.9% | 0.720 | 0.420-1.233 | 0.231 | 0.653 | 0.342-1.245 | 0.195 |
RP: radiation pneumonitis; HR: hazard ratio; GSTP1: Glutathione S-transferase P1; XRCC1: X-ray repair cross- complementing group 1; Ile: isoleucine; Val: valine; Arg: arginine; Gln: glutamine.
†Multivariate analyses in this table were adjusted for all factors listed in Table 3.
‡Either GSTP1 or XRCC1 was independently entered into a multivariate model that adjusted for all factors listed in Table 3.
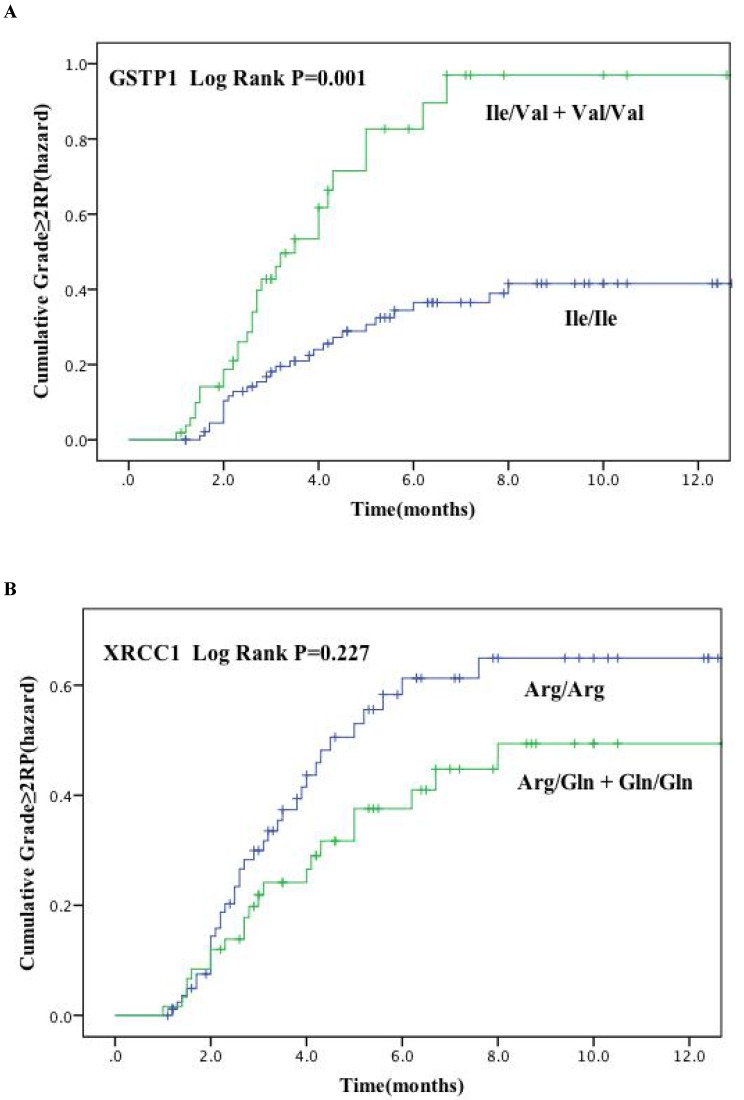
Figure. 1 plots the incidence of RP grade ≥ 2 as a function of time since radiation therapy according to these two genetic polymorphisms. Kaplan-Meier estimates indicated that patients with GSTP1 105Ile/Val and Val/Val variants had significantly higher risk of RP of grade ≥ 2 (P = 0.001) (Figure. 1A). The cumulative incidence of RP (grade ≥ 2) at 3 months after radiotherapy for patients with the variant versus wild-type genotypes were 30.6% and 16.6%, and 59.2% and 34.8% at 6 months after radiotherapy, respectively. However, no significant findings were observed for the XRCC1 Arg399Gln polymorphisms (Log rank P = 0.277) (Figure. 1B).
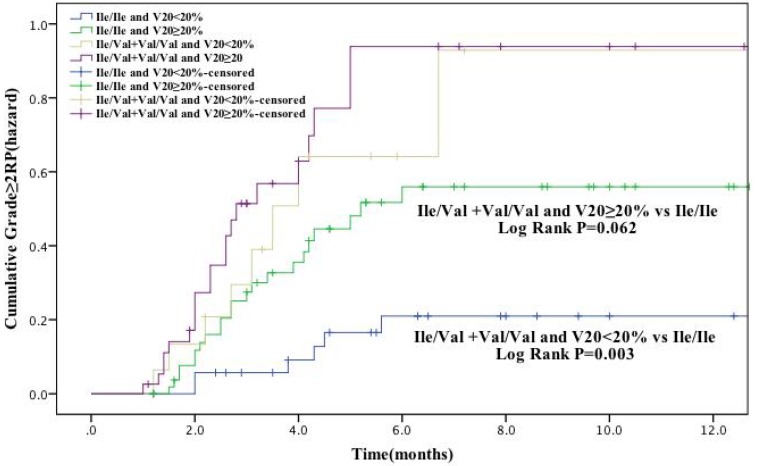
In our study population, V20 was identified as a factor significantly associated with RP risk (grade ≥ 2). Therefore, we further analyzed the cumulative RP incidence (grade ≥ 2) as a function of time according to genotype and V20 (Figure. 2). Patients with GSTP1 105Ile/Val and Val/Val genotypes and received V20 <20% had a remarkable higher RP incidence than Ile/Ile genotype carriers with V20 <20% (P = 0.003). Moreover, there was also difference in RP incidence between these two patient groups with V20 ≥20% (P = 0.062), although with a borderline statistical significance. Altogether, these results suggest that the risk of development of RP (grade ≥ 2) in lung cancer receiving radiotherapy is associated with several factors, including patient age and V20, as well as genetic polymorphisms, including GSTP1 Ile105Val and possibly XRCC1 Arg399Gln.
Discussion
It has been well established that radiation therapy exerts its cytotoxic effects either directly, by attacking cellular macromolecules such as DNA, or indirectly by inducing reactive oxygen species (ROS) production and consequent oxidative damage 29. Therefore, the expression levels and activities of key players in the DNA damage repair and anti-oxidant defense networks are crucial in determining the response of irradiated tissues. An important aspect of gene function, gene polymorphism, has been suggested to regulate radiosensitivity in either a generalized or a tissue- or cell-type-specific manner 30. Radiation-induced injuries are mostly localized to normal tissues and organs exposed to irradiation, suggesting the significance of tissue- or cell-type-specific modulation of radiosensitivity by genetic polymorphisms in lung cancer patients 31. Lots of publications have focused on studying the relationship between SNPs and lung cancer, such as radiotherapy or chemotherapy resistance 13-16, but few of them tried to clarify association of SNPs and radiation pneumonitis (RP). In this study, our data revealed that GSTP1 Ile105Val polymorphism might be a biomarker for predicting the risk of radiation pneumonitis among lung cancer patients in Chinese population.
GSTs play a vital role in the detoxification of and protection against radiation-induced oxidative damages in biological molecules, including DNA and proteins 17. Glutathione Stransferase-P1 (GSTP1) is the most abundantly expressed GST, accounting for 83% of all GST isoenzymes, and functions as the most important detoxification enzyme in the lungs 18. The encoding gene, GSTP1, is a polymorphic gene located to chromosome 11, and has been implicated in the regulation of cell proliferation, apoptosis, stress response, phase II metabolism, oncogenesis, tumor progression and drug resistance.
To date, two variants of this gene have been discovered, as results of A1578G transition (exon 5, A313G, rs1695) and C2293T transition (exon 6, C341T, rs1138272), leading to amino acid residue substitutions Ile105Val and Ala114Val, respectively. Both amino acid residues are located within the active site of GSTP1 17-18,32. Moreover, GSTP1 exon 5 polymorphism was found to dampen GSTP1 enzymatic activity, whereas the exon 6 polymophism had no apparent effects on enzyme activity 32-33. Compared with the wide-type Ile/Ile genotype of GSTP1, the Ile105Val variant exhibited a reduced enzymatic activity by 50%-70% 32, thereby putting this SNP under the limelight in several recent studies. Accumulating data has suggested that GSTP1 Ile105Val polymorphism may contribute to the risks of lung cancer and multidrug resistance 27, 34. However, the results are still not conclusive. As Ile105Val allelic variant, with decreased anti-oxidant activity, may lead to increased susceptibility to oxidative DNA damage. It is therefore conceivable that the GSTP1 Ile105Val SNP can be associated with the normal tissue response to radiation, especially in the lung. However, there has been so far no report on the relationship of genetic polymorphisms of GSTP1 and the risk of RP in lung cancer patients. Our finding suggests that the low activity variant of GSTP1 is strongly associated with a significantly increasing risk for RP in Han Chinese patients with lung cancer, which is in consistent with previous reports in breast cancer 19,35-37. In addition, our novel findings also suggest the potential application of GSTP1 in customizing radiotherapy regimens for lung cancer patients. At the same time, most studies were carried on within western population, while our samples were uniquely collected from Chinese Han population. Plenty of researches have proved that the prevalence of many diseases including RP among different races varies significantly, which may be caused by genetic diversity, and interestingly the frequency of GSTP1 Ile105Val polymorphism is quite different among races according to Exome Aggregation Consortium (ExAC) database which may partially explain why the prevalence of RP is different between east and west.
XRCC1 plays a prominent role in base excision repair (BER) to efficiently repair DNA damage induced by ionizing radiation, oxidative stress, and DNA alkylating agents 38. XRCC1 is mapped to human chromosome 19q13.2-13.339, and shows three relatively common polymorphisms at codons 194 (Arg/Trp), 280 (Arg/His), and 399 (Arg/Gln) 40. Arg399Gln (rs25487, G28152A) is the most frequently observed among these SNPs 41, and has been increasingly characterized as to its association with the risk of normal tissue injury after radiotherapy 11-12. However, similar to GSTP1, there are contradicting reports regarding the validity of this association and its potential as a predictor. In a retrospective study conducted by Yin et al. 11, the XRCC1 399Gln variant was associated with a decreased risk of RP in patients with non-small-cell lung cancer (HR 0.48, P = 0.041). In another report, Kelsey et al. performed a prospective study to examine the correlation between XRCC1 and radiation-induced lung injury in lung cancer patients receiving definitive radiotherapy. The results also suggested association between the 399Arg allele and increased radiosensitivity (P = 0.01). Consistently with these reports, in this study, we found that patients carrying XRCC1 399Gln variant had a lower risk for developing RP (HR 0.653; 95% CI 0.342-1.245) 11-12, 23-24, although the results did not establish statistical significance, in concert with the findings reported by Cheuk et al 24. The possible reason for this discrepancy may be partly attributed to large ethnic differences in the frequency of XRCC1 Arg399Gln alleles.
Several considerations during conducting this study contributed to increase the clinical relevance of this study and our findings, including standardized data collection on side effects at defined time points during and after radiotherapy, and standardized RP classification system. Furthermore, patients received radiotherapy administered using image-guided IMRT, a state-of-the-art technology that allowed radiation delivery with more precise dosage and area targeting. Moreover, a few confounding factors, including tumor-, patient-, treatment-, and dosimetric-related factors, were taken into consideration in the Cox regression analysis in identifying the impact of the genetic components. Together, standardized experiment design and advanced radiotherapy technology help maximize the confidence and relevance of conclusions from this investigation. However, there are certain caveats in this study. For instance, the sample size is relatively small (149 patients), and a larger-scale study is warranted to confirm our findings in the future. Also, we selected GSTP1 and XRCC1 SNPs based on previous reports. A more comprehensive survey of more SNPs in other genes across the genome would most likely benefit the discovery of the association between risk of RP and other SNPs.
Conclusion
In this prospective study, through analyzing the genotyping and following-up data of a total of 149 patients, we found a significant association between the GSTP1 Ile105Val polymorphism and the risk of RP. This association, which has never been reported, suggests potential clinical application of this SNP as a predictive biomarker for RP susceptibility in lung cancer patients. In addition, XRCC1 Arg399Gln appears to be associated with RP risk, although the results were not statistically significant. Together, these associations may help clinicians in optimizing radiotherapy regimen for individual patient with minimized risk of developing severe or even lethal radiation-induced toxicities. This novel, promising discoveries warrant confirmation by further investigations with large sample size.
Figure 1.
Cumulative probability of grade ≥ 2 RP as a function of time from the start of radiation therapy by genotypes. (A) GSTP1 rs1695: Ile105Val; (B) XRCC1 rs25487: Arg399Gln. The Ile/Val and Val/Val genotypes of GSTP1 were associated with a statistically significant higher incidence of RP compared with other genotypes.
Figure 2.
The effects of single nucleotide polymorphism at GSTP1 Ile105Val with volume of normal lung receiving 20Gy or more radiation (V20) < 20% the cumulative incidence of grade ≥ 2 RP. Patients with the Ile/Val and Val/Val genotypes of GSTP1 and V20 < 20% had a statistically significant higher incidence of grade ≥ 2 RP compared with Ile/Ile genotype. These association between GSTP1 rs1695: Ile105Val and risk grade ≥ 2 RP is independent of the V20.
Acknowledgments
This study was supported by the National Key Projects of Research and Development of China (2016 YFC 0904600), the Central Military Commission of the Ministry of Health (16BJZ17), and the Chinese PLA General Hospital Clinical Support Fund (2016FC-TSYS-2017).
Abbreviations
- SNPs
single-nucleotide polymorphisms
- RP
radiation pneumonitis
- IMRT
intensity modulated radiation therapy
- GST
glutathione S-transferase
- GSTP1
Glutathione Stransferase-P1
- XRCC1
cross-complementing group 1
- KPS
Karnofsky performance status
- CTCAE
Common Toxicity Criteria for Adverse Events
- ADL
activities of daily living
- COPD
chronic obstructive pulmonary disease
- FEV1
force expiratory volume in 1 second
- DLCO
diffusion capacity for carbon monoxide of the lung
- MLD
median mean lung dose
- FVC
forced vital capacity
- HR
hazard ratio
- ROS
reactive oxygen species
- BER
base excision repair.
References
- 1.Chen W, Zheng R, Baade PD. et al. Cancer Statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel R. et al. Global cancer statistics 2012. CA cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Medhora M, Gao F, Fish BL. et al. Dose-modifying factor for captopril for mitigation of radiation injury to normal lung. J Radiat Res. 2012;53:633–40. doi: 10.1093/jrr/rrs004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo CX, Wang J, Huang LH. et al. Impact of single-nucleotide polymorphisms on radiation pneumonitis in cancer patients. Mol Clin Oncol. 2016;4:3–10. doi: 10.3892/mco.2015.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsoutsou PG, Koukourakis MI. Radiation pneumonitis and fibrosis: Mechanisms underlying its pathogenesis and implications for future research. Int J Radiat Oncol Biol Phys. 2006;66:1281–93. doi: 10.1016/j.ijrobp.2006.08.058. [DOI] [PubMed] [Google Scholar]
- 6.Das SK, Zhou S, Zhang J. et al. Predicting lung radiotherapy-induced pneumonitis using a model combining parametric Lyman probit with nonparametric decision trees. Int J Radiat Oncol Biol Phys. 2007;68:1212–21. doi: 10.1016/j.ijrobp.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schallenkamp JM, Miller RC. et al. Incidence of radiation pneumonitis after thoracic irradiation: Dose-volume correlates. Int J Radiat Oncol Biol Phys. 2007;67:410–6. doi: 10.1016/j.ijrobp.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Bentzen SM, Overgaard J. Patient-to-patient variability in the expression of radiation-induced normal tissue injury. Semin Radiat Oncol. 1994;4:68–80. doi: 10.1053/SRAO00400068. [DOI] [PubMed] [Google Scholar]
- 9.Yuan X, Liao Z, Liu Z. et al. Single nucleotide polymorphism at rs1982073:T869C of the TGF beta 1 gene is associated with the risk of radiation pneumonitis in patients with non-small-cell lung cancer treated with definitive radiotherapy. J Clin Oncol. 2009;27:3370–8. doi: 10.1200/JCO.2008.20.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Bi N. TGF-beta1 gene polymorphisms for anticipating radiation-induced pneumonitis in non-small-cell lung cancer: Different ethnic association. J Clin Oncol. 2010;28:e621–2. doi: 10.1200/JCO.2010.31.0458. [DOI] [PubMed] [Google Scholar]
- 11.Yin M, Liao Z, Liu Z. et al. Functional polymorphisms of base excision repair genes XRCC1 and APEX1 predict risk of radiation pneumonitis in patients with non-small cell lung cancer treated with definitive radiation therapy. Int J Radiat Oncol Biol Phys. 2011;81:e67–73. doi: 10.1016/j.ijrobp.2010.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelsey CR, Jackson IL, Langdon S. et al. Analysis of single nucleotide polymorphisms and radiation sensitivity of the lung Assessed with an objective radiologic endpoint. Clin Lung Cancer. 2013;14:267–74. doi: 10.1016/j.cllc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosseinimehr SJ. The protective effects of trace elements against side effects induced by ionizing radiation. Radiat Oncol. 2015;33:66–74. doi: 10.3857/roj.2015.33.2.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo L, Wang Y, Feng Q. et al. Recombinant protein glutathione S-transferase P1 attenuates inflammation in mice. Mol Immunol. 2009;46:848–57. doi: 10.1016/j.molimm.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Cantlay AM, Smith CA, Wallace WA. et al. Heterogeneous expression and polymorphic genotype of glutathione S-transferases in human lung. Thorax. 1994;49:1010–4. doi: 10.1136/thx.49.10.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moradi M, Mojtahedzadeh M, Mandegari A. et al. The role of glutathione-S-transferase polymorphisms on clinical outcome of ALI/ARDS patient treated with N-acetylcysteine. Respir Med. 2009;103:434–41. doi: 10.1016/j.rmed.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 17.HayesJD. PulfordDJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 18.WatsonMA StewartRK, SmithGB et al. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275–80. doi: 10.1093/carcin/19.2.275. [DOI] [PubMed] [Google Scholar]
- 19.Falvo E, Strigari L, Citro G. et al. SNPs in DNA repair or oxidative stress genes and late subcutaneous fibrosis in patients following single shot partial breast irradiation. J Exp Clin Cancer Res. 2012;31:7–15. doi: 10.1186/1756-9966-31-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnett GC, Coles CE, Elliott RM. et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: A prospective analysis study. Lancet Oncol. 2012;13:65–77. doi: 10.1016/S1470-2045(11)70302-3. [DOI] [PubMed] [Google Scholar]
- 21.Zschenker O, Raabe A, Boeckelmann IK. et al. Association of single nucleotide polymorphisms in ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with clinical and cellular radiosensitivity. Radiother Oncol. 2010;97:26–32. doi: 10.1016/j.radonc.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Mangoni M, Bisanzi S, Carozzi F. et al. Association Between Genetic Polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT Genes and Radiosensitivity in Breast Cancer Patients. Int J Radiat Oncol Biol Phys. 2011;81:52–8. doi: 10.1016/j.ijrobp.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Chang-Claude J, Popanda O, Tan XL. et al. Association between polymorphisms in the DNA Repair genes, XRCC1, APE1, and XPD and acute side effects of radiotherapy in breast cancer patients. Clin Cancer Res. 2005;11:4802–9. doi: 10.1158/1078-0432.CCR-04-2657. [DOI] [PubMed] [Google Scholar]
- 24.Cheuk IW, Yip SP, Kwong DL. et al. Association of XRCC1 and XRCC3 gene haplotypes with the development of radiation-induced fibrosis in patients with nasopharyngeal carcinoma. Mol Clin Oncol. 2014;2:553–8. doi: 10.3892/mco.2014.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gravs PR, Siddiqui F, Anscher MS. et al. Radiation pulmonary toxicity: from mechanisms to management. Semin Radiat Oncol. 2010;20:201–7. doi: 10.1016/j.semradonc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Z, He J, Zeng T. et al. A multiplexer liquidchip technology for detecting single nucleotide polymorphisms from metabolism of anti-thrombotic drugs in dried blood spots. Acta Biochim Biophys Sin. 2014;46:522–5. doi: 10.1093/abbs/gmu028. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Ren BU, Zhang L. et al. Correlation between metabolic enzyme GSTP1 polymorphisms and susceptibility to lung cancer. Exp Ther Med. 2015;10:1521–7. doi: 10.3892/etm.2015.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao B, Wang H, Zhou K. et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64:4378–84. doi: 10.1158/0008-5472.CAN-04-0372. [DOI] [PubMed] [Google Scholar]
- 29.Ho AY, Atencio DP, Peters S. et al. Genetic predictors of adverse radiotherapy effects: the Gene-PARE project. Int J Radiat Oncol Biol Phys. 2006;65:646–55. doi: 10.1016/j.ijrobp.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Andreassen CN, Alsner J, Overgaard J. Does variability in normal tissue reactions after radiotherapy have a genetic basis-Where and how to look for it? Radiother Oncol. 2002;64:131–40. doi: 10.1016/s0167-8140(02)00154-8. [DOI] [PubMed] [Google Scholar]
- 31.Chang-Claude J, Popanda O, Tan XL. et al. Association between polymorphisms in the DNA Repair genes, XRCC1, APE1, and XPD and acute side effects of radiotherapy in breast cancer patients. Clin Cancer Res. 2005;11:4802–9. doi: 10.1158/1078-0432.CCR-04-2657. [DOI] [PubMed] [Google Scholar]
- 32.Ali-Osman F, Akande O, Antoun G. et al. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase P1 gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem. 1997;272:10004–12. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- 33.Cote ML, Chen W, Smith DW. et al. Meta-and pooled analysis of GSTP1 polymorphism and lung cancer: a HuGE-GSEC review. Am J Epidemiol. 2009;169:802–4. doi: 10.1093/aje/kwn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–75. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ambrosone CB, Tian C, Ahn J. et al. Genetic predictors of acute toxicities related to radiation therapy following lumpectomy for breast cancer: a case-series study. Breast Cancer Res. 2006;8:40–6. doi: 10.1186/bcr1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terrazzino S, La Mattina P, Gambaro G. et al. Common variants of GSTP1, GSTA1, and TGF beta1 are associated with the risk of radiation-induced fibrosis in breast cancer patients. Int J Radiat Oncol Biol Phys. 2012;83:504–11. doi: 10.1016/j.ijrobp.2011.06.2012. [DOI] [PubMed] [Google Scholar]
- 37.Cordoba EE, Abba MC, Lacunza E. et al. Polymorphic Variants in Oxidative Stress Genes and Acute Toxicity in Breast Cancer Patients Receiving Radiotherapy. Cancer Res Treat. 2016;48:948–54. doi: 10.4143/crt.2015.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Luo Z, Yang L. et al. The association between four SNPs of X-ray repair cross complementing protein 1 and the sensitivity to radiotherapy in patients with esophageal squamous cell carcinoma. Oncol Lett. 2016;11:3508–14. doi: 10.3892/ol.2016.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohrenweiser HW, Carrano AV, Fertitta A. et al. Refined mapping of the three DNA repair genes, ERCC1, ERCC2, and XRCC1, on human chromosome 19. Cytogenet Cell Genet. 1989;52:11–4. doi: 10.1159/000132829. [DOI] [PubMed] [Google Scholar]
- 40.Shen MR, Jones IM, Mohrenweiser H. Nonconser-vative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–8. [PubMed] [Google Scholar]
- 41.Horton JK, Watson M, Stefanick DF. et al. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]