Abstract
Viruses are the smallest known microbes, yet they cause the most significant losses in human health. Most of the time, the best-known cure for viruses is the innate immunological defense system of the host; otherwise, the initial prevention of viral infection is the only alternative. Therefore, diagnosis is the primary strategy toward the overarching goal of virus control and elimination. The introduction of a new class of nanoscale materials with multiple unique properties and functions has sparked a series of breakthrough applications. Gold nanoparticles (AuNPs) are widely reported to guide an impressive resurgence in biomedical and diagnostic applications. Here, we review the applications of AuNPs in virus testing and detection. The developed AuNP-based detection techniques are reported for various groups of clinically relevant viruses with a special focus on the applied types of bio-AuNP hybrid structures, virus detection targets, and assay modalities and formats. We pay particular attention to highlighting the functional role and activity of each core Au nanostructure and the resultant detection improvements in terms of sensitivity, detection range, and time. In addition, we provide a general summary of the contributions of AuNPs to the mainstream methods of virus detection, technical measures, and recommendations required in guidance toward commercial in-field applications.
Keywords: Gold nanoparticles, virus, infection, detection, sensing
1. Introduction
Viruses are remarkable pathogens that are causing prominently increasing morbidity and mortality worldwide. Their highly contagious nature and the absence of immediate and efficient control systems are the main reasons behind their potential health impacts. Currently, viral infections and associated diseases are major causes of death in mankind, and under the present context of industrialization and immigration, they continue to emerge at a rapid pace, causing significant human, social, and financial costs 1. Additionally, gaps in currently applied detection systems potentially contribute to increasing the likelihood of international incidences and outbreak of viral infections 2-5. The implementation of highly sensitive and specific diagnostic tools has the potential to rapidly identify viral infections, initiate and guide judicious controls, and subsequently curtail their dissemination. Toward this endeavor and beyond the pitfalls of current immunological and molecular techniques commonly applied to virus detection, several new approaches based on nanoparticles (NPs) have recently been developed. AuNPs are widely described to be suitable for numerous biosensing functions and applications. Their unique photonic, electric, and catalytic properties, coupled with the molecular interaction specificity of various biomolecules (e.g., antibodies, single-stranded (ss) DNA, and RNA aptamers, among others), represent the design principles of a wide range of virus detection systems 6-8. The advantages of being simple, rapid, and sensitive and facilitating quantitative detection with excellent multiplexing capabilities have greatly promoted these systems to be envisioned as state-of-the-art technologies for virus detection 9, 10. However, there are no available reviews of the applications of bio-AuNP hybrid structures for sensing and detecting viruses. In this article, we provide a current review of AuNP-based virus detection at the level of virus type, including the types and structures of the applied AuNPs. We highlight their role in enhancing sensory and detection performance in comparison with current techniques in terms of analytical sensitivity, detection range, and time. Furthermore, this review specifically summarizes detection designs, formats, functions, and contributions, which are of special importance to both scientific and applied research.
2. Viruses: nanoscale pathogens with a global burden
Viruses are particulate in nature and usually exist in different morphological forms that generally range in size from 20 to 900 nm 11-13. Their intact, mature infectious particles are typically composed of definite units of proteins and nucleic acid that self-assemble to form nanoparticulate structures called virions. The viral proteins are usually arranged in a surface layer called the capsid and sometimes in an outer envelope surrounding an inner core of nucleic acids. This core of viral nucleic acids can be single- or double-stranded (ds), DNA or RNA, one or several molecules, linear or circular in shape, and a few thousands of nucleotides to one million base pairs in size.
The nanoscale size and the relatively simple structure of viruses tend to impose technical difficulties in establishing wide-use and long-term systems for virus detection. The small size of viruses increases the difficulty of their isolation and visualization compared with other microbes, such as bacteria and fungi that can be readily examined using ordinary light microscopes. Only electron microscopy (EM) with a high-magnification power of ~100,000× can allow the direct visualization of viruses and the study of their structures 14, 15. Therefore, EM remains crucial for many purposes in virus research. However, EM is certainly inappropriate for routine clinical diagnosis because of the required time and cost, as well as many safety concerns. Furthermore, although the simple structure of virus particles allows the features of diagnostic relevance to be easily defined and tested, it presents limitations on the use of their characteristics for practical applications, especially with the increasing number of discovered viruses and recorded viral infections 14, 16, 17. In addition, this structural simplicity confers viruses with rapid rates of spontaneous adaptation and evolution that may occur through direct genetic mutation, genetic substitution, or recombination. In this way, viruses not only outpace our attempts to develop sustainable control strategies but also raise more questions about the appropriateness and validity of current diagnostic techniques for long-term use 18, 19.
Along with these limitations in virus detection and control, the possibility of viral infection emergence or reemergence has become increasingly prevalent, and severe pandemics and epidemics have occurred around the world. In the past, many outbreaks of viruses, such as human immunodeficiency virus (HIV), severe acute respiratory syndrome (SARS), influenza virus (H5N1 and H1N1) and Zika virus, started as local events and then expanded to have global consequences. HIV was discovered in Central Africa as a new virus causing acquired immune deficiency syndrome (AIDS) three decades ago, and it now represents one of the most significant public health threats worldwide 20, 21. The highly contagious respiratory virus SARS has fueled global fears of pandemics since its first appearance in China 22, 23. In late 2012, the World Health Organization (WHO) raised a global alert for a new SARS-like respiratory coronavirus, which is now called Middle East Respiratory Syndrome (MERS) coronavirus. MERS is now classified as one of the most prominent viral threats to the Middle East and has caused hundreds of deaths and thousands of infections in a short time. Moreover, in the past few years, the world witnessed the worst Ebola outbreak ever that started to hit West Africa in early 2014 and the pandemic reemergence of Zika virus in 2016 11, 12, 24. With these evident possibilities of massive outbreaks and implications, the situation is rapidly growing more serious, and the demand for the development of rapid diagnostics and effective control strategies is becoming more urgent.
3. Past and current virus detection options
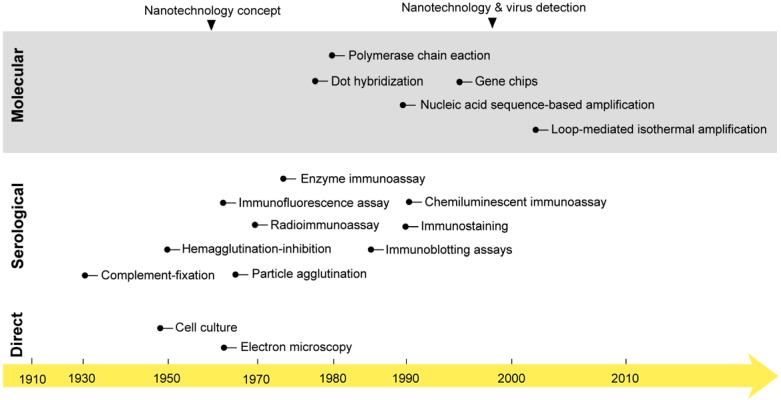
The first studies on virus isolation and detection were started early in the 1950s when the first cell culture system and electron microscope were developed 25, 26. For decades, these techniques represented the main tools for studying and investigating the biochemical and morphological properties of viruses that remain the main foundation of all known classification and detection systems. However, their practical use in virus detection has remained greatly debated due to several considerations, including laboratory equipment expenses and time and safety concerns 27, 28. In the early 1980s, the field of diagnostic virology was boosted with two other major developments: 1) the birth of various immunoassays; and 2) the invention of polymerase chain reaction (PCR). This was followed by the development of a very wide range of serological and molecular detection techniques, which rapidly evolved to constitute the mainstream approaches of both laboratory research and the clinical diagnosis of viruses (Fig. 1) 24, 29.
Figure 1.
The onset of nanotechnology in virus detection applications compared with the development of the most common virus detection techniques. Cell culture and electron microscopy techniques that are now commonly applied in the direct testing for and detection of viruses were discovered in the mid-20th century 25. Then, different serological and molecular techniques were developed. The serological detection of viruses with immunoassays was first reported in 1970: a radioimmunoassay was applied for the detection of the Australia antigen, later called hepatitis B virus surface antigen 218. PCR was discovered in the 1980s and first reported in virus detection in 1988 for acquired immune deficiency syndrome detection 219. Later, many molecular techniques, including amplification- and nonamplification-based techniques, were reported in virus detection. The concept of nanotechnology was envisioned as early as 1959 by the renowned physicist Richard Feynman 36. However, nanotechnology was only applied to virus detection in 1997, when gold nanoparticles were employed for the detection of single-copy human papillomavirus 39. Nanotechnology has recently come to represent one of the most outstanding trends in virus detection and diagnosis via the wide variety of assays described in this review.
Serology remains the standard method for virus detection. It primarily relies on testing for the presence of specific viral antigens or the corresponding antibody responses of the immune system. The most common types of serological tests include the neutralization assay, complement-fixation test, immunoprecipitation assay (IPA), hemagglutination-inhibition (HAI) assay, enzyme immunoassay (EIA), radioimmunoassay (RIA), chemiluminescent immunoassay (CIA), particle agglutination, immunostaining, immunofluorescence assay, single radial hemolysis, immunoblotting assay (IBA), and immunochromatographic test (ICT). The main working principle of these techniques is the use of specific antibodies in conjugation with different signal reporting systems, such as red blood cells, enzymes, and radioactive or fluorescent materials 30, 31. Most of these techniques are relatively easy to perform and flexible in their timing of specimen collection, and most of the needed reagents are usually commercially available. Furthermore, they are generally unrestricted by the practical limitations of virus isolation and propagation usually involved with other direct detection methods, such as cell culture and EM 9, 32. Therefore, serological methods are widely accepted and recognized techniques for simple, safe, and cheap virus detection 25, 30. In addition, they are usually described as the first choice for large-scale testing in epidemiological studies and for evaluating antiviral therapies and vaccinations. However, the accuracy and reliability of serological methods are usually challenged by the cross-reactivity of the used antibodies, and the risk of false-positive results is usually very high. In addition, most immune responses can only be detected for a period of time after the initial virus infection; thus, serological detection does not normally benefit patients.
Molecular techniques are attracting more interest and have found an increasing number of applications in virus detection. The discovery of the genetic enzyme systems involved in the cellular machinery of nucleic acid replication and the stunning invention of an in vitro nucleic acid amplification system, commonly called PCR, by Mullis in the early 1980s, opened new frontiers in nucleic acid-based detection 33. In addition, the known high specificity of nucleic acid hybridization and the absolute availability of its synthesis and modification guided the development of many detection and genotyping techniques known for the rapid and specific detection of viruses. These techniques can be classified as amplification-or nonamplification-based molecular detection techniques. Amplification-based molecular techniques usually employ one or more forms of nucleic acid amplification to allow the indirect detection of the target virus. These techniques represent the majority of molecular techniques applied in virus detection and include various types of target amplification techniques (e.g., PCR, loop-mediated isothermal amplification (LAMP), transcription-mediated amplification, and nucleic acid sequence-based amplification), signal amplification techniques (e.g., branched DNA and hybrid capture), and probe amplification techniques (e.g., ligase chain reaction and strand-displacement amplification). Nonamplification-based techniques are mainly applied to the direct testing for the presence of a specific virus in clinical samples (e.g., in situ hybridization, southern blot hybridization, and dot blot hybridization).
Generally, molecular methods are relatively rapid and more sensitive than immunoassays and can be applied either in a simple form for the manual detection of viruses or as an embedded component of more advanced systems. Numerous fully automated and high-throughput detection systems are now available and widely used in clinical settings. In fact, the development of advanced molecular detection systems has revolutionized the way in which diagnostic tests are delivered and greatly enhanced the control of many viral infections, whether in hospitals or communities. In addition, these systems have eliminated the biosafety and time concerns usually associated with the clinical and academic study of viruses. However, despite these wide and promising applications, most molecular techniques still have several potential limitations in repeatability, accuracy, sensitivity, and specificity that are mainly caused by the high genetic variability of some viruses. Furthermore, these assays are expensive and time consuming and typically call for specialized laboratory instruments and skilled personnel 34, 35.
4. Nanotechnology and virus detection
The concept of nanotechnology was introduced in early 1959 36. Subsequently, nanotechnology was realized via various types of new materials that rapidly emerged as promising tools for biological and chemical analyses. Nanomaterials are known to possess multiple unique optical, electronic, magnetic, and mechanical properties enabling very attractive applications, especially in the fields of biomedical imaging and clinical diagnosis 37, 38. The first reported application of nanomaterials in the detection of viruses was attempted in the late 1990s: AuNPs were coupled with silver staining and applied for the detection of human papillomavirus in cervical carcinoma cells (Fig. l) 39. Currently, there is a very wide range of nanomaterials, including metal NPs, carbon nanotubes, silica NPs, quantum dots (QDs), upconversion NPs, and polymeric NPs, that are being heavily investigated for virus detection 37, 38, 40. One of the most common approaches for exploiting these nanostructures in virus detection is the development of nanobio hybrid systems that contain one or more biomolecules derived from viruses (e.g., DNA, RNA, antibody, pentabody, antigen, or peptide) conjugated to the surface of different NP forms. These systems leverage the significant labeling properties and signal transduction functions of NPs and the specific activity of the conjugated biomolecules to act as multivalent-NP probes 37, 38, 41. Such virus-specific NP probes have surprisingly been used to build up various optical, fluorometric, electrochemical, and electrical assays that have been extensively reported for single and multiple detection modes (Table 1). The results of most of these studies clearly demonstrate the inherent potential of these probes, along with numerous advantages over traditional approaches, in terms of size, performance, specificity, signal sensitivity, and stability. Additionally, these studies have extensively described their application to allow simple, rapid, highly sensitive and label-free detection.
Table 1.
Gold nanoparticle based detections of viruses
| Group/Virusa | Targetb | AuNP systemc | Assayd | Detection limit |
Detection range |
Detection platform | Time (minutes) | Ref | ||
|---|---|---|---|---|---|---|---|---|---|---|
| AuNPs | Biomolecule | |||||||||
| Shape | Size | |||||||||
| Bunyaviridae | ||||||||||
| HTNV | NC protein | Spherical | 30 nm | DNA | IPCR | 10 fg/mL | 105 ‒ 1 fg/mL | Microtitre plate | 125 | 96 |
| RVFV | Capsid antigen | Spherical | 60 nm | Antibody | SERS | 5 fg/mL | 50 - 0.005 pg/mL | Glass slide | >60 | 49 |
| Filoviridae | ||||||||||
| EBOV | NP gene | Spherical | 13 nm | DNA | Scanometric | 20 fM | 20 fM | Chip | 330 | 50 |
| Spherical | 13 nm | DNA | DLS | 20 fM | 20 fM | Chip | 330 | 50 | ||
| Coronaviridae | ||||||||||
| SARS | PP1ab gene | Spherical | 13 nm | None | Colorimetric | 60 fmol | ND | Tube | 5 | 54 |
| NC protein | Spherical | 70 nm | DNA | Electrochemical | 2.5 pM | 50 - 2.5 pM | Electrodes | >120 | 72 | |
| Flaviviridae | ||||||||||
| HCV | HCV antigen | Spherical | 12 nm | DNA Antibody |
Electrical | 1 pg/μL | 10 ng/μL ‒ 1 pg/μL | Electrodes | 245 | 69 |
| C gene | Spherical | 8-15 nm | DNA | DLS | 0.36 pM | 0.3 μM - 0.3 pM | Chip | 75 | 80 | |
| Spherical | 8-15 nm | DNA | Colorimetric | 0.36 pM | 0.3 μM - 0.3 pM | Chip | 75 | 80 | ||
| HCV antibody | Spherical | 15 nm | SPA | Scanometric | 3 ng/mL | 3 μg/mL - 3 ng/mL | Chip | 10 | 88 | |
| 5' UTR gene | Spherical | 40 nm | DNA | Colorimetric | 2 fmol | 30 ‒ 2 fmol | Stripe | 50 | 55 | |
| Spherical | 10-15 nm | DNA | Electrochemical | ~1 pM | 2.0 - 0.01 nM | Electrodes | 60 | 78 | ||
| Full genome | Spherical | 15 nm | None | Colorimetric | 50 copies | ND | Tube | 110 | 56 | |
| Spherical | 8 nm | DNA | Fluorometric | 300 fM | 550 - 15 pM | ND | 184 | 60 | ||
| DENV | E gene | Spherical | 13 nm | DNA | QCM | 2 PFU/mL | 2×106 ‒ 2 PFU/mL | Chip | 90 | 93 |
| Spherical | ND | DNA | ICP-MS | 1.6 fM | 5 pM - 5 fM | Tube | 90 | 94 | ||
| WNV | E antigen | Spherical | 60 nm | Antibody | SERS | 5 fg/mL | 50 - 0.005 pg/mL | Tube | 49 | |
| Hepadnaviridae | ||||||||||
| HBV | anti-HBV | Spherical | 15 nm | SPA | Scanometric | 3 ng/mL | 3 μg/mL - 3 ng/mL | Chip | 10 | 88 |
| HBeAg | Rod | L = 46 nm D = 13 nm |
Antibody | Fluorometric | 8.3 ng/mL | Up to 264 ng/mL | ND | ND | 62 | |
| HBsAg | Spherical | 10, 50, and 100 nm | Antibody | DLS | 0.005 IU/mL | 1 - 0.005 IU/mL | Tube | <60 | 51 | |
| Rod | L = 68 nm D = 30 nm |
Antibody | LSPR | 0.01 IU/mL | 1 - 0.01 IU/mL | Tube | ND | 52 | ||
| Spherical | 16 nm | Antibody | Electrochemical | 0.1 ng/mL | 650 ‒ 0.5 ng/mL | Electrodes | 65 | 76 | ||
| Spherical | ~10 nm | Antibody | 2.3 pg/mL | 1.0 - 0.01 ng/mL | Electrodes | 60 | 77 | |||
| Spherical | 16 nm | Antibody | 87 pg/mL | 1500 - 0.1 ng/mL | Electrochemical cell | ~50 | 75 | |||
| Rod | L = 46 nm D = 13 nm |
Antibody | Fluorometric | 9 ng/mL | Up to 288 ng/mL | ND | 62 | |||
| Spherical | 5.5 nm | Peptide | 0.1 pg/mL | 0.1 - 0.0001 ng/mL | Microtitre plate | 30 | 65 | |||
| HBV DNA | Rod | ND | DNA | Fluorometric | 15 pM | 6.0 - 0.045 nM | ND | ~60 | 61 | |
| S gene | Spherical | 15-8 nm | DNA | Colorimetric | 0.36 pM | 0.3 μM - 0.3 pM | Chip | 75 | 80 | |
| Spherical | 13 nm | DNA | Scanometric | 20 fM | At 20 fM | 330 | 50 | |||
| Spherical | 5 nm | Avidin | Electrochemical | 0.7 ng/mL | 1.47 - 0.7 ng/mL | Electrochemical cell | 105 | 220 | ||
| C gene | Spherical | 10 nm | DNA | Scanometric | 1 fM | 10-11 - 10-15 | Chip | 90 | 85 | |
| Particles | Spherical | 5.5 nm | Peptide | Fluorometric | 100 - 1 particle/μL | 1000 - 1 particle/μL | Microtitre plate | 30 | 65 | |
| Herpesviridae | ||||||||||
| HEV | ORF1gene | Spherical | 14 nm | DNA | Colorimetric | 100 fM | 1x104 - 50 fM | Chip | 110 | 82 |
| Herpesviridae | ||||||||||
| KSHV | KSHV DNA | Spherical | 15 nm | DNA | Colorimetric | ~1 nM | 1 mM - 10 pM | Tube | 60-30 | 57 |
| HCMV | Full genome | Spherical | 20 nm | DNA | Electrochemical | 5 pM | 5000 - 5 pM | Electrochemical cell | 780 | 68 |
| HSV-2 | anti-HSV-2 | Spherical | ND | Antibody | Colorimetric | ND | ND | Strip | 20 | 59 |
| Orthomyxoviridae | ||||||||||
| H1N1 | Anti-H1N1 | Spherical | 20 nm | Antibody | DLS | <100 TCID50/mL | 1.4x106 - 5.5x103 TCID50/mL 4.8x105 - 7.2x101 TCID50/mL |
Tube | 30 | 53 |
| HA antigen | Spherical | 25 nm | Protein A | Fluorometric | 13.9 pg/mL | 800 - 12.5 ng/mL | ND | ND | 63 | |
| NA gene | Spherical | ND | DNA | SERS | 25 nM | 50 - 25 nM | ND | ND | 64 | |
| Spherical | ND | DNA | Fluorometric | 25 nM | 50 - 25 nM | ND | ND | 64 | ||
| M gene | Spherical | ND | Avidin | Electrochemical | 577 pM | 3 - 0.001 pmol | Electrodes | 80-50 | 70 | |
| Rod | L = <30 nm D = <10 nm |
None | Colorimetric | 1pg | 0.1 ng - 0.01 pg | Tube | ~ 60 | 67 | ||
| H5N1 | HA gene | Spherical | 32 nm | Antibody | Colorimetric | 40 - 0.1 ng | 100 - 0.1 ng | Stripe | ND | 66 |
| Spherical | ND | DNA | Electrochemical | 0.4 pM | 1.0 nM - 5.0 pM | Electrodes | 20 | 71 | ||
| Spherical | 3 nm | Daunorubicin | Scanometric | 10 pM | 10 pM - 100 nM | Chip | 20 | 83 | ||
| Spherical | 15 nm | DNA | 100 fM 103 TCID50 |
ND | Chip | 150 | 86 | |||
| Spherical | 1.4 nm | None | 10 pM | 10 pM - 100 nM | Chip | 90 | 87 | |||
| NA gene | Spherical | 15 nm | DNA | Scanometric | 100 fM 103 TCID50 |
ND | Chip | 150 | 86 | |
| Particles | Spherical | 22 nm | Antibody | Fluorometric | 0.09 ng/mL | 12 - 0.27 ng/mL | Strip | 30 | 58 | |
| Spherical | 10 nm | Pentabody | Colorimetric | 10 ng/mL | 10‒1 - 10-4 μg/mL | Tube | 35 | 92 | ||
| Papillomaviridae | ||||||||||
| HPV | L1 gene | Spherical | ND | Antibody | Electrical | 30 pM | 5 nM - 100 pM | Chip | 155 | 89 |
| Spherical | 16 nm | DNA | Fluorometric | 1 fM | 100 - 0.001 pM | Microbead array | 155 | 98 | ||
| Picornaviridae | ||||||||||
| HAV | VP1gene | Spherical | 14 nm | DNA | Scanometric | 100 fM | 0.3 ‒ 0.03 nM | Chip | 240 | 221 |
| Vall7 gene | Spherical | 13 nm | DNA | Scanometric | 20 fM | At 20 fM | Chip | 330 | 50 | |
| Spherical | 13 nm | DNA | DLS | 20 fM | At 20 fM | Chip | 330 | 50 | ||
| Retroviridae | ||||||||||
| HIV | p24 antigen | Spherical | 15 nm | Avidin DNA |
DLS | 0.2 pM | 31.4 pM - 1.6 fM | Chip | >170 | 81 |
| Spherical | 15 nm | Avidin DNA |
Scanometric | 0.1 pg/mL | 500 - 0.1 pg/mL | Microtiter plate | ~360 | 84 | ||
| Spherical | 20 nm | Antibody | AFM | 0.025 pg/mL | 3500 - 50 copies/mL | Chip | <360 | 95 | ||
| Spherical | 30 nm | Antibody/ DNA |
IPCR | 1 pg/mL | 10000 - 1 pg/mL | Microtiter plate | >125 | 97 | ||
| Spherical | 30 nm | Antibody/ DNA |
0.1 pg/mL | 1000 - 0.1 pg/mL | Microtiter plate | >85 | 97 | |||
| gag gene | Spherical | 60 nm | DNA | Colorimetric | ~11 log10 copies/mL | 11 - 12.5 log10 copies | Stripe | 30 | 91 | |
| pol gene | Spherical | 3 nm | DNA | Electrochemical | 0.34 fM | 1.0 µM - 0.1 pM | Electrodes | 50 | 73 | |
| Spherical | 13 nm | DNA | DLS | 10 fM | 10 nM - 1 fM | Microtiter plate | 80 | 90 | ||
a DENV: dengue virus; EBOV: Ebola virus; H1N1: influenza A virus subtype H1N1; H5N1: influenza A virus subtype H5N1; HAV: hepatitis A virus; HBV: hepatitis B virus; HCMV: human cytomegalovirus; HCV: hepatitis C virus; HEV: hepatitis E virus; HIV: human immunodeficiency virus; HPV: human papilloma virus; HSV-2: herpes simplex virus-2; HTNV: hantaan virus; KSHV: Kaposi's sarcoma-associated herpesvirus; RVFV: Rift Valley fever virus; SARS: severe acute respiratory syndrome; WNV: west Nile virus.
b 5 UTR: five prime untranslated region; C gene: core gene; E gene: envelop gene; gag gene: group-specific antigen gene; HA gene: hemagglutinin gene; HBeAg: hepatitis B e antigen; HBsAg: hepatitis B surface antigen; L1 gene: late expressed gene1; M gene: matrix gene; NA gene: neuraminidase gene; NC protein: nucleocapsid protein; NP gene: nucleoprotein gene; ORF1: open reading frame1; P24 antigen: P24 capsid protein antigen; pol gene: polymerase gene; PP1ab: ployprotein1ab; S gene: surface gene; Vall7 gene: hepatitis A virus Vall7 polyprotein gene; VP1 gene: viral protein1 gene.
c SPA, staphylococcal protein A.
d AFM: atomic force microscopy; DLS: dynamic light scattering; ICP-MS: inductively-coupled plasma mass spectrometry; LSPR: localized surface plasmon resonance; QCM: quartz crystal microbalance; SERS: surface enhanced Raman scattering.
5. AuNPs in virus detection
AuNPs are a leading class of metal nanostructures that is widely known for its chemical stability, water solubility, and broad size and shape controllability. AuNPs can range from 1 to 800 nm in size and have different morphological shapes, including spheres, rods, prisms, tetrapods, dog bones, cubes, shells and several hollow structures 42, 43. The synthesis of AuNPs can be performed using different methods such as chemical reduction of salts, ultraviolet irradiation, lithography, aerosol technologies, laser ablation, ultrasonic fields, photochemical reduction of Au, and biological synthesis 9, 32. AuNPs possess a high surface density of free electrons that results in inherent optical, electrical, and catalytic properties. The excitation of AuNPs with light can cause these free surface electrons, i.e., “plasmons,” to oscillate to one side away from the atomic core, which remains as a positive charge on the other side, thereby creating a dipole or plasmon polariton 44, 45. This dipole plasmon can change its direction in accordance with the frequency of incident light, and the resonance condition is reached when their frequency is approximately the same. This condition has been referred to as surface plasmon resonance (SPR). The most widely used AuNPs exhibit intense SPR bands that usually exist between 510-1100 nm and are known to be weakly dependent on the size of the AuNPs and the refractive index of the surrounding media but very sensitive to both the shape of the NPs and the interparticle distance 46. The SPR of AuNPs has been observed to cause intense enhancing or quenching effects upon interactions with nearby photon emitters. These distance-dependent coupling effects are dipole-dipole interactions that usually include surface plasmon-mediated energy nanotransfer processes similar to fluorescence resonance energy transfer (FRET) and may be either destructive (resulting in quenched emission) or constructive (resulting in enhanced emission).
Compared to other types of nanomaterials, metal nanoparticles, particularly AuNPs, constitute ideal tools in virus detection for numerous reasons, including the ease of synthesis, characterization, and surface modification, outstanding stability, biocompatibility, and exceptionally high absorption coefficients 47, 48. Furthermore, as labeling agents, AuNPs are easily visualized due to their intense colors and are known to form stable and highly active bioconjugates with common targeting biomolecules, such as DNA and proteins, thereby enabling highly sensitive and specific sensing and detection applications 48.
6. Key functions of AuNPs in virus detection
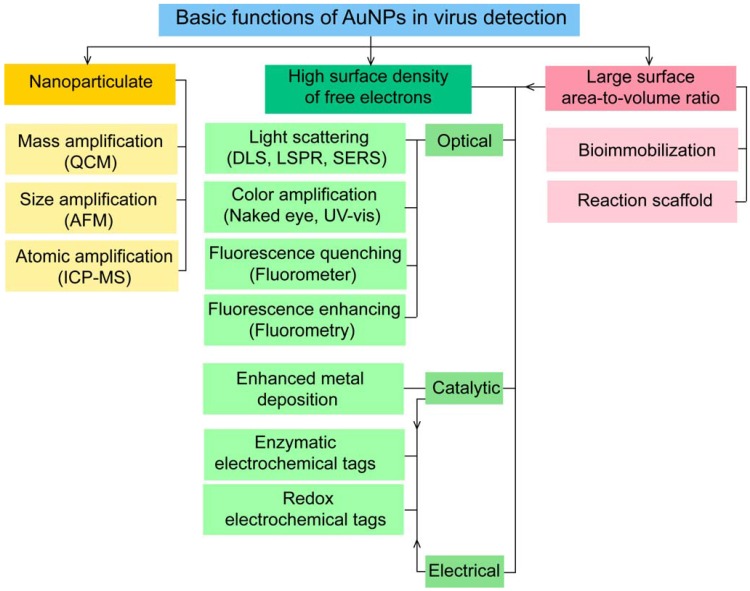
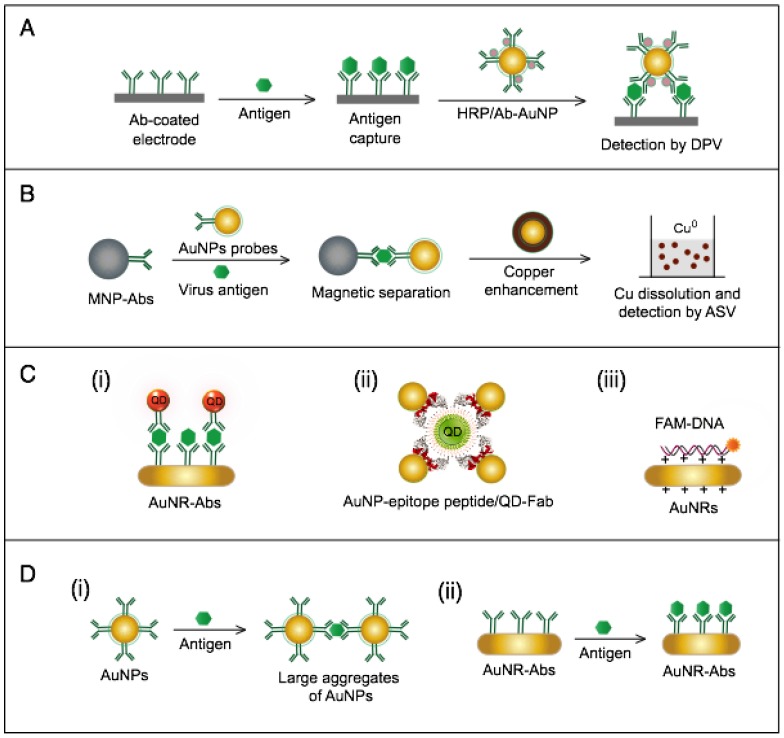
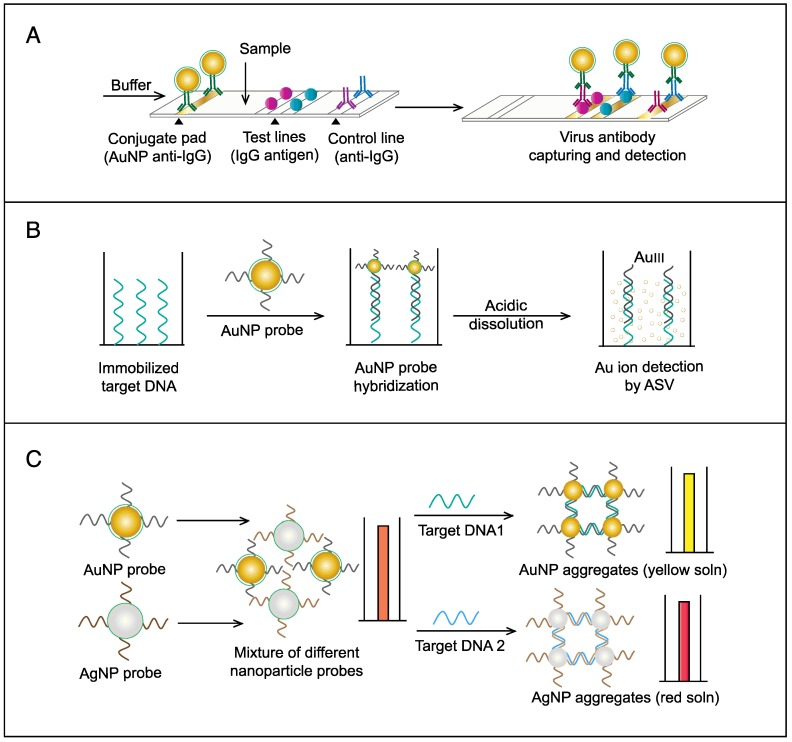
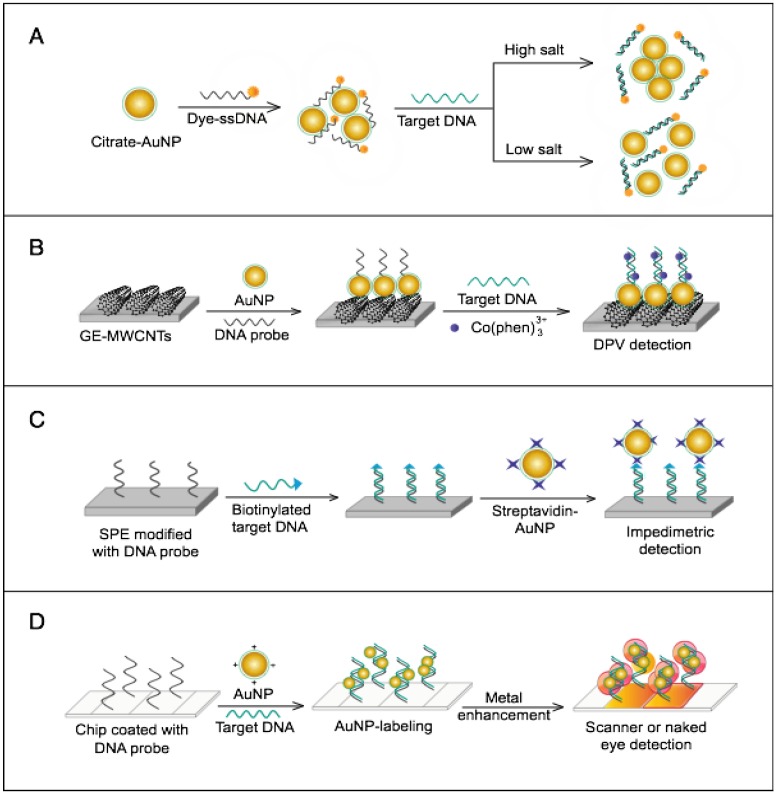
AuNPs have been preferentially employed to perform numerous optical signal transduction functions in virus detection, such as resonance light scattering 49-53, color amplification 54-59, and fluorescence quenching or enhancing 60-65 (Fig. 2). Resonance light-scattering-based detections usually involve measuring the amount of light scattered by different light spectroscopy techniques, including localized surface plasmon resonance (LSPR) 52, Raman spectroscopy 49, 50 and dynamic light scattering (DLS) 51, 53. The colorimetric detection of viruses using AuNPs usually relies on two main techniques: 1) a color amplification technique in which AuNPs are applied to act as direct coloring labels with their characteristic, intense red color 55, 58, 59, 66; and 2) color change technique in which a color change from red to purple occurs in response to particle aggregation. AuNP aggregation can be noncrosslinked aggregation caused by the target-triggered removal of stabilizing ligands from the surfaces of AuNPs 54, 56 or interparticle-crosslinked aggregation caused by the binding of ligands on modified AuNPs with target analytes 57, 67. Analogous to FRET, AuNP-based fluorescence quenching is a distance-dependent process in which AuNPs act to reduce the radiative rate of fluorophores in their proximity. Thus, as efficient acceptors, AuNPs have been paired with dye- and QD-based fluorophores in different analyte-induced donor-acceptor crosslinking and coupling protocols 60-62, 64, 65.
Figure 2.
A generalized scheme for the basic characteristics and functions of AuNPs applied in virus detection. The size and metal nature of AuNPs are the key characteristics that enable them to directly transduce multiple types of signals, including optical, catalytic and electrical signals that can be detected by dynamic light scattering (DLS), localized surface plasmon resonance (LSPR), surface-enhanced Raman scattering (SERS), ultraviolet-visible (UV-vis) spectroscopy, fluorometry and other electrochemical analysis techniques. In addition, AuNPs act as nanoparticulate tags to enhance the signal detection of several analytical techniques, such as quartz crystal microbalance (QCM), atomic force microscopy (AFM), and inductively coupled plasma mass spectrometry (ICP-MS). Their large surface area-to-volume ratio is known to permit them to function as an excellent scaffold for bioimmobilization and target-probe interactions with highly enhanced specificity and sensitivity.
AuNPs are widely described as electroactive and catalytic tags in various electrochemical assays applied to virus detection (Fig. 2). Based on their redox and electrical properties, AuNPs can directly act as electrochemical tags detected either by their acid dissolution followed by electrochemical stripping measurements of gold ions 68 or by their direct deposition on the surface of electro transducers, allowing an enhanced electrical conduction and resistance change 69-73. AuNPs can indirectly perform electrochemical transduction functions based on their catalytic activity toward some chemical reduction reactions of metal ions (e.g., silver and copper) 72, 74, 75 or other species, such as H2O2 76-78. This potential to catalyze metal deposition is preferentially called NP-metal enhancement amplification, and it represents the basic function of multiple scanometric 50, 70, 79-88, photoelectric 89, light-scattering 90, and colorimetric schemes 91 for virus detection. Additionally, AuNP-based catalysis for the oxidation-reduction reaction of hydroquinone has been reported in another simple, colorimetric-based virus detection method 92.
Other studies have exploited AuNPs to enhance the detection sensitivity of some bioanalytical techniques, such as quartz crystal microbalance (QCM) 93, inductively coupled plasma mass spectrometry (ICP-MS) 94, and atomic force microscopy (AFM) 95 in virus detection. Through these schemes, AuNPs have been applied as effective nanoparticulate amplification tags to enhance mass, elemental, and topographic signal transduction, respectively (Fig. 2).
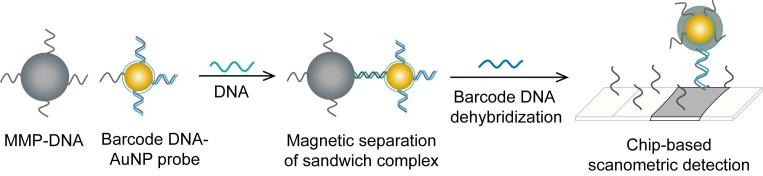
Interestingly, the enhanced surface area-to-volume ratio of AuNPs has allowed them to further act as exquisite scaffolds for biorecognition and detection reactions (Fig. 2); in addition, it has granted these structures unprecedented capabilities for bioimmobilization, which is a basic principle of most AuNP-based diagnostic designs. By coupling these bioimmobilization and signal transduction functions, AuNPs can achieve virus detection with highly improved analytical sensitivity and specificity. Furthermore, the direct increase in loading efficiency allows new possibilities of controlled multifunctionalization with different biomolecules. Based on this principle, developed AuNP tags are characterized by enhanced reactivity and stability. Increased loading efficiency also allows the design of new probes modified with multiple structures for biotargeting (e.g., antibodies and DNA), together with other signal amplification structures (e.g., enzymes and DNA oligonucleotides) 69, 76, 77, 96-98, both types of which are stridently included in the development of new diagnostic schemes and designs. For example, the AuNP-barcoding assay and its descendent technique of immuno-PCR universally rely on AuNP tags modified with antibodies and DNA oligonucleotides 81, 96, 97, and AuNP-based enzymatic electrochemical assays are based on using AuNPs dually modified with DNA and antibodies together with an enzyme to induce a chemical reduction reaction allowing electrochemical signal transduction 76, 77.
7. Detection of specific viruses using AuNPs
Several recent studies have attempted to harness the unique properties of AuNPs to develop various advanced schemes for virus detection. The developed assays are greatly variable in design and underlying principle. However, the utilization of AuNPs conjugated with specific virus-targeting biomolecules is a key component in most of these assays. Various AuNP bioconjugates have been widely employed in many colorimetric, scanometric, electrochemical, and fluorometric systems for the detection of many groups of well-known human viruses (Table 1 and Fig. 3). Each of these groups will be discussed in more detail in the following sections.
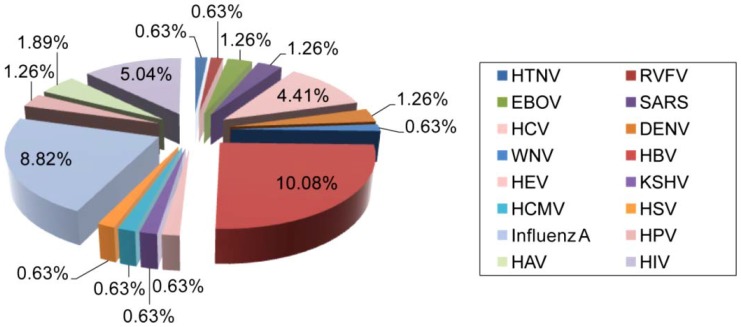
Figure 3.
Distribution of AuNP applications for the detection of different human viruses. The clinical significance, prevalence, and infectivity of viruses are important factors affecting the number of specific detection schemes developed. For instance, HBV is the most reported and investigated virus among studies focusing on the utilization of AuNPs for virus detection, which might be in part due to its highly contagious nature and global prevalence 65. HIV, IAV and HPV are also among the most widely reported viruses. The data analysis is based on a related-terms search of www.ncbi.nlm.nih.gov/pubmed. DENV: dengue virus; EBOV: Ebola virus; HAV: hepatitis A virus; HBV: hepatitis B virus; HCMV: human cytomegalovirus; HCV: hepatitis C virus; HEV: hepatitis E virus; HIV: human immunodeficiency virus; HPV: human papilloma virus; HSV: herpes simplex virus; HTNV: Hantaan virus; KSHV: Kaposi's sarcoma-associated herpesvirus; RVFV: Rift Valley fever virus; SARS: severe acute respiratory syndrome; WNV: West Nile virus.
7.1. Bunyaviridae
The Bunyaviridae family comprises more than 300 members that are primarily organized into four main genera: Hantavirus, Orthobunyavirus, Phlebovirus and Nairovirus 99. Bunyaviruses are spherical, enveloped RNA viruses 80-100 nm in size. Their genome is composed of three segments of negative-sense, ssRNA: a large segment (L, 6.3-12 kb) that encodes RNA polymerase, a medium segment (M, 3.5-6 kb) that encodes viral glycoprotein, and a small segment (S, 1-2.2 kb) that encodes nucleocapsid protein 100. These viruses are diverse in their host range and are frequently involved in a wide range of diseases in plants, animals, and humans. Human pathogenic bunyaviruses, such as Crimean-Congo hemorrhagic fever virus, Hantaan virus, La Crosse virus, Oropouche virus, Rift Valley fever virus, and Toscana virus, continue to present increasingly important health concerns worldwide 101, 102.
7.1.1. HantaanVirus (HTNV)
The genus Hantavirus was recently expanded to include more than 24 antigenically and genetically distinct HTNVs 99. Among them, rodent-borne HTNVs can cause serious diseases in humans, including hemorrhagic fever with renal syndrome, and hantavirus cardiopulmonary syndrome, and hospitalizes more than 150,000 persons each year with a mortality rate reaching 10%. Recently, the health impacts of HTNVs are expected to dramatically increase in the near future due to the increasing number of reports on newly discovered HTNVs 102-104.
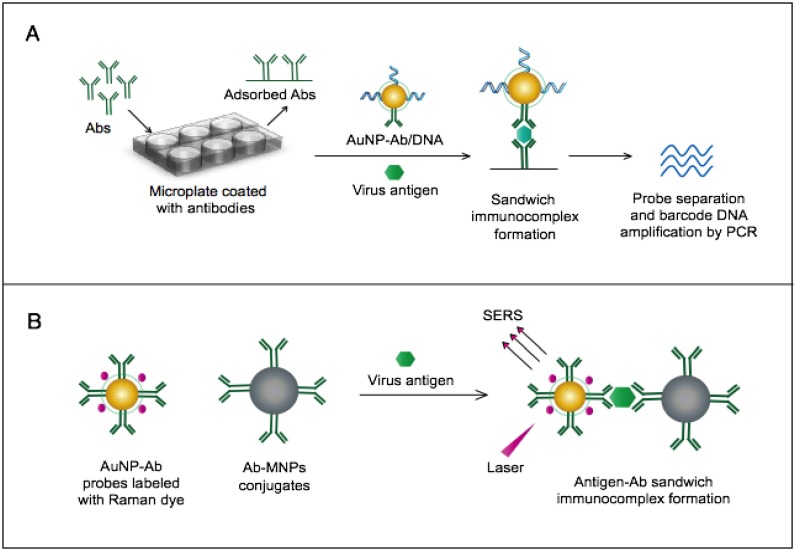
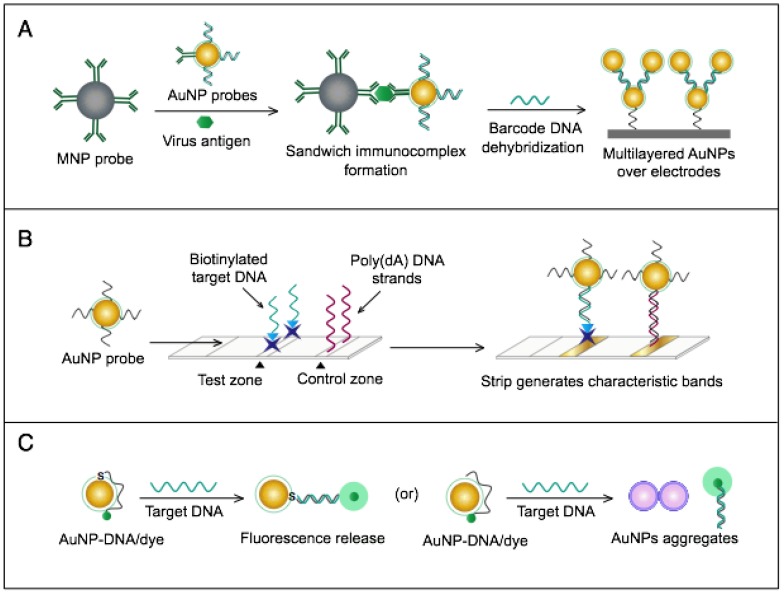
AuNPs were utilized to develop a novel immuno-PCR assay for the immunological detection of HTNV nucleocapsid protein 96. This assay mainly relies on the enhanced surface area of AuNPs to prepare dually functionalized antibody-oligonucleotide conjugates that exceptionally carry specific monoclonal antibodies to label the target HTNV antigen; the assay also involves barcoding DNA for signal amplification (Fig. 4A). This assay could optimally detect concentrations as low as 200 aM of purified or spiked antigen samples, which is ~7 orders of magnitude more sensitive than conventional ELISA. The high detection sensitivity together with the targeting of HTNV nucleocapsid protein 105, which is the most abundant viral component synthesized post virus infection, makes this assay a promising candidate method for the early diagnosis and control of HTNV.
Figure 4.
AuNP-based immunoassays for the detection of hantaan virus (HTNV) and Rift Valley fever virus (RVFV). (A) Immuno-PCR assay for HTNV detection using AuNP probes dually functionalized with antibody (Ab) and double-stranded (ds)DNA. Au-nanoprobes are directly applied to label virus antigens precaptured on a microplate. The AuNPs are carrying dsDNA that includes barcode single-stranded (ss) DNA for signal amplification. The barcode DNA is separated, amplified and detected by gel electrophoresis 96. Additionally, this assay can be modified in a more complex detection scheme that has been reported for human immunodeficiency virus detection 81. (B) Surface-enhanced Raman spectroscopy (SERS)-based assay for detection of RVFV using Raman reporter dye-coated AuNPs and magnetic NPs (MNPs). AuNPs and MNPs are conjugated with a polyclonal Ab specific for the target virus antigen forming AuNP/virus antigen/MNP complexes; then, a 785 nm laser excites the magnetically concentrated AuNP/virus antigen/MNP complexes. The presence of the target antigen yields a reduction in the intensification of Raman dye signature spectrum peaks, thereby providing an estimation of its concentration 49. This assay has been applied for the detection of WNV infection 49.
7.1.2. Rift Valley Fever Virus (RVFV)
RVFV is an arthropod-borne pathogen primarily known to affect animals and later discovered to infect humans. RVFV infection in humans can progress to serious hemorrhagic fevers that often lead to death, with prominent mortality rates of up to 10-12%. Infected persons can also develop other clinical manifestations, including retinitis, encephalitis, and paralysis 106, 107. RVFV is historically endemic to Africa and has had a long history of outbreaks ranging from its reemergence in Egypt in 1977 to the most recent outbreak in South Africa in 2011 108, 109. Thus far, RVFV outbreaks are unpredictable and usually associated with very high socio-economic and public health consequences 110, 111. Therefore, diagnostic methods that can rapidly identify the virus have become crucial not only for avoiding and preventing such outbreaks but also for monitoring cross-border dissemination.
A novel immunoassay based on AuNPs coupled with a surface-enhanced Raman scattering (SERS) technique has been developed for the detection of RVFV capsid antigen 49. In this assay, AuNPs are applied as both immobilization scaffolds for the target-capturing antibodies and metal promoters to enhance the Raman reporter dyes. The target antigen is first captured by antibody-magnetic particle conjugates and then labeled with dye/antibody-AuNP conjugates, forming a three-component immunocomplex that is magnetically concentrated and finally detected by SERS (Fig. 4B). This approach has shown an outstanding sensitivity level down to 5 fg/mL of the target antigen. This high sensitivity with the possibility for the direct detection of RVFV in complex media or samples allowed by magnetic particles support the application of this assay for the rapid and sensitive detection of RVFV and the control of any future outbreaks.
7.2. Coronaviridae
7.2.1. Severe Acute Respiratory Syndrome (SARS)
The Coronaviridae family comprises two subfamilies, Coronavirinae and Torovirinae, that were recently expanded to include many viruses. Coronavirinae are classified into four genetically distinct genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus) 112. Coronaviruses are enveloped RNA viruses that are pleomorphic in shape (spherical and 120-160 nm or bacilliform and 170-200 nm by 75-88 nm). They have a relatively large, positive-sense, ssRNA genome of 27-32 kb that encodes four to five structural proteins (S, the spike glycoprotein; M, the membrane glycoprotein; N, the nucleocapsid interrupt phosphoprotein; E, the envelope protein; and HE, the hemagglutinin-esterase glycoprotein) and 2 nonstructural polyprotein precursors (pp1a, polyprotein 1a; and pp1ab, polyprotein 1ab), which are later processed into several other nonstructural proteins and viral polymerase 113, 114.
Coronaviruses are clinically significant to humans and usually associated with different respiratory, intestinal, hepatic, or neurological diseases 115. HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 are among the most disseminating coronaviruses that usually infect the upper respiratory tract in humans, causing mild common cold-like diseases 116-119. Other coronaviruses, such as SARS-CoV identified in China in 2002 and MERS-CoV discovered in the Middle East in 2012, are notorious pathogens able to cause widespread outbreaks of pneumonia and pneumonia-like conditions with very high morbidity and mortality rates of up to 35% 120, 121. Due to their high clinical significance and contagious nature, there has recently been great interest in testing and developing advanced SARS detection methods.
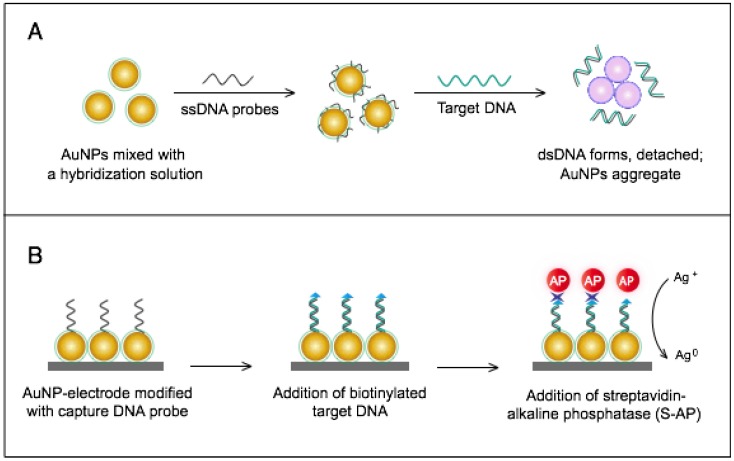
SARS detection using AuNPs is primarily focused on developing rapid and specific molecular detection through two main assays: 1) a colorimetric assay for pp1ab gene detection; and 2) an electrochemical assay for nucleocapsid protein gene detection (Table 1).
The colorimetric assay involves the ability of AuNPs to preferentially adsorb ssDNA over dsDNA and specifically sense the presence of the target DNA 54. Specific, short ssDNA probes adsorb to the surface of AuNPs, resulting in an increased particle colloidal stability and an increased ability to withstand slightly elevated salt concentrations without significant aggregation or color changes. Upon addition, the target DNA specifically forms dsDNA with the adsorbed ssDNA probes, which then easily detach, leaving the AuNPs to aggregate due to the salt. The particle aggregation causes the red color of the solution to change to blue, indicating the presence of target SARS nucleic acids; this change can be directly assessed by the naked eye or precisely quantified by ultraviolet-visible (UV-vis) spectroscopy in correlation to the target DNA concentration (Fig. 5A). In the electrochemical assay, AuNPs are applied to enhance the electrode conductivity and increase the surface area available for detection probe immobilization 72. SARS-specific DNA-capturing probes are first immobilized on the surface of an electrode structured with AuNPs; then, they are allowed to hybridize with the biotinylated targets. Then, streptavidin-labeled alkaline phosphatase is applied to catalyze the indirect reduction and deposition of silver ions, which are eventually measured by anodic stripping voltammetry in correlation with the target DNA concentration (Fig. 5B).
Figure 5.
AuNP-based nucleic acid assays for the detection of severe acute respiratory syndrome (SARS). (A) Colorimetric detection of SARS using AuNPs stabilized by single-stranded (ss)DNA probes. In the presence of the target DNA sequence, double-stranded (ds)DNA is formed and desorbs from the surface of citrate-reduced AuNPs, leaving them to aggregate. This aggregation results in a color change from red to blue, indicating the presence of target nucleic acids 54, 56. (B) Enzymatic electrochemical detection of SARS by anodic stripping voltammetry using a AuNP-screen-printed carbon electrode. The AuNP-modified electrode is modified with capture DNA probes that are allowed to hybridize with biotinylated targets. Streptavidin-alkaline phosphatase (S-AP) is then applied for the indirect reduction of silver ions in the solution into a metallic deposit. The formed silver metals are then electrochemically measured to determine the target virus DNA concentration 72.
The AuNP-based assays developed for the molecular detection of SARS are relatively rapid and simple; especially the colorimetric assay, which completely eliminates the need for instrumentation or trained personnel and yields results exclusively in the liquid phase that can be rapidly identified as positive/negative by the naked eye within 5 min. This technique can allow the detection of target SARS nucleic acids with a sensitivity limit down to 100 fM; thus, it can be very beneficial for the early diagnosis of the SARS virus, which is critical for such a contagious pathogen. In addition, the simplicity and very high sensitivity of AuNP-based colorimetric assays for nucleic acids have promoted the application of AuNPs for the detection of other viruses either using slightly modified procedures or in combination with other sensing characteristics of AuNPs 56.
7.3. Filoviridae
7.3.1. Ebola Virus (EBOV)
The family Filoviridae can be classified into two main genera, Ebolavirus and Marburgvirus. The genus Ebolavirus includes five different species, Zaire ebolavirus, Sudan ebolavirus, Reston ebolavirus, Taï Forest ebolavirus, and Bundibugyoebolavirus, while Marburgvirus is a single-species genus comprising Marburgvirus marburgvirus, albeit consisting of two divergent viruses, Marburg virus and Ravn virus 119, 122. Filoviruses are filamentous, enveloped RNA viruses 80 nm in diameter and approximately 800-1000 nm in length. Their genome is nonsegmented, negative-sense, ssRNA 19 kb in size that encodes for at least 7 proteins (nucleoprotein, viral proteins VP24, VP30, VP35, and VP40, glycoprotein, and polymerase protein) 123.
Most EBOVs primarily originate from Africa, where they are frequently associated with large outbreaks of Ebola hemorrhagic fever (EHF), which is now known to be one of the most deadly and virulent diseases affecting humans, with a case fatality approaching 90% 124. The early outbreaks of EHF were confined to Sudan and its neighbor, Democratic Republic of the Congo, from 1976 to 1978, followed by several independent outbreaks in different countries: Côte d'Ivoire in 1994; Gabon in 1994, 1996, 1997 and 2001-2002; Sudan in 2004; Republic of the Congo in 2001-2002, 2003 and 2005; Democratic Republic of the Congo in 2007-2008, 2008-2009; and Uganda in 2000, 2007, and 2011 124, 125. Most of these outbreaks were caused by ZEBOV and SEBOV, while some were caused by new species later named CIEBOV and BEBOV 125. These heavy series of EHF outbreaks are continuing, and most recently, multiple countries in West Africa have struggled against a massive outbreak of EBOV beginning in March 2014. Ebola is expected to remain a major global public health threat, especially with the current absence of effective treatments; therefore, it is necessary to have flexible rapid-detection schemes in place to diagnose and control EBOV outbreaks 126.
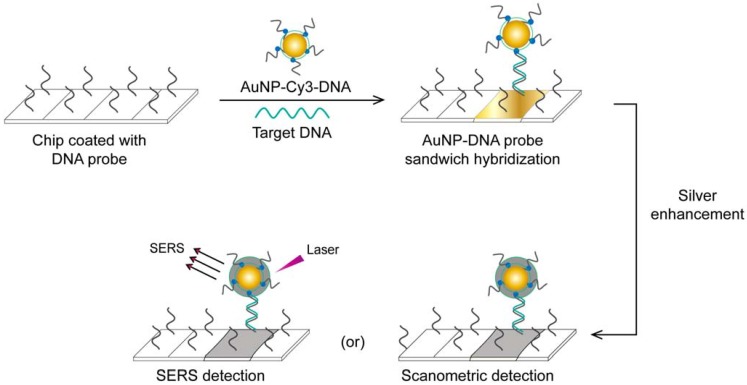
A very interesting AuNP-based molecular assay has been developed for the bimodal scanometric and SERS-based detection of EBOV by using AuNPs to promote silver staining on their large surface area 50. The target EBOV DNA sequence is first captured on chip surfaces by specific, short DNA probes and labeled with AuNP-DNA-Cy3 probes. Then, the formed 3-component hybridization complexes are subjected to a silver enhancement step. The deposited silver metal eventually enables scanometric detection (based on a silver-caused dark color) and SERS detection (based on using Cy3 as a Raman-active tag) (Fig. 6).
Figure 6.
AuNP-based scanometric and surface-enhanced Raman scattering (SERS) for the detection Ebola virus (EBOV). It is a chip-based detection scheme in which the target DNA is captured by immobilized, specific DNA oligonucleotides and then directly labeled by AuNP-DNA probes, followed by silver enhancement. The AuNPs enhance the surface area available for silver deposition and DNA immobilization to achieve highly sensitive scanometric detection signals. Modification of the AuNP probes with Cy3 allows the additional SERS-based detection of target DNA. This scheme has been reported for the detection of a wide range of viral nucleic acids, including those of HCV, HBV, and HAV, which belong to different virus groups 50. Furthermore, the scanometric detection reported in this scheme has been considered a universal step in other detection schemes described for other viruses, such as HEV 82 and HIV 81, 84.
This assay has additional benefits beyond its high specificity and sensitivity that reach down to 20 fM of the target RNA. Because a very large number of probes can be designed based on using different Raman tags, this assay can be readily adapted for multiplex detection, allowing the simultaneous detection of different diagnostic targets of the same virus or even completely different viruses. Furthermore, it does not require DNA amplification and eliminates the need for specific thermal cyclic equipment. In addition, the capability for bimodal detection simplifies evaluation of the results, as the output can be positive/negative either by traditional scanner systems or by SERS when the probes are labeled with Raman dyes. Such simplicity based on bimodal detection without the need for complicated thermal amplification equipment, multiplexing, and high sensitivity supports the application of this technique for EBOV, which is very contagious and usually necessitates the detection of several targets.
This assay has been reported by the same authors for multiplex detection of hepatitis A virus (HAV), hepatitis B virus (HBV), HIV, and variola virus (smallpox, VV), which confirms the broad capability of this assay for virus detection 50.
7.4. Flaviviridae
Flaviviridae is a large virus family comprising three main genera: Flavivirus (53 species, type species is yellow fever virus), Hepacivirus (one species, hepatitis C virus; HCV), and Pestivirus (four species, type species is bovine virus diarrhea) 127. Flaviviruses are spherical, enveloped RNA viruses 40-65 nm in size. Their genome is nonsegmented, positive-sense ssRNA approximately 10-11 kb in size and includes one open reading frame (ORF) encoding three structural proteins (C capsid; prM, pre-membrane; and E, envelope), and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). Several well-known human pathogens are included in this family, such as dengue virus (DENV), HCV, yellow fever virus, and West Nile virus (WNV), Japanese encephalitis virus, and Tick-borne encephalitis virus. These viruses have become increasingly serious and have been frequently involved in causing many endemics and epidemics around the world 128, 129.
7.4.1. Dengue Virus (DENV)
DENV is the most rapidly spreading vector-borne virus in the world. DENV is now known to be epidemic in more than 100 subtropical and tropical countries, threatening up to 2.5 billion people. DENV causes more than 350 million new annual infections, including 90 million with symptoms varying from flu-like, mild, undifferentiated fever to classic dengue fever (DF) or DF with hemorrhagic manifestations 130, 131. Although many of these infections are self-limiting and resolve without hospitalization, some progress to severe disease that needs to be quickly identified and treated 132, 133. Currently, there are no specific treatments or protective vaccines available against DENV, and as a result of its rapid expansion, accurate and sensitive diagnostic techniques have become crucial for effective control and treatment applications 134.
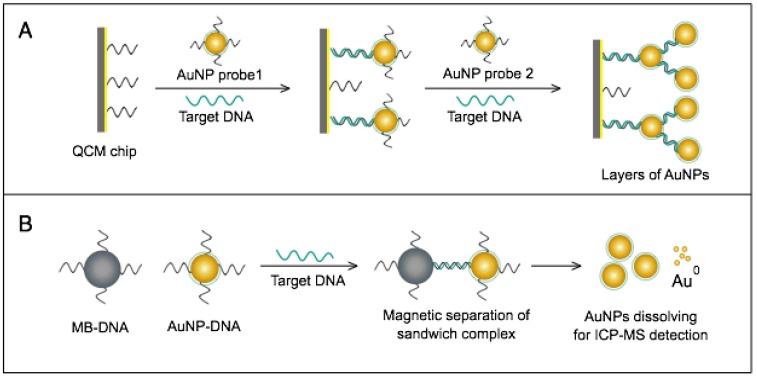
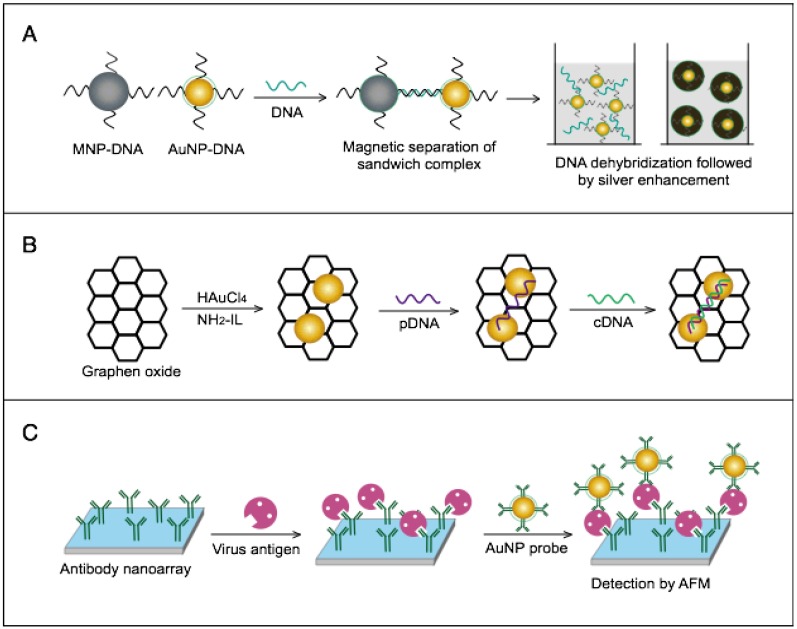
AuNPs have been combined with the well-known analytical techniques QCM and ICP-MS to develop two novel molecular assays for DENV detection. For the first time, these assays implemented AuNPs to increase the mass of target DNA to allow its detection by QCM in a highly quantitative and sensitive manner (Fig. 7A) or to amplify the detection signal by releasing many gold ions during ICP-MS analysis of the target DNA (Fig. 7B). In the AuNP-based QCM assay, the presence of target DNA initiates a layer-by-layer hybridization of the target DNA and several specific multivalent AuNP-DNA probes, resulting in significant mass changes detected by QCM 93 (Fig. 8A). Whereas, in the ICP-MS-based assay, two types of probes prepared from specific AuNP-DNA and MNP-DNA conjugates are applied to specifically recognize and sandwich the target virus DNA sequences. The MNP probes then help to magnetically separate the formed sandwich complexes, and the target DNA concentration is indirectly estimated by detection of the Au concentration existing in the sample 94 (Fig. 8B).
Figure 7.
AuNP-based assays for hepatitis C virus (HCV) detection. (A) Bio-barcode-amplification (BCA) enhanced electrochemical immunoassay using AuNPs dually modified with bio-barcode DNA and antibodies. AuNPs probes are applied to label target HCV antigen captured by magnetic NP (MNP) probes. The bio-barcode ssDNA is released and employed to establish multilayered AuNPs over the electrodes to enhance the produced electric current 72. (B) Dry-reagent strip-based DNA assay using AuNP-oligo (dT) reporters. AuNPs with poly (dT) are applied to label biotinylated target DNA modified with a poly (dA)-tail pre-captured by immobilized streptavidin in the test zone of the strip and generate characteristic red bands 55. (C) Single-particle fluorometric detection of HCV using Cy3-tagged ssRNA that is electrostatically adsorbed or covalently coupled through thiol (SH) chemistry to the surface of AuNPs. The coupling of AuNPs and ssRNA probes brings Cy3 in the vicinity of the AuNP surface and eventually results in emission quenching; upon hybridization with the target RNA, the quenched emission is released 55.
Figure 8.
AuNP-based nucleic acid assays dengue virus type 1 (DENV-1) detection. (A) Quartz crystal microbalance (QCM)-based detection through target-induced assembly of AuNPs multilayers on a QCM chip. On the surface of a QCM chip coated with a gold film (yellow layer), the target DNA is captured and labeled by two types of virus-specific AuNP probes to amplify the detection signal by AuNP-based mass enhancement 93. (B) Inductively coupled plasma mass spectrometry (ICP-MS)-based detection using magnetic separation and labeling with AuNPs-DNA probes. Magnetic bead (MB)-DNA and AuNP-DNA sandwich the target DNA sequence, forming 3-component hybridization complexes, which are magnetically separated and analyzed by ICP-MS 94.
It is worth noting that both techniques are among the most novel applications of AuNPs in virus detection. The implementation of AuNPs extends the ability of QCM and ICP-MS to detect DNA. Moreover, AuNP-based ICP-MS is among the most sensitive molecular DNA assays applied for virus detection, with a detection limit down to 80 zmol, which can greatly help to control DENV and other viruses 94.
7.4.2. Hepatitis C Virus (HCV)
HCV is one of the leading causes of chronic liver diseases in humans. Approximately 3% of the global population has experienced HCV infection. Currently, there are 170 million chronic carriers, and more than three million new infections occur globally each year 135, 136. Chronic HCV infection is frequently associated with marked hepatic injuries, including cirrhosis, liver failure or hepatocellular carcinoma. These types of injuries can be very severe and often end in death or liver transplantation 135, 137. HCV infection is generally resilient and usually requires treatment that combines numerous medications. The most popular treatment regimen for HCV is a combination of PEGylated interferon alfa (PEG-IFNα) and ribavirin (RBV) 138. Other three-drug regimens based on the addition of the serine protease inhibitor telaprevir (TVR) or boceprevir (BEC) to the standard PEG-IFNα/RBV treatment have also been described 139. In addition, several recently developed oral drugs, such as simeprevir and sofosbuvir, have been approved by the FDA for more effective care and shorter treatment duration 140-143. Despite these numerous drugs and due to the lack of efficient vaccines against HCV, reliable and sensitive detection methods are essential for viral infection prevention and therapeutic responses.
Numerous AuNP-based scanometric, fluorometric, electrochemical, and colorimetric assays have been reported for the molecular detection of HCV (Table 1). In these assays, AuNPs are utilized for different sensing functions: 1) AuNPs have been modified with poly(dT)-tailed DNA and directly applied to label target DNA sequences terminally modified with a poly(dA)-tail in a LFA assay to allow the colorimetric detection of HCV (Fig. 7B) 55. 2) AuNPs electrostatically or covalently modified with Cy3-tagged ssRNA sequences have been applied for the fluorometric detection of target HCV sequences, in which the fluorophores are quenched in a process analogous to FRET. The presence of Cy3 in the vicinity of the surface of AuNPs eventually results in Cy3-emission quenching. Upon hybridization with target viral RNA, the electrostatically adsorbed probes labeled with Cy3 are detached from the surface of the particles, releasing the quenched fluorescence. The covalently bound probes form rigid dsRNA with a linear configuration, causing Cy3 to move away from the particle surface and release the quenched emission 60 (Fig. 7C). 3) AuNPs have also been applied to enhance electrical conduction and catalyze a reduction reaction on the surface of detection electrodes. A specifically designed hairpin DNA probe terminally modified with AuNPs and bound to the surface of a glassy carbon electrode is utilized to detect the amplified viral RNA. The presence of target DNA results in the formation of rigid dsDNA, and the hairpin conformation consequently changes, moving the AuNPs away from the electrode surface, which eventually appears as a detectable decline in the current value in proportion to the target DNA concentration 78. 4) AuNPs decorated with a specific nucleic acid probe were used for colorimetric detection of HCV. The assay is based on using cationic AuNPs to induce the aggregation of AuNPs probes. In the absence of HCV, the cationic AuNPs electrostatically bind to negatively charged AuNPs-DNA probes causing their aggregation and change of the solution color to blue. The presence of HCV nucleic acid protects the probes and prevents their aggregation by cationic AuNPs and the solution color remains red. This assay was specific and showed a sensitivity of 93.3% and a detection limit of 4.57 IU/μL 13.
In the immunological detection of HCV, AuNPs dually modified with capturing antibodies and barcoding DNA, similar to those described for the immuno-PCR technique, in combination with magnetic NP (MNP)-antibody conjugates, have been applied to capture and separate the target antigen. Subsequently, the barcode DNA is further allowed to act as a bridge, guiding the formation of a multilayer of AuNP-DNA probes over the surface of a nanogap electrode (Fig. 7A). The formation of such barcode DNA-guided AuNP multilayers significantly increases the detected electrical current and is utilized to quantitatively measure the target HCV antigen concentration in applied solutions. The results of this immunoassay indicate a detection limit of 1 pg/µL 69. In a similar protocol, the barcoding DNA was further applied to HCV core antigen detection based on an enzyme to release the barcode DNAs to be quantified with RT-PCR. This assay showed a detection limit of 1 fg/mL, which is one magnitude greater than the standard ELISA (2 ng/mL) 14.
It is worth mentioning that the primary focus of the developed AuNP-based assays was the detection of HCV RNA rather than HCV antigens or antibodies. This is due to the broader potential of molecular assays for the detection and management of active HCV infections.
7.5. Hepadnaviridae
7.5.1. Hepatitis B Virus (HBV)
Hepadnaviridae comprises two main genera, Orthohepadnavirus and Avihepadnavirus. The genus Orthohepadnavirus includes 4 species (HBV, woodchuck hepatitis virus, ground squirrel hepatitis virus, and woolly monkey HBV) that infect mammals, while the genus Avihepadnavirusincludes2 species (duck HBV and heron HBV) infect birds 144. Hepadnaviruses are spherical, enveloped DNA viruses 40-48 nm in size. They possess a unique gapped genome of 3.2-kb circular dsDNA, which contains 4 partly overlapping ORFs encoding the different viral proteins: the core protein (C), e antigen, polymerase protein (Pol), envelope proteins, and transcriptional transactivator protein 144, 145. The prototype member of the family Hepadnaviridae, HBV, is a health threat to ~88% of the global population 145, 146. HBV is the etiological agent of hepatitis type B in humans. It is estimated that nearly 2 billion people around the world have been exposed to HBV infection, and there are more than 360 million chronic carriers suffering from the serious risk of developing liver cirrhosis and cancer 146, 147. The availability of an effective protective vaccine against HBV has greatly reduced the number of new infections. In addition, there have been significant advances in the available treatments for chronic HBV infection. Viral DNA polymerase inhibitors and PEG-IFNα are now well known to significantly control HBV infection and prevent its progression to chronic hepatitis B-associated liver failure and hepatocellular carcinoma 148, 149. However, chronic HBV infections remain highly contagious, and the virus can easily be transmitted to other persons through any close contact allowing the transfer of infected bodily fluids. The highly contagious nature of HBV and its ability to spread among people are the main reasons why this pathogen continues to threaten the public. Therefore, diagnostic methods that are highly sensitive to low concentrations of virus are needed to curtail this virus and prevent its dissemination.
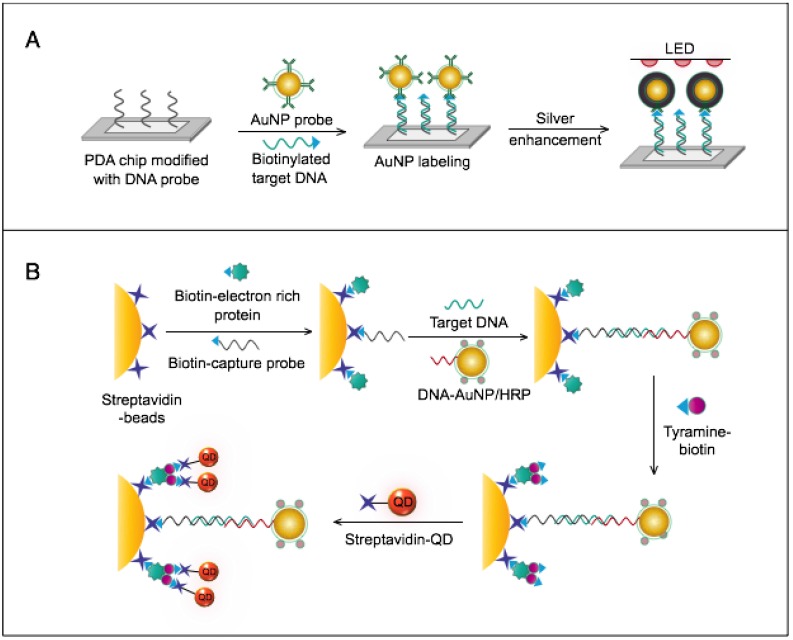
HBV is by far the most common virus reported in the published literature regarding the applications of AuNPs in virus detection (Fig. 3). Interestingly, the developed assays cover most of the known diagnostic markers of HBV through many different electrochemical, scanometric, fluorometric, light-scattering, colorimetric, and SERS detection schemes (Table 1 and Fig. 6, 14-18).
Several AuNP-based electrochemical assays have been developed for the detection of HBV antigens and DNA. Two enzyme-amplified electrochemical assays have been reported for the detection HBV surface antigen (HBsAg) using conjugates of AuNPs and horseradish peroxidase (HRP)-labeled antibodies against HBV surface antigen (HBsAbs) 76, 77. The target HBsAg is captured and labeled with AuNP-HBsAb/HRP as secondary antibody conjugates. Then the reduction of H2O2 catalyzed by the bound HRP is measured either by using an AuNP/thionin/DNA-modified Au electrode coupled with a cyclic voltammeter 76 or through using a nanoporous Au electrode coupled with differential pulse voltammeter 77 (Fig. 9A). Alizadeh et al. reported AuNPs modified with horseradish peroxidase mimicking DNAzyme to label HBsAg magnetically captured and concentrated on the surface of a Au sheet-like electrode. Due to the efficient catalytic activity of HRP-mimicking DNAzyme, the proposed immunoassay allowed quantitative detection of HBsAg with a linear concentration range of 0.3-1000 pg/mL and detection limit of 0.19 pg/mL 15. Other studies described the electrochemical detection of HBV based on AuNP-metal enhancement amplification, including copper and silver metal enhancement 74, 75. Using copper metal enhancement, HBsAg is sandwiched by MNP-HBsAb conjugates and AuNP-HBsAb probes to form a 3-component immunocomplex, which is magnetically separated and stained through a copper enhancement step (Fig. 9B). After copper acidic dissolution, the resulting ions are measured by anodic stripping voltammetry (ASV) 75. Using silver enhancement, HBV DNA is detected using probes prepared of streptavidin-modified AuNPs. Biotinylated HBsAg gene sequences are magnetically pre-separated and concentrated using magnetic bead-DNA conjugates. The deposited silver ions are then analyzed by an electrochemical stripping technique to detect the target HBV DNA quantitatively 74.
Figure 9.
AuNP-based immunoassays for hepatitis B virus (HBV) detection. (A) Enzyme-catalyzed electrochemical immunoassay. In this assay, the target HBsAg is captured and labeled with AuNP-Ab/HRP, and the detection signal is measured by using an Ab-modified Au electrode to assess the reduction of H2O2 catalyzed by the bound HRP using differential pulse voltammetry (DPV) technique 76. (B) Copper (Cu)-enhanced electrochemical immunoassay. AuNPs serve as a scaffold for a horseradish peroxidase (HRP) enzyme that catalyzes a redox reaction in the first scheme or enhances metal deposition in the second scheme 75. (C) Fluorometric detection by the FRET-induced quenching of fluorophores on the surface of AuNPs. AuNPs and gold nanorods (AuNRs) are applied to quench the fluorescence of fluorophores, such as (i and ii) quantum dots (QDs) 62, 65 and (iii) fluorescein amidite (FAM) 61, through a FRET-based interaction in the presence of the target virus. A similar AuNP FRET-based scheme has been reported for the detection of H1N1, which belongs to the family Orthomyxoviridae 63. (D) Light-scattering assays using AuNP-antibody probes. (i) Light-scattering immunoassay of virus antigen based on using AuNPs with different sizes (10 nm and 50 nm) conjugated with antibodies (Abs) to sandwich target antigens and enhance the dynamic light scattering 51. Additionally, this scheme has been reported with the use of AuNPs of the same size for H1N1 detection 53. (ii) Plasmonic immunoassay based on allowing AuNR-Ab conjugates to interact with the target antigen, thus changing the AuNR localized surface plasmon resonance (LSPR) behavior 52.
HBV detection through AuNP-based scanometric assays mainly relies on the well-described AuNP-promoted silver-staining protocol, in which AuNP probes are applied to facilitate the reduction of silver ions into metallic silver that is deposited as visible black spots, indicating the presence of HBV. Following this protocol, AuNPs modified with staphylococcal protein A (SPA) 88 or specific DNA probes 50, 80 have been utilized in multiple chip-based assays to permit the detection of different HBV antibodies and DNA (Fig. 6). In addition, Wang et al. combined this AuNP-promoted silver-staining protocol integrated with a bio-barcode-amplification (BCA) technique to allow an enhanced scanometric detection of HBV DNA 85. This BCA-based scanometric assay employs two different sets of DNA conjugates to capture and detect a specific HBV sequence. The first set is a group of AuNPs modified with the short dsDNA of a signal amplification DNA strand (barcode DNA) partially complemented with a detection probe strand, while the second set is a group of magnetic microparticles (MMPs) modified with ssDNA specific to the target HBV DNA. The presence of the complementary target sequences of DNA guides the assembly of AuNP/DNA/MMP sandwich hybrids. Subsequently, the sandwiched hybrids are magnetically isolated and washed, and the barcode DNA is eventually released from the AuNPs. The barcode DNA is then added to the surface of a chip modified with specific DNA-capturing probes and directly labeled with AuNP-DNA conjugates, followed by silver staining to further amplify the detection signal (Fig. 10).
Figure 10.
AuNP-based nucleic acid assay for hepatitis B virus (HBV) detection. HBV DNA was detected using a scanometric detection that integrates magnetic separation and AuNPs-based Bio-barcode-amplification method. AuNPs provide a large surface area for silver deposition and DNA immobilization, thereby achieving highly sensitive scanometric detection signals through a chip-based silver deposition scheme 85.
The AuNPs-fluorometric assays for HBV are performed through different schemes that basically depict the quenching efficiency of AuNPs in a manner remarkably similar to the traditional FRET protocol. Zheng et al. developed a multiplex detection system based on applying gold nanorods (AuNRs) as acceptors and QDs of different colors as donors to simultaneously detect HBsAg and HBV e antigen 62. This assay follows the direct sandwich immunoassay format, and the presence of target antigens is manifested by the FRET-induced quenching of QD fluorescence in the formed sandwich nanostructure of AuNR-Ab1/Ag/QD-Ab2 (Fig. 9C). Draz et al. further coupled multiple AuNP-peptide conjugate-acceptors with single-core QD-antibody Fab conjugate-donors, forming a preassembled hybrid nanocluster plasmonic resonator complex for HBsAg and HBV particle detection (Fig. 9C). This scheme follows a competitive assay format, and the addition of HBV target surface antigen or particles to the preassembled nanocluster releases the quenched fluorescence signal of the QDs in proportion to the target concentration 65. Furthermore, Lu et al. developed a fluorometric assay for HBV DNA detection in which positively charged AuNRs are employed to chelate fluorescein amidite (FAM)-tagged ssDNA electrostatically onto their surface, forming FAM-ssDNA-AuNR ternary complexes 61. The formation of these complex results in the FRET-based quenching of FAM fluorescence, and when complementary target DNA is present in the system, a FAM-ssDNA/cDNA duplex is formed. This DNA duplex is comparatively more negative than the FAM-ssDNA and can mediate stronger electrostatic interactions with AuNRs, leading to increased FRET efficiency, which manifests as a further decline in the fluorescence intensity of FAM. This change in fluorescence intensity is utilized to sense and estimate the exact concentration of the added target DNA (Fig. 9C).
AuNP-based light-scattering assays have been described for the detection of HBsAg through two main schemes: the first scheme applies spherical AuNPs of different diameters (10 nm and 50 nm) modified with HBsAb to sandwich HBsAg, and the presence of HBsAg results in specific immuno-based aggregations that can be estimated by measuring the increases in hydrodynamic diameter using DLS (Fig. 9D) 47, 51; the second scheme relies on the LSPR characteristic behavior of AuNRs to directly sense the binding of HBsAg to the antibodies conjugated to their surface by measuring LSPR peak shifts via UV-vis spectroscopy (Fig. 9D) 52.
A AuNPs-based colorimetric lateral flow assay (LFA) assay was developed for the detection of HBsAg. This assay relies on the characteristic red color of different sized AuNPs to label HBsAg on an immunochromatographic test strip. A strip composed of a sample pad, conjugation pad, control line, test line and absorbent pad was used to capture HBsAg labeled with AuNPs and the test line becomes red in color. The results showed that the signal visibility of LFA could be improved by increasing the diameter of AuNPs, and ~43 nm AuNP enabled LFA assay with a detection limit of 100 ng/mL 16.
A AuNPs-based SERS assay for HBV DNA detection was developed following a sandwich assay format performed on a heat-responsive hybrid silicon substrate modified with AuNPs. The target HBV DNA is captured on the surface of the silicon substrate and forms a sandwich complex with specifically designed AuNPs modified with DNA-indocyanine green proximally to the surface, leading to high SERS signals. This heat-responsive hybrid silicon substrate-based SERS platform can detect remarkably low HBV DNA concentrations of ~0.44 fM at 25 °C and ~0.14 fM at 37 °C 17.
AuNP-based fluorometric detection is the most sensitive among the reported methods for HBV detection, with a detection limit of 1 particle/μL. Due to the highly infectious nature of HBV, this limit of detection can be very useful for exploiting these assays for efficient control and management of HBV infection. In addition, the simplicity of scanometric and colorimetric detections is very interesting, allowing flexibility in the detection design and target, and was reported for single and multiple target detection for more efficient and accurate screening of HBV. On the other hand, the developed DLS assay is very novel to HBV research, simple and sensitive; however, the need for bulky equipment for DLS measurement remains the major challenge for its wide use and real application in HBV detection.
7.6. Hepeviridae
7.6.1. Hepatitis E Virus (HEV)
The family Hepeviridae is monogeneric with one exclusive member called HEV, which is a spherical, small, nonenveloped RNA virus with a ~7.2 kb ssRNA genome 150. The viral RNA is a positive-sense molecule organized into 3 partially overlapping ORFs flanked by short untranslated regions at both ends. The first ORF encodes 4 mature nonstructural proteins (methyltransferase, protease, helicase and replicase), the second ORF encodes 2 structural proteins (C and VP1), and the third ORF encodes a small protein of unknown function 150. HEV is known for causing human liver inflammation called hepatitis E, which is widely disseminated in developing countries with poor sanitation conditions and has recently become apparently endemic in some industrialized countries, including the USA 151.
One AuNP-based colorimetric assay has been reported for the molecular detection of HEV 152. This assay targets a conservative fragment of HEV in ORF1 that is pre-amplified by one-step RT-PCR/PCR amplification, and the generated cDNA is labeled by AuNP-DNA conjugates, forming three-component sandwich hybrids in a manner very similar to that previously depicted in Fig. 6. This hybridization is followed by a silver-staining step to allow the scanometric detection of target DNA either by the naked eye or using simple scanners, with a detection limit of 100 fM of HEV amplicon.
One of the major advantages of this HEV detection assay is its sensitivity, which can help control HEV. However, this assay suffers from the same disadvantages of conventional PCR because of the requirements of the amplification step.
7.7. Herpesviridae
Herpesviridae is a taxonomically large family of viruses, including 3 major subfamilies (alphaherpesvirinae, betaherpesvirinae and gammaherpesvirinae), which together comprise nearly 9 genera with over 100 viruses 153. Herpesviruses are spherical, enveloped DNA viruses approximately 200 nm in size. They have a large, linear dsDNA genome of 125-240 kbp encoding approximately 70-165 genes. This genome consists of unique long (UL) and unique short (US) regions bound by inverted repeats. The rearrangements of these repeated sequences result in different genome isomers. In Epstein-Barr virus (EBV) and Kaposi sarcoma-associated herpes virus (KSHV), from the subfamily gammaherpesvirinae, there are repeated sequences at the termini of their genomes that differ from those of other human pathogenic herpesviruses, including herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2), human cytomegalovirus (HCMV), and varicella zoster virus (VZV); these viruses are characterized by the definite presence of US and UL regions in their genomes 153-155. Herpesviruses can infect humans and many animals. They account for significant and recurrent human cutaneous diseases, including oral herpes, genital herpes, and many cancers. Eight Herpesviridae family members are commonly involved in human infection: HSV-1 (human herpesvirus1; HHV-1), HSV-2 (HHV-2), VZV (HHV-3), EBV (HHV-4), HCMV (HHV-5), HHV-6, HHV-7, and KSHV (HHV-8).
7.7.1. Herpes Simplex Virus (HSV)
HSVs are among the most prevalent viruses in humans. There are two serotypically and genetically different HSVs, types 1 (HSV-1) and 2 (HSV-2) 156. HSV-1 is estimated to infect up to one-third of the world population, while HSV-2 infects nearly 500 million people around the globe, with more than 20 million new cases occurring every year 157-159. Both HSV types are renowned for infecting epithelial cells of the skin or mucosal surfaces and then infecting the central nervous system, causing lifelong, incurable infections in humans. These infections are primarily asymptomatic but can progressively result in serious health complications. For example, HSV-1 infection is the main cause of infectious blindness in the world and has emerged as an important cause of genital disease in the developed world, and HSV-2 infection is the leading cause of genital ulcers worldwide 159, 160. Nevertheless, these viruses are involved in many other clinical conditions, such as encephalitis, conjunctivitis, zosteriform skin lesions, pneumonia, and systemic infections that compromise vital organs in the body 161. Therefore, desperate efforts have been directed toward developing an effective vaccine against HSVs. However, there are no protective vaccines against HSVs, and only a few anti-herpes drugs, such as acyclovir, valacyclovir, and famciclovir, are commercially available 162-164. These drugs are only helpful to control symptoms and signs of infection and do not eradicate the virus or even decrease the frequency or severity of infection recurrence and due to the drug resistance rapidly developed by HSVs during treatment, the efficacy of anti-herpes drugs is increasingly hampered over time. Thus, the definitive diagnosis of HSV virus infection is essential for effectively controlling its severity and applying treatment regimens.
With their characteristic red color, AuNPs have been exploited as colorimetric labels in a lateral-flow immunochromatographic assay (LFIA) to allow the immunological detection of HSV antigen. In this assay, the LFIA strip is preloaded with the labeling AuNP-anti-human IgG conjugates onto the conjugate pad, and the capturing IgG-1 and IgG-2 antigens, along with the anti-IgG antibody, are immobilized onto separate test and control lines directly next to the sampling area (Fig. 11A). The sample is loaded and allowed to migrate across the strip with the chase buffer to react with the capturing antigens (IgG-1 and IgG-2) loaded on the test lines, as well as the anti-IgG antibody loaded on the control line. As a result, HSV antibodies are captured on the test lines and labeled with the loaded AuNP-DNA conjugates, causing characteristic red lines to appear. According to the color of the test lines, the samples can be qualitatively determined to be positive or negative within 20 min 59.
Figure 11.
AuNP-based assays for herpes simplex virus (HSV), human cytomegalovirus (HCMV), and Kaposi's sarcoma-associated herpesvirus (KSHV). (A) Lateral-flow immunochromatographic assay of HSV-2 antibodies using AuNP anti-IgG probes. Sample addition followed by chase buffer addition causes gold nanoparticle (AuNP)-anti-IgG conjugates and the sample to migrate, contacting the test lines sequentially and resulting in the capture of type-specific antibodies to HSV-2. The concentration of HSV-2 antibody-AuNP complexes on the test line causes a corresponding pink color to form 59. This scheme is similar to that reported for the detection of H5N1 58 and HIV-1 91. (B) Electrochemical detection of HCMV by the acidic dissolution of AuNPs. HCMV DNA is directly labeled with specific AuNP-DNA probes. After the acidic dissolution of AuNPs, AuIII ions are measured by anodic stripping voltammetry (ASA) 68. (C) Colorimetric one-pot NP-based assay of KSHV. This assay has been reported for the differential diagnosis of KSHV DNA and DNA from a frequently confounding Bartonella species. Upon the addition of sample DNA to a mixture of silver NP (AgNP)-DNA and AuNP-DNA conjugates specific for KSHV and Bartonella DNA, respectively, complementary DNA hybridization triggers the aggregation of bare NP conjugates and the subsequent decrease or disappearance of its corresponding color 60.
7.7.2. Human Cytomegalovirus (HCMV)
HCMV/HHV-5 is the largest known endemic virus in humans. It persistently infects over 70% of the population worldwide 165. HCMV can infect a broad range of cells, including endothelial cells, epithelial cells, smooth muscle cells, fibroblasts, neuronal cells, hepatocytes, and many types of immune cells 165, 166. Nearly all HCMV infections are asymptomatic, except in immunocompromised individuals and neonates, and can lead to severe symptoms. HCMV infection is the leading cause of congenital malformations in live births worldwide, while in immunocompromised individuals HCMV is usually opportunistic, causing very serious clinical manifestations, such as pneumonia, hepatitis, encephalitis, colitis, and retinitis 167, 168. Moreover, HCMV has been described to be associated with several human malignancies, especially in the brain, breast, prostate, skin and colon 169. Numerous HCMV vaccine candidates have been developed and tested, but a vaccine has yet to be licensed. Antiviral therapy with several anti-HCMV drugs, such as ganciclovir, valganciclovir, foscarnet, and cidofovir, which target the viral DNA polymerase, is currently the main strategy for controlling HCMV infection 166, 170, 171. Unfortunately, these drugs result in substantial hematotoxicity and nephrotoxicity in patients, and HCMV remains a considerable public health threat without an efficient treatment or control strategy. In such conditions, the need for new diagnostic methods that can help to control the dissemination of this pathogen is very important.
A novel AuNP-based electrochemical assay for the molecular detection of HCMV DNA has been reported 68. In this assay, AuNPs are applied to amplify the detected electrochemical signal by releasing many Au ions upon exposure to acid dissolution. Typically, the immobilized target HCMV ssDNA is initially labeled with oligonucleotide-modified AuNP probes followed by a metal dissolution step, releasing many gold ions. The concentration of solubilized gold ions is then indirectly assessed by ASV (Fig. 11B). This assay allows the highly sensitive detection of target DNA concentrations down to 5 pM 68.
7.7.3. Kaposi's Sarcoma-associated Herpesvirus (KSHV)
KSHV/HHV-8 is a leading cancer-causing virus. It is commonly responsible for inducing a considerable range of neoplastic diseases in immunocompromised patients, such as Kaposi sarcoma, primary effusion lymphoma, and some kinds of multicentric Castleman disease 172, 173. Owing to the dramatically increasing rates of immunocompromised patients over the past two decades, the clinical significance of KSHV is clearly increasing; additionally, KSHV has a prevalence of 3-40% in this class of patients worldwide 173, 174. While KSHV is a relatively newly discovered virus, there are no standard diagnostic approaches, and the detection of this virus through traditional serological and molecular methods remains challenging. This difficulty is mainly due to the inconsistency observed among serological assays targeting different antigens, and the low levels of viral DNA that are usually present in the peripheral blood of infected individuals 175, 176. Hence, there is a continued interest in developing new reliable and sensitive methods for KSHV detection.
Through using a mixture of AuNPs and silver NPs (AgNPs), a colorimetric assay has been established for the multiplex, one-pot molecular detection of KSHV and its frequently confounding disease bacillary angiomatosis 57. In this assay, two types of probes are prepared from AuNPs and AgNPs separately functionalized with KSHV and Bartonella-specific DNA-capturing DNA probes, respectively, and mixed with the target DNA. Then, the color of the reaction mixture changes in a manner dependent on the type of target DNA added to the reaction solution: the addition of KSHV DNA causes AuNP conjugates to aggregate and the solution to appear murky yellow-orange, i.e., the specific color of the nonaggregated AgNPs, while the addition of Bartonella DNA causes AgNP conjugates to aggregate and the solution to appear pink, i.e., the color of nonaggregated AuNPs (Fig. 11C). In addition to being simple, this multicolor-change system successfully allows the multiplex detection of targets present in nanomolar concentrations.
7.8. Orthomyxoviridae
7.8.1. Influenza A Virus (IAV)
The family Orthomyxoviridae includes five genera: InfluenzavirusA, Influenzavirus B, Influenzavirus C, Isavirus, and Thogotovirus 177. The first three genera are widely known to infect birds, humans, and other mammals, causing influenza. The InfluenzavirusA genus contains many strains and can be further divided into several antigenic subtypes according to the antigenic properties of their envelope glycoproteins, i.e., hemagglutinin (HA) and neuraminidase (NA). In contrast, the InfluenzavirusB genus has no distinct antigenic subtypes of hemagglutinin or neuraminidase. InfluenzavirusC is similar to types A and B but less common and lacks the NA protein. The genus Isavirus is known to infect salmon, while thogotoviruses usually cause infection in some vertebrates and invertebrates 177, 178.
Orthomyxoviruses can be spherical (80-120 nm) or filamentous (20 nm in diameter and 200-300 nm long) in shape, and their genome contains 7 or 8 segments of linear negative-sense ssRNA with a total length of 12-15 kb encoding different structural and nonstructural proteins, including nucleoprotein, three large proteins (PB1, PB2, and PA), matrix proteins (M1 and M2), the glycoproteins HA and NA, and two nonstructural proteins (NS1 and NS2) 177, 179, 180.
Several strains of the genus InfluenzavirusA cause seasonal and occasional acute respiratory diseases called influenza, which is profoundly and rapidly spread in humans. Influenza is reported to affect up to 5% of adults and 20% of children of the global population each year 181, 182. Over the past decade, the world witnessed two major influenza outbreaks caused by newly emerging InfluenzavirusA strains: H5N1 in 2003 (avian influenza), and H1N1 in 2009 (swine influenza). Both were severe enough to incite the WHO to issue a pandemic warning alert, which reached the highest level, i.e., “phase 6,” for H1N1 in 2009 21, 183. The severity of influenza infection depends on both the virus strain and the age of the infected person. In elderly persons, influenza can result in hospitalization or death, while in younger adults, influenza-associated morbidity usually results in restricted activity, impaired work performance, and increased health care use 184, 185. Most people with the flu recover within one to two weeks without treatment. However, serious complications usually require medications to reduce the severity and duration of infection. Some antiviral drugs, such as oseltamivir and zanamivir, can effectively treat or prevent influenza 186. On the other hand, immunization with vaccine preparations of the most-circulating virus strains (trivalent or quadrivalent vaccines) remains the key strategy for preventing both influenza and its serious complications 187, 188. In fact, both the treatment and management of influenza infection, either by antivirals or vaccination, depend on the influenza virus strains and the timing and severity of the infection. Therefore, the rapid and accurate testing and typing of influenza is critical for treating infection and plays an important role in public health measures.
The application of AuNPs in IAV detection has primarily been focused on developing assays that enable the direct detection of whole IAV particles 53, 58, 92 or viral RNA 64, 66, 67, 70, 71, 83, 86 through numerous AuNP-based fluorometric, SERS, scanometric, light-scattering, colorimetric, and electrochemical detection schemes (Table 1).
The AuNP-based fluorometric assays developed for H5N1 detection have simply exploited the intense plasmonic-based quenching properties of AuNPs through different detection formats. One of these assays targets the detection of the whole virus particle through a lateral-flow immunoassay format that is similar to that previously described for the detection of HSV-2, as shown in Fig. 11A. In this assay, the virus is first captured on a lateral-flow test strip by pre-immobilized antibodies; then, AuNP-antibody conjugates are applied to label the captured virus particles. After washing, the AuNPs are collected and dissolved to release gold ions that are eventually utilized to quench the fluorescence of QDs in an additional, separate step 58. In addition, Ganbold et al. have developed a bimodal assay for the fluorometric and SERS detection of viral RNA using AuNPs modified with ssDNA-dye probes 64. The surface ssDNA-dye probes are weakly adsorbed to the surface of AuNPs by electrostatic interactions, and with the addition of the complementary target DNA, perfectly matched dsDNA forms and desorbs from the surface of AuNPs, resulting in measurable changes in the conjugated dye fluorescence and Raman fingerprint that are more substantial than those resulting from either mismatched dsDNA or ssDNA (Fig. 12A).
Figure 12.
AuNP-based assays for the detection of influenza A virus (IAV). (A) Fluorometric detection based on FRET-induced quenching of fluorophores by AuNPs. Single-stranded (ss)DNA probes modified with a fluorescent dye are attracted to the surface of AuNPs, resulting in quenched emission. The presence of target DNA sequences results in the formation of dsDNA in the suspension of citrate-reduced AuNPs and increases the fluorescence recovery intensities due to the desorption of the dsDNA-dye conjugates from the surface of the AuNPs 64. (B) Electrochemical assays by AuNP-enhanced differential pulse voltammetry (DPV). Specific DNA aptamers immobilized on a gold electrode (GE) modified with a layer of multiwall carbon nanotubes (MWCNTs), polypyrrole nanowires (PPNWs) and AuNPs are collectively applied as an electrochemical platform for the detection of DNA hybridization using DPV 71. (C) Impedimetric electrochemical assay using streptavidin-AuNPs probes. The direct/sandwich hybridization of biotinylated target DNA with capture probes immobilized on the surface of a screen-printed electrode (SPE) and additional conjugation with streptavidin-AuNPs results in impedimetric signal enhancement 70. (D) Label-free scanometric detection using positively charged AuNPs. The electrostatic interaction between positively charged AuNPs and negatively charged DNA is utilized to visualize the double-stranded (ds)DNA formed by the hybridization of single-stranded probe DNA and complementary target DNA on the array of chips through AuNP-mediated metal staining 83, 86, 87.
AuNP-based colorimetric assays have been developed for IAV detection through four main detection schemes: 1) The first scheme involves the application of AuNPs conjugated with anti-digoxigenin as colorimetric labels to detect the target viral DNA premodified with digoxigenin and electrophoresed through a membrane-based lateral-flow technique 66. 2) The second scheme is simply based on the electrostatic interactions between the negatively charged phosphate groups of the target DNA amplified with LAMP and the positively charged CTAB bilayer on AuNRs, resulting in the aggregation of AuNRs and a color change from red to purple 67. 3) The third scheme relies on a magnetic-based hydroquinone oxidation reaction involving two metal NP bioconjugates: IAV-specific pentabody-MNPs as capture probes and anti-AIV monoclonal antibody-AuNPs as detection probes. In the presence of virus particles, an immunocomplex forms and is separated by a magnetic field; after washing, AuNPs in the separated complex act to catalyze the oxidation of hydroquinone, which is detected optically in correlation with the AIV sample concentration 92. 4) The fourth scheme relies on the peroxidase-like catalytic activity of Au NP films and colloidal AuNPs toward TMB-H2O2 mixtures. The virus is captured on a 96-well plate modified with AuNP film biofunctionalized with HA antibody. Then it is labeled with AuNPs modified with NA antibody for signal generation. TMB-H2O2 is added, and rapid color changes are observed because of the oxidation of peroxidase substrate TMB. Using H1N1, the response of the developed assay was linear in the range from 10 pg/mL to 10 μg/mL and the limit of detection was 50.5 pg/mL, while with H3N2, the optical density of the developed color was dependent on the virus concentration in the range of 10-50,000 PFU/mL and the limit of detection was 24.3 PFU/mL 22.
AuNP-based electrochemical assays rely on using AuNPs to directly modify the target DNA or electron transducer surfaces, allowing enhanced electrical conduction and increased surface area for probe immobilization and target interaction. Following this protocol, Liu et al. 71 successfully detected AIV HA gene sequences using a DNA aptamer immobilized onto an electrode; its surface was modified with a composite of carbon nanotubes, polypyrrole nanowires and AuNPs (Fig. 12B). Furthermore, Bonanni et al. 70 developed an impedimetric electrochemical assay for the detection of AIV M gene sequences based on measuring changes in the impedimetric behavior of the electrode when the target DNA hybridizes with the capture DNA probes immobilized onto its surface and is subsequently labeled by AuNPs via streptavidin/biotin interaction (Fig. 12C).
AuNP-based scanometric assays for detecting AIV are mainly performed using the well-described AuNP-promoted metal staining protocol shown in Fig. 12D. In this protocol, the target nucleic acid is recaptured on the surface of glass chips modified with specific capture DNA probes 86. Then, the captured probes are labeled with AuNPs either modified with specific DNA probes or with the renowned dsDNA intercalator daunorubicin 83 to mediate the metal staining. Following this labeling step with AuNP probes, the presence of the target nucleic acids is visually determined with an additional silver- or gold-staining step.
The light-scattering detection of H1N1 virus using AuNPs relies on DLS to measure the aggregation of AuNP-antibody conjugates induced by the addition of target virus particles (similar to Fig. 9D). The magnitude of AuNP aggregation and the corresponding increase in their hydrodynamic size is correlated with the concentration of virus particles 53.
Finally, the immunological detection of H1N1 hemagglutinin (HA) antigen has only been reported through a novel LSPR-based immunoassay. In this assay, the presence of target antigen induces a sandwich immunoreaction between the fluorophore-labeled detection antibodies and the AuNP-capturing antibody conjugates, directly resulting in enhanced fluorescence caused by the coupling of fluorophore emission with the localized surface plasmons of AuNPs in a manner similar to that shown in Fig. 9C. The sensitivity of this method has been described to be 103-fold, improved over that of the conventional ELISA technique, with a reported limit of detection of 13.9 pg/mL 63.
7.9. Papillomaviridae
7.9.1. Human Papilloma Virus (HPV)
Papillomaviridae is a widely diverse virus family that includes hundreds of recognized viruses classified into 16 taxonomically distinct genera designated with specific Greek alphabet letters, from alpha to pi. The first five genera (Alphapapillomavirus, Betapapillomavirus, Gammapapillomavirus, Mupapillomavirus and Nupapillomavirus) include the human papillomaviruses, while the remaining genera specifically include other mammalian and avian papillomaviruses 189, 190. Papillomaviruses are spherical, nonenveloped DNA viruses 40-55 nm in size. Their genome consists of a single molecule of circular, supercoiled, dsDNA 5.3-8 kbp in size and encodes 6 early expressed (E) and 2 late (L) expressed proteins, in addition to a noncoding long control region 189, 191. HPV infections are the most epidemic sexually transmitted viral infections worldwide 192, 193. HPVs are highly epitheliotropic pathogens that usually infect the deepest layer of the skin or the genital surfaces. Most HPV infections cause no symptoms and usually self-clear without causing clinical problems, but persistent infections lead to serious clinical complications 194. This persistence typically depends on the type of HPV causing the infection. Among more than 100 types of HPVs, approximately 40 types infect the mucous membranes and can be either high- or low-risk HPVs. High-risk types, such as HPVs 16, 18, 31, 51 and 52, are usually associated with many cancers of the cervix, vagina, vulva, penis, anus, neck, and head, as well as oropharyngeal cancer, while low-risk types, such as HPVs 6 and 11, are mainly related to anogenital warts and recurrent respiratory papillomatosis 195, 196. Currently, HPV is considered the most important cancer-causing virus, as it is associated with ~4.8% of all human cancers 197. Despite this high clinical significance of HPV, there are no available treatments, and vaccination using the bivalent vaccine against HPVs 16 and 18 or the quadrivalent vaccine against HPVs 6, 11, 16, and 18 remains the first and only choice for controlling the global HPV disease burden 198-200. In fact, these vaccines are relatively expensive, and their efficacy is very restricted to specific HPV types. Therefore, the development of accurate diagnostic techniques for HPV detection and typing is essential for acquiring surveillance data and initiating appropriate vaccination programs.
AuNP-based assays have been developed for the molecular detection of HPV through different photoelectric, fluorometric, electric and colorimetric schemes 89, 98. The photoelectric assay essentially leverages AuNPs to enhance the deposition of silver metal on their surfaces, as previously described for several other assays. However, the enhanced precipitation of silver metal in this assay is used to block the light generated from a photodiode array (PDA), and the target DNA concentration is measured in proportion to the decrease in light intensity as electrically detected using a bipolar integrated circuit PDA chip 89. Specifically, PCR-amplified biotinylated target DNA is captured on the surface of the PDA chip, and then labeled with AuNP-biotin antibody conjugates; subsequently, a silver enhancement step decreases the light intensity, which is quantitatively translated into an electrical signal depending on the amount of target DNA (Fig. 13A). On the other hand, a fluorometric assay integrating AuNPs and a microfluidic bead-based array has been developed as a rapid and controlled HPV DNA detection system 98. In this assay, AuNPs dually functionalized with HRP enzyme and capture DNA strands are applied to label the target DNA captured on the surface of microbeads (Fig. 4A), causing HRP to be in proximity to biotin-tyramine loaded on the surface of the microbeads and catalyze their oxidation by hydrogen peroxide. This process eventually increases the deposition of biotin moieties on the surface of the beads, subsequently increasing the labeling efficiency with QD-streptavidin conjugates and yielding an enhanced fluorescence signal that varies significantly based on the target DNA concentration (Fig. 13B).
Figure 13.
AuNP-based nucleic acid assays for human papillomavirus (HPV) detection. (A) Photodiode array (PDA)-biochip system for HPV detection. AuNPs modified with anti-biotin IgG are applied to label biotinylated target DNA captured on the PDA chip. After a silver enhancement step, the precipitated silver metal at the surfaces of the AuNPs blocks the light irradiated from above by light-emitting diodes (LEDs) and decreases the resulting photocurrent 89. (B) Microfluidic bead-based enzyme-AuNP amplification method for the fluorometric detection of HPV. Streptavidin-coated beads are dually functionalized with biotin-electron-rich protein and biotin-DNA capture probes. The captured target DNA first reacts with horseradish peroxidase (HRP)-functionalized AuNP-DNA labels, bringing HRP close to the surface of the microbeads. The oxidation of tyramine-biotin by hydrogen peroxide results in the deposition of multiple biotin moieties onto the surface of the beads. This deposition is markedly increased in the presence of immobilized electron-rich proteins. Streptavidin-labeled quantum dots (QDs) are then allowed to bind to the deposited biotin moieties and display amplified fluorescence signals in relation to the target DNA concentration 98.
In the electrical detection of HPV, AuNPs were used as an immobilization scaffold for DNA immobilization on a silicon substrate carrying nanogapped interdigitated electrodes. This AuNPs deposition technique was shown to reduce the size of the nanogaps, allowing enhanced electric detection of the target DNA captured on the silicon substrate by specific HPV DNA probes. The results demonstrated that this assay was able to specifically detect HPV DNA within 30 min and at concentrations down to 1 pM 201.
Based on the color change of AuNPs upon aggregation, AuNPs carrying HPV DNA probe were coupled with the LAMP technique for colorimetric detection of HPV. LAMP reaction was performed at a temperature of 65°C for 20 min and the generated products were hybridized with AuNP-DNA probes for 5 min. Then the presence of the target DNA was detected by the addition of magnesium salt. The addition of the salt causes AuNPs aggregation and the color changes from red to blue. The presence of LAMP amplicons can protect AuNPs from aggregation and prevents AuNPs aggregation. The sensitivity of the developed LAMP-AuNP assay was greater than the LAMP turbidity assay by up to 10-fold with detection limits of 102 and 100 copies for HPV16 and HPV18, respectively 27.
7.10. Picornaviridae
7.10.1. Hepatitis A Virus (HAV)
The family Picornaviridae is taxonomically very diverse and has recently been expanded to involve 12 genera: eight are originally identified genera (Aphthovirus, Cardiovirus, Enterovirus, Erbovirus, Hepatovirus, Kobuvirus, Parechovirus, and Teschovirus), and four newly proposed genera (Avihepatovirus, Sapelovirus, Senecavirus, and Tremovirus) 202. Picornaviruses are spherical, relatively small (22-30 nm), nonenveloped RNA viruses. Their genome is positive-sense ssRNA that is 7.0-9.5 kb in size and terminally bonded to a noncapsid viral protein (VPg) at its 5' end and to a polyadenylated tail at its 3' end. The expression of this genome produces a single large polyprotein that undergoes a cascade of cleavage and processing reactions to form 10-12 final structural (VP1-4) and nonstructural (2A-C and 3A-D) proteins 203. Picornaviruses can infect a broad spectrum of hosts, including birds, humans, and other mammals 202, 204. The genera Enterovirus, Hepatovirus, Parechovirus, Kobuvirus, and Cardiovirus include several important species that affect humans. Particularly, the genus Hepatovirus encompasses HAV, which is the most likely causal agent of acute viral hepatitis in humans. HAV infection is usually asymptomatic and self-limiting, and the related symptoms can be mild or sporadically progress to fulminant hepatic failure 205, 206. The availability of an effective vaccine against HAV has substantially reduced the number of people who become infected with HAV 207, 208. However, the ability of this virus to survive harsh environmental conditions and remain in seawater, fresh water, wastewater, and soil for extended periods increases the possibility of outbreaks, especially in developing countries where sanitation is not typically available 205. The transmission of the virus usually occurs through the consumption of contaminated materials. As such, the virus does not replicate in the environment, water, or foods, and traces of viral contamination are difficult to identify, which remains a challenge for controlling HAV. The need for sensitive assays for HAV remains an urgent public health demand.
HAV detection using AuNPs was accomplished through scanometric and SERS-based assays. Both assays rely on the extensively described virtue of AuNPs to enhance silver staining, thereby allowing the molecular detection of HAV DNA in a microarray chip format (Table 1 and Fig. 6). In the scanometric detection scheme, AuNP-DNA conjugates are applied as specific detection probes to label target HAV RNA captured by specific VP1 probes on the surface of a microarray chip. A silver metal enhancement step is subsequently used for signal amplification, followed by the visual detection of nucleic acids 79. In the SERS detection, the captured HAV DNA is labeled by AuNP-DNA probes loaded with the Raman reporter dye Cy3; this approach has been applied as previously described for EBOV and depicted in Fig. 6 of this review 50. These assays are very simple and can detect target concentrations ranging from 3.37×10-7 - 3.37×10-8 M, with 100 fM as a limit of detection, which can be superior to the relatively expensive PCR-based approaches currently used for HAV detection and management.
7.11. Retroviridae
7.11.1. Human Immunodeficiency Virus (HIV)
The family Retroviridae is broadly classified into 7 genera (Alpharetrovirus, Betaretrovirus, Deltaretrovirus, Epsilonretrovirus, Gammaretrovirus, Lentivirus, and Spumavirus), which include nearly 51 viruses 209. Retroviruses are spherical, small (80-100 nm), enveloped RNA viruses. Their viral genome is typically bipartite and composed of two positive-sense ssRNA molecules (10 kb in total length). They comprise 4 major genes that encode the different structural and nonstructural proteins of the virus: gag encodes the large protein form matrix, capsid, and nucleocapsid; pro encodes the protease enzyme; pol encodes the reverse transcriptase and integrase enzymes; and env encodes the envelope glycoproteins and transmembrane proteins 209-211. Retroviruses infect a wide variety of vertebrates, including humans. Their infection in humans is usually very serious and can potentially cause oncogenesis and variable hematopoietic and neurological conditions. Among retroviruses, HIV is a well-known, formidable human pathogen responsible for the most devastating viral pandemic that has ever occurred in human history 212. HIV infection is usually correlated with the progression of what is clinically called AIDS, in which the immune system is harshly destroyed, and the entire body becomes vulnerable to terrible, life-threatening opportunistic infections and cancers. People with AIDS also suffer from other systemic symptoms, such as fevers, sweats, swollen glands, chills, weakness, and weight loss 213. Currently, there is no effective vaccine against HIV, and most of the treatment strategies rely on using antiretroviral drugs, which in most cases maintain the viral load at low levels to prevent the formation of acute HIV infection rather than cure the virus 214. Therefore, the accurate detection of HIV is a key determinant for monitoring HIV treatment and reducing its transmission and dissemination.
Currently, AuNPs have been extensively tested for the immunological and molecular detection of HIV through various scanometric, colorimetric, electrochemical, IPCR, and AFM assays (Table 1). The scanometric assays target the detection of the HIV p24 antigen and HIV nucleic acid using the silver metal enhancement protocol. For HIV p24, the target antigen is precaptured by magnetic particles 81 or isolated in a microplate 84, followed by a silver enhancement step for the quantitative visual immunological detection of HIV (Fig. 4A, B and Fig. 6). For HIV nucleic acid detection, AuNP-DNA probes specific to the HIV pol gene are applied to detect HIV nucleic acids precaptured and isolated by magnetic particles, followed by a silver metal enhancement step (Fig. 14A). The deposition of silver on the AuNP surface increases the NP size and scattering efficiency and additionally allows the target detection via DLS 90. In AuNP-based colorimetric detection of HIV, the large surface area of AuNPs was exploited to amplify the detection signal by enhancing the gold metal staining on their surfaces, allowing naked-eye detection of HIV nucleic acids (HIV gag gene) in a lateral-flow format, similar to the scheme presented in Fig. 11A 91. On the other hand, the ability of AuNPs to enhance electron transfer has been applied to develop an electrochemical assay for HIV detection (Fig. 14B). In this electrochemical assay, the target HIV genome is detected in correlation with the change in electrical impedance of graphene sheets decorated with spherical AuNP-DNA probes specific to the HIV pol gene 73. The developed AuNP-based RNA detection of HIV using the AuNP-LFA technique has a sensitivity limit of ~11 log10 copies/mL, while the best DNA detection can be achieved through the light-scattering scheme with a sensitivity limit of 10 fM. Furthermore, AFM coupled with AuNPs as nanoparticulate labels has been applied for the detection of HIV p24 as follows: the target antigen is precaptured on the surface of a nanoarray specifically coated with anti-p24 and labeled with AuNP-antibody conjugates that significantly changes the surface topography of the array, allowing the detection of p24 (Fig. 14C) 95.
Figure 14.
AuNP-based assays for human immunodeficiency virus (HIV) testing. (A) Colorimetric assay of HIV based on silver metal enhancement in solution. Magnetic NP (MNP)-DNA probes scavenge target HIV sequences, which are then magnetically collected and labeled with AuNP-DNA probes. After repetitive washing, AuNPs are released into solution by a dehybridization step and subjected to silver metal enhancement, resulting in particle growth and increased scattering 90. (B) Electrochemical impedance assay of HIV RNA. Graphene oxide sheets decorated with AuNPs are applied for the immobilization of DNA detection probes. Hybridization with specific DNA sequence results in increased impedance 73. (C) Atomic force microscopy (AFM)-based immunoassay of HIV. AuNP-antibody probes form a three-component sandwich immunocomplex with the target virus antigen captured on the surface of a specific antibody nanoarray; eventually, the increase in the surface height of the array is detected by AFM 95.
So far the developed AuNPs-based assays for HIV are among the most innovative in virus detection and offer a significantly improved detection limit down to attomolar range, which is hundreds of times lower than that of conventional techniques used in HIV detection, including PCR and ELISA. Therefore, AuNPs assays can be very beneficial in detecting HIV early, preventing its dissemination, and treatment monitoring. Furthermore, simple detection methods such as lateral flow and scanometric assays can be easily incorporated into the point-of-care detection of HIV, which is currently of great interest and more suitable for developing countries.
8. Contributions of AuNPs to virus detection
AuNPs have made a tremendous impact in the detection of viruses, as demonstrated by the numerous assays for most of the clinically relevant groups of human viruses (Fig. 3). Coupled with different biomolecules, AuNPs have enabled the development of a completely new class of probes that are specifically composed of a AuNP core (spherical or rod-like in shape) for signal readout and a surface layer of one or multiple types of biomolecules for target recognition and interaction. These probes have mostly been incorporated as a new set of functional and highly interactive components in different traditional techniques (e.g., EIAs and PCR) and have derived substantial improvements in design, detection performance, sensitivity, specificity and stability. This created a new trend in applying AuNPs to overcome the limitations of traditional techniques. AuNP-based immunological and serological detection methods are the most common and have been accomplished through multiple scanometric, colorimetric, DLS, immuno-PCR, electrochemical, electrical, SERS, LSPR, fluorometric, and AFM-based approaches (Table 1). AuNP-based DLS and LSPR assays are especially novel, simple and among the most sensitive reported assays for the immunodetection of viruses with a detection limit that is hundreds of times lower than that of the standard ELISA technique 51, 52. In contrast, the developed AuNP-based molecular assays are not as sensitive as the conventional PCR technique or other PCR-based molecular techniques. However, the addition of AuNPs greatly simplifies the detection procedures of viral nucleic acids, and largely eliminates the need for complex procedures and expensive thermal cycling machines, which greatly reduces the detection time and total cost. In particular, colorimetric assays based on the intense red color of AuNPs represent an interesting qualitative shift in nucleic acid detection due to the complete elimination of the PCR amplification step and the possibility of direct and rapid detection of the target virus within a few minutes. Other assays rely on well-known analytical techniques, such as ICP-MS and QCM, which have been introduced to virus nucleic acid detection for the first time through the application of AuNPs and have achieved detection sensitivity down to femtomolar concentrations 93, 94. Aside from these immunological and molecular assays, AuNPs have also been described for viral load testing by detecting intact virus particles, which are of special interest as a substitution to traditional cell culture technique 58, 65, 92. In this context, the small size of AuNPs probes that is comparable to virus particles, allows rapid, and highly dynamic interaction with viruses for gaining information about infectivity.
9. Technical challenges and recommendations
The application of AuNPs, especially in biology and medicine, require specific structural and physicochemical characteristics, and any subtle alterations to the intended characteristics can compromise the function and performance of the whole system. For bioanalytic and diagnostic applications, AuNPs exhibiting homogeneous morphological and fine structural characteristics are important specifically for effective and accurate response to target detection. Therefore, many pre-analytical factors related to NP synthesis and surface modification are critical to the development of an efficient virus detection assay.
The synthesis of AuNPs commonly relies on methods that basically employ wet chemistry reduction techniques, as presented in other reviews 9, 32. Although these techniques are generally simple and fast and can support the large-scale production of AuNPs, the synthesis of uniform AuNPs with a discrete size and shape remains challenging 215. Recently, some approaches have been reported for the controlled synthesis and modification of AuNPs. However, these methods remain unsuitable either due to the protocol complexity or difficulty of particle separation, which might require additional steps or specific surface modifications. Therefore, the development of more stringent and advanced methods for the facile and controlled synthesis of uniform AuNPs with the option of performing multiple surface modifications is strongly recommended.
Because of the direct relationship between AuNP size and shape and their function as labels, such characteristics are also of inherent importance to both the performance of Au nanoprobes and their efficiency for diagnostic applications. For instance, small AuNPs are preferred in colorimetric and light scattering-based assays due to their SPR band sensitivity. In contrast, large AuNPs are preferentially employed in assays such as BCA, immuno-PCR, and enzymatic-based electrochemical detection techniques, which rely on the surface functionalization of AuNPs with targeting or signaling biomolecules. Additionally, a large NP size is highly desirable in assays that depend on AuNPs as metal labels, such as electrochemical stripping, QCM, and ICP-MS detection methods. In addition, spherical AuNPs are more commonly applied than rod-shaped NPs. However, the latter exclusively enable a very interesting class of LSPR-based detection methods that have shown an unprecedented detection sensitivity range.
Since AuNPs by themselves are “blind” with respect to sensing the target, the addition of specific biomolecules to their surfaces is a key step for detecting viruses 37, 38, 40. Unfortunately, most biomolecules have been reported to be very sensitive to harsh chemical modifications that can adversely affect their reactivity and specificity. Thus, the type of targeting biomolecule and the surface conjugation reaction method applied are crucial and require optimization. Short oligonucleotides and other DNA sequences are physically rigid and more stable than protein biotargeting structures, including antibodies, which usually have several secondary and tertiary conformations that are important for their full, specific interaction with a target molecule. Furthermore, the full reactivity of surface biomolecules via a suitable conjugation reaction is critical and should be retained and confirmed with additional assessment steps before use in virus detection. While the application of traditional conjugation techniques, such as electrostatic adsorption, is still widely reported, it is strongly recommended to be avoided due to the incident loss of conjugated biomolecule activity and large inequality in loading efficiency, which directly result in decreased assay sensitivity and reproducibility 216. In this regard, AuNPs have an enhanced surface area-to-volume ratio that greatly improves the efficacy of current conjugation techniques, and several directional and controlled conjugation methods based on covalent and noncovalent interactions have been thoroughly described in the literature 9, 32, 35.
Although AuNPs are widely employed in various sensing functions and formats, their use for monitoring of virus infection is challenged by long-term stability and reusability of the developed AuNP-based sensors. The reusability of sensing platforms has a crucial role in developing applications that are reliable, economic and sustainable. Nonetheless, the reusability of AuNPs in virus detection is limited to electrical and electrochemical assays, in which the main function of AuNPs is limited to enhancing the performance of the modified electrodes 68, 70-73, 75-78. The long-term stability of AuNPs is reported and well investigated in numerous studies. However, their conjugates to different biomolecules (i.e., DNA, RNA, antibody, enzymes) are sensitive to storage time and conditions and the reusability of these conjugates is usually hampered by the loss of activity of surface molecules and irreversible aggregations. It was demonstrated that exposure to elevated temperatures, high salt concentrations, and reducing agents such as dithiothreitol (DTT), mercaptoethanol, and glutathione (commonly existing in biological fluids) results in the loss of stability and activity of AuNP-conjugates 9, 41, 43. Thus, the development of new chemistries and novel conjugation strategies for the synthesis of stable AuNP-conjugates becomes very crucial for practical use of AuNPs in virus detection.
Considering these limitations and challenges, important analytical parameters, such as the use of positive and negative controls, follow-up patient specimens, and internal reference tests, are recommended to confirm the overall accuracy and practicality of developed schemes. A restricted set of measures related to the assay detection sensitivity, specificity, and performance is required for the quality assurance and assessment of these applications 217. Due to the rapid development rate of AuNP-based assays, international data analysis measures and standards are also critical to reflect the true assay sensitivity and specificity and to allow the evaluation of their ability and potential to substitute current detection techniques. Furthermore, more efforts are needed to test the practicality and generalizability of these assays through clinical sample testing and in-field evaluation of the newly developed AuNP-based methods along with the conventional methods using standard evaluation systems and protocols.
10. Conclusion
AuNPs are a powerful addition to the range of techniques available for virus testing and control. When used as probes, AuNPs enable simple, rapid, and sensitive detection with an unprecedented multiplexing capability. Importantly, AuNP-based probes have matured from tools being added to improve traditional techniques, such as PCR, ELIZA, and many others, to the basis of completely novel techniques developed and introduced for the first time for virus detection. With AuNPs, virologists can create an unlimited number of detection assays that match the diagnostic needs of each virus in terms of simplicity, speed and cost with sensitivity levels that reach zeptomolar concentrations. AuNPs can be further developed to become components of more routine techniques, and they will likely engage the interest of scientists from all the disciplines, whether fundamental, environmental, clinical or industrial, in the future.
Acknowledgments
We would like to thank Frank Fanqing Chen, Athe Tsibris, and Anish Vasan. Research reported in this publication was partially supported by the National Institute of Health under award numbers R01AI118502, R21HD092828 and P30ES000002; Harvard T.H. Chan School of Public Health, Harvard Center for Environmental Health through Harvard NIEHS Grant; and American Board of Obstetrics and Gynecology, American College of Obstetricians and Gynecologists, American Society for Reproductive Medicine, Society for Reproductive Endocrinology and Infertility through ASRM Award, and Harvard University Center for AIDS Research (CFAR) under award number 5P30AI060354-14.
Abbreviations
- 5' UTR
five prime untranslated region
- AFM
atomic force microscopy
- AgNPs
silver nanoparticles
- ASV
anodic stripping voltammetry
- AuNPs
gold nanoparticles
- AuNRs
gold nanorods
- BCA
bio-barcode-amplification
- BEBOV
Bundibugyo ebolavirus
- BEC
boceprevir
- cDNA
complementary DNA
- CIA
chemiluminescent immunoassay
- CIEBOV
Côte d'Ivoire ebolavirus
- CoV
coronavirus
- CTAB
cetyltrimethyl ammonium bromide
- DENV
dengue virus
- DF
dengue fever
- DLS
dynamic light scattering
- DNA
deoxyribonucleic acid
- DPV
differential pulse voltammetry
- dsDNA
double stranded deoxyribonucleic acid
- DTT
dithiothreitol
- E gene
envelope gene
- EBOV
Ebola virus
- EBV
Epstein-Barr virus
- EHF
Ebola hemorrhagic fever
- EIA
enzyme immunoassay
- ELISA
enzyme-linked immunosorbent assay
- EM
electron microscopy
- FAM
fluorescein amidite
- FDA
Food and Drug Administration
- FRET
fluorescence resonance energy transfer
- gag gene
group-specific antigen gene
- GE
gold electrode
- HA gene
hemagglutinin gene
- HAI
hemagglutination-inhibition
- HAV
hepatitis A virus
- HBeAg
hepatitis B virus E antigen
- HBsAbs
hepatitis B virus surface antigen antibodies
- HBsAg
hepatitis B virus surface antigen
- HBV
hepatitis B virus
- HCMV
human cytomegalovirus
- HCV
hepatitis C virus
- HEV
hepatitis E virus
- HIV
human immunodeficiency virus
- HPV
human papilloma virus
- HRP
horse radish peroxidase
- HSV
herpes simplex virus
- HSV-1
herpes simplex virus type 1
- HSV-2
herpes simplex virus type 2
- HTNV
hantaan virus
- IAV
influenza A virus
- IBA
immunoblotting assay
- ICP-MS
inductively coupled plasma mass spectrometry
- ICT
immunochromatographic test
- IPA
immunoprecipitation assay
- IPCR
immuno-polymerase chain reaction
- KSHV
Kaposi's sarcoma-associated herpes virus
- L1 gene
late expressed gene1
- LAMP
loop-mediated isothermal amplification
- LFA
lateral flow assays
- LFIA
lateral-flow immunochromatographic assay
- LSPR
localized surface plasmon resonance
- M gene
matrix gene
- MERS
middle east respiratory syndrome
- MMPs
magnetic microparticles
- MNP
magnetic nanoparticles
- MWCNT
multiwall carbon nanotubes
- NA gene
neuraminidase gene
- NC protein
nucleocapsid protein
- NP gene
nucleoprotein gene
- NPs
nanoparticles
- ORF
open reading frame
- P24 antigen
P24 capsid protein antigen
- PCR
polymerase chain reaction
- PDA
photodiode array
- PEG-IFNα
pegylated interferon alfa
- pol gene
polymerase gene
- PP1ab
polyprotein1ab
- PPNW
polypyrrole nanowires
- QCM
quartz crystal microbalance
- QD
quantum dot
- RBV
ribavirin
- RIA
radioimmunoassay
- RNA
ribonucleic acid
- RT-PCR
reverse transcriptase-polymerase chain reaction
- RVFV
Rift Valley fever virus
- S gene
surface gene
- S-AP
streptavidin-alkaline phosphatase
- SARS
severe acute respiratory syndrome
- SEBOV
Sudan ebolavirus
- SERS
surface-enhanced Raman scattering
- SPA
Staphylococcal protein A
- SPE
screen-printed electrode
- SPR
surface plasmon resonance
- ssDNA
single-stranded DNA
- TMB
3: 3': 5: 5'-Tetramethylbenzidine
- TVR
telaprevir
- UL
unique long
- US
unique short
- USA
United States of America
- UV-Vis
ultraviolet-visible
- Vall7 gene
hepatitis A virus Vall7 polyprotein gene
- VP1 gene
viral protein1 gene
- VPg
noncapsid viral protein
- VV
variola virus
- VZV
varicella zoster virus
- WHO
World Health Organization
- WNV
West Nile virus
- ZEBOV
Zaire ebolavirus.
References
- 1.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–9. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellamy C. Globalization and infectious diseases in women. Emerg Infect Dis. 2004;10:2022–4. doi: 10.3201/eid1011.040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fidler DP. Globalization, international law, and emerging infectious diseases. Emerg Infect Dis. 1996;2:77–84. doi: 10.3201/eid0202.960201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frenk J, Gomez-Dantes O, Knaul FM. Globalization and infectious diseases. Infect Dis Clin North Am. 2011;25:593–9. doi: 10.1016/j.idc.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gushulak BD, MacPherson DW. Globalization of infectious diseases: the impact of migration. Clin Infect Dis. 2004;38:1742–8. doi: 10.1086/421268. [DOI] [PubMed] [Google Scholar]
- 6.Dykman L, Khlebtsov N. Gold nanoparticles in biomedical applications: recent advances and perspectives. Chem Soc Rev. 2012;41:2256–82. doi: 10.1039/c1cs15166e. [DOI] [PubMed] [Google Scholar]
- 7.Arvizo R, Bhattacharya R, Mukherjee P. Gold nanoparticles: opportunities and challenges in nanomedicine. Expert Opin Drug Deliv. 2010;7:753–63. doi: 10.1517/17425241003777010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao H, Nehl CL, Hafner JH. Biomedical applications of plasmon resonant metal nanoparticles. Nanomedicine (Lond) 2006;1:201–8. doi: 10.2217/17435889.1.2.201. [DOI] [PubMed] [Google Scholar]
- 9.Zeng S, Yong K-T, Roy I, Dinh X-Q, Yu X, Luan F. A Review on Functionalized Gold Nanoparticles for Biosensing Applications. Plasmonics. 2011;6:491–506. [Google Scholar]
- 10.Baaske M, Vollmer F. Optical resonator biosensors: molecular diagnostic and nanoparticle detection on an integrated platform. Chemphyschem. 2012;13:427–36. doi: 10.1002/cphc.201100757. [DOI] [PubMed] [Google Scholar]
- 11.Passi D, Sharma S, Dutta SR, Dudeja P, Sharma V. Ebola virus disease (the killer virus): another threat to humans and bioterrorism: brief review and recent updates. J Clin Diagn Res. 2015;9:LE01–8. doi: 10.7860/JCDR/2015/13062.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rojek A, Horby P, Dunning J. Insights from clinical research completed during the west Africa Ebola virus disease epidemic. Lancet Infect Dis. 2017;17:e280–e92. doi: 10.1016/S1473-3099(17)30234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shawky SM, Awad AM, Allam W, Alkordi MH, El-Khamisy SF. Gold aggregating gold: A novel nanoparticle biosensor approach for the direct quantification of hepatitis C virus RNA in clinical samples. Biosens Bioelectron. 2017;92:349–56. doi: 10.1016/j.bios.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H-q, Ji C-f, Yang X-q, Wang R, Yang S, Zhang H-q. et al. An improved gold nanoparticle probe-based assay for HCV core antigen ultrasensitive detection. J Virol Methods. 2017;243:142–5. doi: 10.1016/j.jviromet.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Alizadeh N, Hallaj R, Salimi A. A highly sensitive electrochemical immunosensor for hepatitis B virus surface antigen detection based on Hemin/G-quadruplex horseradish peroxidase-mimicking DNAzyme-signal amplification. Biosens Bioelectron. 2017;94:184–92. doi: 10.1016/j.bios.2017.02.039. [DOI] [PubMed] [Google Scholar]
- 16.Kim DS, Kim YT, Hong SB, Kim J, Huh NS, Lee M-K. et al. Development of Lateral Flow Assay Based on Size-Controlled Gold Nanoparticles for Detection of Hepatitis B Surface Antigen. Sensors. 2016;16:2154. doi: 10.3390/s16122154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zengin A, Tamer U, Caykara T. SERS detection of hepatitis B virus DNA in a temperature-responsive sandwich-hybridization assay. J Raman Spectrosc. 2017;48:668–72. [Google Scholar]
- 18.Parrish CR, Holmes EC, Morens DM, Park E-C, Burke DS, Calisher CH. et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72:457–70. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figlerowicz M, Alejska M, Kurzynska-Kokorniak A. Genetic variability: The key problem in the prevention and therapy of RNA-based virus infections. Med Res Rev. 2003;23:488–518. doi: 10.1002/med.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morse SS. Factors in the emergence of infectious-diseases. Emerg Infect Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson N, Mueller J. Updating the accounts: global mortality of the 1918-1920 "Spanish" influenza pandemic. Bull Hist Med. 2002;76:105–15. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed SR, Kim J, Van Tan Tran TS, Neethirajan S, Lee J, Park EY. In situ self-assembly of gold nanoparticles on hydrophilic and hydrophobic substrates for influenza virus-sensing platform. Sci Rep. 2017;7:44495. doi: 10.1038/srep44495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–67. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuaib W, Stanazai H, Abazid AG, Mattar AA. Re-emergence of Zika virus: a review on pathogenesis, clinical manifestations, diagnosis, treatment, and prevention. Am J Med. 2016;129:879.. doi: 10.1016/j.amjmed.2016.02.027. e7-e12. [DOI] [PubMed] [Google Scholar]
- 25.Stephenson JR, Warnes A. Diagnostic virology protocols. New Jersey, USA: Humana Press; 1998. [Google Scholar]
- 26.Kudesia G, Wreghitt TG. Clinical and diagnostic virology. Cambridge, UK: Cambridge University Press; 2009. [Google Scholar]
- 27.Kumvongpin R, Jearanaikool P, Wilailuckana C, Sae-ung N, Prasongdee P, Daduang S. et al. High sensitivity, loop-mediated isothermal amplification combined with colorimetric gold-nanoparticle probes for visual detection of high risk human papillomavirus genotypes 16 and 18. J Virol Methods. 2016;234:90–5. doi: 10.1016/j.jviromet.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimarães KP. et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–71. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis JS, Zambon MC. Molecular diagnosis of influenza. Rev Med Virol. 2002;12:375–89. doi: 10.1002/rmv.370. [DOI] [PubMed] [Google Scholar]
- 30.Storch GA. Diagnostic virology. Clin Infect Dis. 2000;31:739–51. doi: 10.1086/314015. [DOI] [PubMed] [Google Scholar]
- 31.Greer S, Alexander GJM. Viral serology and detection. Baillieres Clin Gastroenterol. 1995;9:689–721. doi: 10.1016/0950-3528(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 32.Alex S, Tiwari A. Functionalized gold nanoparticles: synthesis, properties and applications—a review. J Nanosci Nanotechnol. 2015;15:1869–94. doi: 10.1166/jnn.2015.9718. [DOI] [PubMed] [Google Scholar]
- 33.Read SJ, Burnett D, Fink CG. Molecular techniques for clinical diagnostic virology. J Clin Pathol. 2000;53:502–6. doi: 10.1136/jcp.53.7.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratcliff RM, Chang G, Kok T, Sloots TP. Molecular diagnosis of medical viruses. Curr Issues Mol Biol. 2007;9:87–102. [PubMed] [Google Scholar]
- 35.Draz MS, Fang BA, Zhang P, Hu Z, Gu S, Weng KC. et al. Nanoparticle-mediated systemic delivery of siRNA for treatment of cancers and viral infections. Theranostics. 2014;4:872–92. doi: 10.7150/thno.9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feynman RP. There's plenty of room at the bottom. Engineering and Science. 1960;23:22–36. [Google Scholar]
- 37.Wang X, Liu L-H, Ramstroem O, Yan M. Engineering Nanomaterial Surfaces for Biomedical Applications. Exp Biol Med. 2009;234:1128–39. doi: 10.3181/0904-MR-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirkin CA, Rosi NL. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–62. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 39.Zehbe I, Hacker GW, Su H, Hauser-Kronberger C, Hainfeld JF, Tubbs R. Sensitive in situ hybridization with catalyzed reporter deposition, streptavidin-Nanogold, and silver acetate autometallography: detection of single-copy human papillomavirus. Am J Pathol. 1997;150:1553–61. [PMC free article] [PubMed] [Google Scholar]
- 40.Alivisatos P. The use of nanocrystals in biological detection. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 41.Dougan JA, Karlsson C, Smith WE, Graham D. Enhanced oligonucleotide-nanoparticle conjugate stability using thioctic acid modified oligonucleotides. Nucleic Acids Res. 2007;35:3668–75. doi: 10.1093/nar/gkm237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boisselier E, Astruc D. Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chem Soc Rev. 2009;38:1759–82. doi: 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]
- 43.Jazayeri MH, Amani H, Pourfatollah AA, Pazoki-Toroudi H, Sedighimoghaddam B. Various methods of gold nanoparticles (GNPs) conjugation to antibodies. Sens Biosensing Res. 2016;9:17–22. [Google Scholar]
- 44.Lu XM, Rycenga M, Skrabalak SE, Wiley B, Xia YN. Chemical Synthesis of Novel Plasmonic Nanoparticles. Annu Rev Phys Chem. 2009;60:167–92. doi: 10.1146/annurev.physchem.040808.090434. [DOI] [PubMed] [Google Scholar]
- 45.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nat Biotechnol. 2005;23:741–5. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 46.Lee KS, El-Sayed MA. Gold and silver nanoparticles in sensing and imaging: Sensitivity of plasmon response to size, shape, and metal composition. J Phys Chem B. 2006;110:19220–5. doi: 10.1021/jp062536y. [DOI] [PubMed] [Google Scholar]
- 47.Lin C-AJ, Lee C-H, Hsieh J-T, Wang H-H, Li JK, Shen J-L. et al. Review: Synthesis of fluorescent metallic nanoclusters toward biomedical application: recent progress and present challenges. J Med Biol Eng. 2009;29:276–83. [Google Scholar]
- 48.Baptista P, Pereira E, Eaton P, Doria G, Miranda A, Gomes I. et al. Gold nanoparticles for the development of clinical diagnosis methods. Anal Bioanal Chem. 2008;391:943–50. doi: 10.1007/s00216-007-1768-z. [DOI] [PubMed] [Google Scholar]
- 49.Neng J, Harpster MH, Wilson WC, Johnson PA. Surface-enhanced Raman scattering (SERS) detection of multiple viral antigens using magnetic capture of SERS-active nanoparticles. Biosens Bioelectron. 2013;41:316–21. doi: 10.1016/j.bios.2012.08.048. [DOI] [PubMed] [Google Scholar]
- 50.Cao YWC, Jin RC, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–40. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Li Y, Quan D, Wang J, Zhang Y, Du J. et al. Detection of hepatitis B surface antigen by target-induced aggregation monitored by dynamic light scattering. Anal Biochem. 2012;428:119–25. doi: 10.1016/j.ab.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Li Y, Wang H, Fu Q, Peng J, Wang Y. et al. Gold nanorod-based localized surface plasmon resonance biosensor for sensitive detection of hepatitis B virus in buffer, blood serum and plasma. Biosens Bioelectron. 2010;26:404–10. doi: 10.1016/j.bios.2010.07.121. [DOI] [PubMed] [Google Scholar]
- 53.Driskell JD, Jones CA, Tompkins SM, Tripp RA. One-step assay for detecting influenza virus using dynamic light scattering and gold nanoparticles. Analyst. 2011;136:3083–90. doi: 10.1039/c1an15303j. [DOI] [PubMed] [Google Scholar]
- 54.Li HX, Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Natl Acad Sci U S A. 2004;101:14036–9. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glynou K, Ioannou PC, Christopoulos TK, Syriopoulou V. Oligonucleotide-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for DNA analysis by hybridization. Anal Chem. 2003;75:4155–60. doi: 10.1021/ac034256+. [DOI] [PubMed] [Google Scholar]
- 56.Shawky SM, Bald D, Azzazy HME. Direct detection of unamplified hepatitis C virus RNA using unmodified gold nanoparticles. Clin Biochem. 2010;43:1163–8. doi: 10.1016/j.clinbiochem.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 57.Mancuso M, Jiang L, Cesarman E, Erickson D. Multiplexed colorimetric detection of Kaposi's sarcoma associated herpesvirus and Bartonella DNA using gold and silver nanoparticles. Nanoscale. 2013;5:1678–86. doi: 10.1039/c3nr33492a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Lu D, Sheng Z, Chen K, Guo X, Jin M. et al. A fast and sensitive immunoassay of avian influenza virus based on label-free quantum dot probe and lateral flow test strip. Talanta. 2012;100:1–6. doi: 10.1016/j.talanta.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 59.Laderman EI, Whitworth E, Dumaual E, Jones M, Hudak A, Hogrefe W. et al. Rapid, sensitive, and specific lateral-flow immunochromatographic point-of-care device for detection of herpes simplex virus type 2-specific immunoglobulin G antibodies in serum and whole blood. Clin Vaccine Immunol. 2008;15:159–63. doi: 10.1128/CVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffin J, Singh AK, Senapati D, Rhodes P, Mitchell K, Robinson B. et al. Size- and Distance-Dependent Nanoparticle Surface-Energy Transfer (NSET) Method for Selective Sensing of Hepatitis C Virus RNA. Chem Eur J. 2009;15:342–51. doi: 10.1002/chem.200801812. [DOI] [PubMed] [Google Scholar]
- 61.Lu X, Dong X, Zhang K, Han X, Fang X, Zhang Y. A gold nanorods-based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst. 2013;138:642–50. doi: 10.1039/c2an36099c. [DOI] [PubMed] [Google Scholar]
- 62.Zeng Q, Zhang Y, Liu X, Tu L, Kong X, Zhang H. Multiple homogeneous immunoassays based on a quantum dots-gold nanorods FRET nanoplatform. Chem Commun. 2012;48:1781–3. doi: 10.1039/c2cc16271g. [DOI] [PubMed] [Google Scholar]
- 63.Chang Y-F, Wang S-F, Huang JC, Su L-C, Yao L, Li Y-C. et al. Detection of swine-origin influenza A (H1N1) viruses using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens Bioelectron. 2010;26:1068–73. doi: 10.1016/j.bios.2010.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ganbold E-O, Kang T, Lee K, Lee SY, Joo S-W. Aggregation effects of gold nanoparticles for single-base mismatch detection in influenza A (H1N1) DNA sequences using fluorescence and Raman measurements. Colloids Surf B Biointerfaces. 2012;93:148–53. doi: 10.1016/j.colsurfb.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 65.Draz MS, Fang BA, Li L, Chen Z, Wang Y, Xu Y. et al. Hybrid nanocluster plasmonic resonator for immunological detection of hepatitis B virus. ACS Nano. 2012;6:7634–43. doi: 10.1021/nn3034056. [DOI] [PubMed] [Google Scholar]
- 66.Wu JC, Chen CH, Fu JW, Yang HC. Electrophoresis-enhanced detection of deoxyribonucleic acids on a membrane-based lateral flow strip using avian influenza H5 genetic sequence as the model. Sensors (Basel) 2014;14:4399–415. doi: 10.3390/s140304399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nikbakht H, Tabarraei A, Niazi A. Nanomolecular detection of human influenza virus type A using reverse transcription loop-mediated isothermal amplification assisted with rod-shaped gold nanoparticles. RSC Advances. 2014;4:13575–80. [Google Scholar]
- 68.Authier L, Grossiord C, Brossier P, Limoges B. Gold nanoparticle-based quantitative electrochemical detection of amplified human cytomegalovirus DNA using disposable microband electrodes. Anal Chem. 2001;73:4450–6. doi: 10.1021/ac0103221. [DOI] [PubMed] [Google Scholar]
- 69.Chang T-L, Tsai C-Y, Sun C-C, Chen C-C, Kuo L-S, Chen P-H. Ultrasensitive electrical detection of protein using nanogap electrodes and nanoparticle-based DNA amplification. Biosens Bioelectron. 2007;22:3139–45. doi: 10.1016/j.bios.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 70.Bonanni A, Pividori MI, del Valle M. Impedimetric detection of influenza A (H1N1) DNA sequence using carbon nanotubes platform and gold nanoparticles amplification. Analyst. 2010;135:1765–72. doi: 10.1039/c000532k. [DOI] [PubMed] [Google Scholar]
- 71.Liu X, Cheng Z, Fan H, Ai S, Han R. Electrochemical detection of avian influenza virus H5N1 gene sequence using a DNA aptamer immobilized onto a hybrid nanomaterial-modified electrode. Electrochim Acta. 2011;56:6266–70. [Google Scholar]
- 72.Martinez-Paredes G, Gonzalez-Garcia MB, Costa-Garcia A. Genosensor for SARS Virus Detection Based on Gold Nanostructured Screen-Printed Carbon Electrodes. Electroanalysis. 2009;21:379–85. [Google Scholar]
- 73.Hu Y, Hua S, Li F, Jiang Y, Bai X, Li D. et al. Green-synthesized gold nanoparticles decorated graphene sheets for label-free electrochemical impedance DNA hybridization biosensing. Biosens Bioelectron. 2011;26:4355–61. doi: 10.1016/j.bios.2011.04.037. [DOI] [PubMed] [Google Scholar]
- 74.Hanaee H, Ghourchian H, Ziaee A. Nanoparticle-based electrochemical detection of hepatitis B virus using stripping chronopotentiometry. Anal Biochem. 2007;370:195–200. doi: 10.1016/j.ab.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 75.Shen G, Zhang Y. Highly sensitive electrochemical stripping detection of hepatitis B surface antigen based on copper-enhanced gold nanoparticle tags and magnetic nanoparticles. Anal Chim Acta. 2010;674:27–31. doi: 10.1016/j.aca.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 76.Wu S, Zhong Z, Wang D, Li M, Qing Y, Dai N. et al. Gold nanoparticle-labeled detection antibodies for use in an enhanced electrochemical immunoassay of hepatitis B surface antigen in human serum. Mikrochim Acta. 2009;166:269–75. [Google Scholar]
- 77.Ding C, Li H, Hu K, Lin J-M. Electrochemical immunoassay of hepatitis B surface antigen by the amplification of gold nanoparticles based on the nanoporous gold electrode. Talanta. 2010;80:1385–91. doi: 10.1016/j.talanta.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 78.Li W, Wu P, Zhang H, Cai C. Catalytic signal amplification of gold nanoparticles combining with conformation-switched hairpin DNA probe for hepatitis C virus quantification. Chem Commun. 2012;48:7877–9. doi: 10.1039/c2cc33635a. [DOI] [PubMed] [Google Scholar]
- 79.Wan Z, Wang Y, Li SS-c, Duan L, Zhai J. Development of array-based technology for detection of HAV using gold-DNA probes. BMB Reports. 2005;38:399–406. doi: 10.5483/bmbrep.2005.38.4.399. [DOI] [PubMed] [Google Scholar]
- 80.Wang YF, Pang DW, Zhang ZL, Zheng HZ, Cao JP, Shen JT. Visual gene diagnosis of HBV and HCV based on nanoparticle probe amplification and silver staining enhancement. J Med Virol. 2003;70:205–11. doi: 10.1002/jmv.10379. [DOI] [PubMed] [Google Scholar]
- 81.Kim E-Y, Stanton J, Korber BTM, Krebs K, Bogdan D, Kunstman K. et al. Detection of HIV-1 p24 Gag in plasma by a nanoparticle-based bio-barcode-amplification method. Nanomedicine. 2008;3:293–303. doi: 10.2217/17435889.3.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu HH, Cao X, Yang Y, Liu MG, Wang YF. Array-based nano-amplification technique was applied in detection of hepatitis E virus. J Biochem Mol Biol. 2006;39:247–52. doi: 10.5483/bmbrep.2006.39.3.247. [DOI] [PubMed] [Google Scholar]
- 83.Cho H, Jung J, Chung BH. Scanometric analysis of DNA microarrays using DNA intercalator-conjugated gold nanoparticles. Chem Commun. 2012;48:7601–3. doi: 10.1039/c2cc32869k. [DOI] [PubMed] [Google Scholar]
- 84.Tang S, Hewlett I. Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J Infect Dis. 2010;201:S59–64. doi: 10.1086/650386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y, Mao H-J, Zang G-Q, Zhang H-L, Jin Q-H, Zhao J-L. Detection of Hepatitis B Virus Deoxyribonucleic Acid Based on Gold Nanoparticle Probe Chip. Chinese J Anal Chem. 2010;38:1133–8. [Google Scholar]
- 86.Zhao J, Tang S, Storhoff J, Marla S, Bao YP, Wang X. et al. Multiplexed, rapid detection of H5N1 using a PCR-free nanoparticle-based genomic microarray assay. BMC Biotechnol. 2010;10:74. doi: 10.1186/1472-6750-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim SK, Cho H, Jeong J, Kwon JN, Jung Y, Chung BH. Label-free and naked eye detection of PNA/DNA hybridization using enhancement of gold nanoparticles. Chem Commun. 2010;46:3315–7. doi: 10.1039/b926940a. [DOI] [PubMed] [Google Scholar]
- 88.Duan LL, Wang YF, Li SSC, Wan ZX, Zhai JX. Rapid and simultaneous detection of human hepatitis B virus and hepatitis C virus antibodies based on a protein chip assay using nano-gold immunological amplification and silver staining method. BMC Infect Dis. 2005;5:53. doi: 10.1186/1471-2334-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baek TJ, Park PY, Han KN, Kwon HT, Seong GH. Development of a photodiode array biochip using a bipolar semiconductor and its application to detection of human papilloma virus. Anal Bioanal Chem. 2008;390:1373–8. doi: 10.1007/s00216-007-1814-x. [DOI] [PubMed] [Google Scholar]
- 90.Xu X, Georganopoulou DG, Hill HD, Mirkin CA. Homogeneous detection of nucleic acids based upon the light scattering properties of silver-coated nanoparticle probes. Anal Chem. 2007;79:6650–4. doi: 10.1021/ac070867g. [DOI] [PubMed] [Google Scholar]
- 91.Rohrman BA, Leautaud V, Molyneux E, Richards-Kortum RR. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PLoS One. 2012;7:e45611. doi: 10.1371/journal.pone.0045611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mu B, Huang X, Bu P, Zhuang J, Cheng Z, Feng J. et al. Influenza virus detection with pentabody-activated nanoparticles. J Virol Methods. 2010;169:282–9. doi: 10.1016/j.jviromet.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 93.Chen S-H, Chuang Y-C, Lu Y-C, Lin H-C, Yang Y-L, Lin C-S. A method of layer-by-layer gold nanoparticle hybridization in a quartz crystal microbalance DNA sensing system used to detect dengue virus. Nanotechnology. 2009;20:215501. doi: 10.1088/0957-4484/20/21/215501. [DOI] [PubMed] [Google Scholar]
- 94.Hsu IH, Chen W-H, Wu T-K, Sun Y-C. Gold nanoparticle-based inductively coupled plasma mass spectrometry amplification and magnetic separation for the sensitive detection of a virus-specific RNA sequence. J Chromatogr A. 2011;1218:1795–801. doi: 10.1016/j.chroma.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 95.Lee KB, Kim EY, Mirkin CA, Wolinsky SM. The use of nanoarrays for highly sensitive and selective detection of human immunodeficiency virus type 1 in plasma. Nano Lett. 2004;4:1869–72. [Google Scholar]
- 96.Chen L, Wei H, Guo Y, Cui Z, Zhang Z, Zhang X-E. Gold nanoparticle enhanced immuno-PCR for ultrasensitive detection of Hantaan virus nucleocapsid protein. J Immunol Methods. 2009;346:64–70. doi: 10.1016/j.jim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 97.Dong H, Liu J, Zhu H, Ou C-Y, Xing W, Qiu M. et al. Two types of nanoparticle-based bio-barcode amplification assays to detect HIV-1 p24 antigen. Virol J. 2012;9:180. doi: 10.1186/1743-422X-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang H, Liu L, Li CW, Fu H, Chen Y, Yang M. Multienzyme-nanoparticles amplification for sensitive virus genotyping in microfluidic microbeads array using Au nanoparticle probes and quantum dots as labels. Biosens Bioelectron. 2011;29:89–96. doi: 10.1016/j.bios.2011.07.074. [DOI] [PubMed] [Google Scholar]
- 99.Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist Å, et al. Bunyaviridae. In: King AMQ, Lefkowitz EJ, Adams MJ, Carstens EB, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. San Diego: Elsevier; 2012. [Google Scholar]
- 100.Fields BN, Knipe DM, Howley PM. Fields virology. 5th ed. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 101.Elliott RM, Weber F. Bunyaviruses and the type I interferon system. Viruses. 2009;1:1003–21. doi: 10.3390/v1031003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem. Emerg Infect Dis. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hart CA, Bennett M. Hantavirus infections: epidemiology and pathogenesis. Microbes Infect. 1999;1:1229–37. doi: 10.1016/s1286-4579(99)00238-5. [DOI] [PubMed] [Google Scholar]
- 104.Bi Z, Formenty PB, Roth CE. Hantavirus infection: a review and global update. J Infect Dev Ctries. 2008;2:3–23. doi: 10.3855/jidc.317. [DOI] [PubMed] [Google Scholar]
- 105.Elliott L, Ksiazek T, Rollin P, Spiropoulou C, Morzunov S, Monroe M. et al. Isolation of the causative agent of hantavirus pulmonary syndrome. Am J Trop Med Hyg. 1994;51:102–8. doi: 10.4269/ajtmh.1994.51.102. [DOI] [PubMed] [Google Scholar]
- 106.Bird BH, Ksiazek TG, Nichol ST, Maclachlan NJ. Rift Valley fever virus. J Am Vet Med Assoc. 2009;234:883–93. doi: 10.2460/javma.234.7.883. [DOI] [PubMed] [Google Scholar]
- 107.Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. Rift Valley fever virus(Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res. 2010;41:61. doi: 10.1051/vetres/2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Archer BN, Thomas J, Weyer J, Cengimbo A, Landoh DE, Jacobs C. et al. Epidemiologic investigations into outbreaks of Rift Valley fever in humans, South Africa, 2008-2011. Emerg Infect Dis. 2013;19:1918–25. doi: 10.3201/eid1912.121527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Archer BN, Weyer J, Paweska J, Nkosi D, Leman P, Tint KS. et al. Outbreak of Rift Valley fever affecting veterinarians and farmers in South Africa, 2008. S Afr Med J. 2011;101:263–6. doi: 10.7196/samj.4544. [DOI] [PubMed] [Google Scholar]
- 110.Rich KM, Wanyoike F. An assessment of the regional and national socio-economic impacts of the 2007 Rift Valley fever outbreak in Kenya. Am J Trop Med Hyg. 2010;83:52–7. doi: 10.4269/ajtmh.2010.09-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dar O, McIntyre S, Hogarth S, Heymann D. Rift Valley fever and a new paradigm of research and development for zoonotic disease control. Emerg Infect Dis. 2013;19:189–93. doi: 10.3201/eid1902.120941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Groot R, Baker S, Baric R, Enjuanes L, Gorbalenya A, Holmes K, Family Coronaviridae. Elsevier Academic Press: Amsterdam, The Netherlands; 2012. p. 806-28. [Google Scholar]
- 113.Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P. et al. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron ciyomicroscopy. J Virol. 2006;80:7918–28. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–50. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635–64. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fouchier RAM, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH. et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101:6212–6. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJM, Wolthers KC. et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Woo PCY, Lau SKP, Chu CM, Chan KH, Tsoi HW, Huang Y. et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–95. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zlateva KT, Coenjaerts FEJ, Crusio KM, Lammens C, Leus F, Viveen M. et al. No novel coronaviruses identified in a large collection of human nasopharyngeal specimens using family-wide CODEHOP-based primers. Arch Virol. 2013;158:251–5. doi: 10.1007/s00705-012-1487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 121.Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Olival KJ, Hayman DTS. Filoviruses in Bats: Current Knowledge and Future Directions. Viruses-Basel. 2014;6:1759–88. doi: 10.3390/v6041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanchez AGT, Feldmann H. Filoviridae: Marburg and Ebola Viruses. In: Knipe DM HP, editor. Fields virology. Pennsylvania, USA: Lippincott Williams and Wilkins; 2007. p. 1409-48. [Google Scholar]
- 124.Li Y, Chen S. Evolutionary history of Ebola virus. Epidemiol Infect. 2014;142:1138–45. doi: 10.1017/S0950268813002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Muyembe-Tamfum JJ, Mulangu S, Masumu J, Kayembe JM, Kemp A, Paweska JT. Ebola virus outbreaks in Africa: Past and present. Onderstepoort J Vet Res; 2012. p. 79. [DOI] [PubMed] [Google Scholar]
- 126.Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. The Lancet; 2015. [DOI] [PubMed] [Google Scholar]
- 127.Simmonds P BP, Collett MS, Gould EA, Heinz FX, Meyers G, Monath T, Pletnev A, Rice CM, Stiasny K, Thiel HJ, Weiner A, Bukh J. Family Flaviviridae. In: King A LE, Adams MJ, Carstens EB, editors. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. San Diego, CA; 2012. p. 1003-20. [Google Scholar]
- 128.Murray CL, Jones CT, Rice CM. Opinion - Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat Rev Microbiol. 2008;6:699–708. doi: 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kuno G, Chang G-JJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Leyssen P, De Clercq E, Neyts J. Perspectives for the Treatment of Infections with Flaviviridae. Clin Microbiol Rev. 2000;13:67–82. doi: 10.1128/cmr.13.1.67-82.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Idrees S, Ashfaq UA. A brief review on dengue molecular virology, diagnosis, treatment and prevalence in Pakistan. Genet Vaccines Ther. 2012;10:6. doi: 10.1186/1479-0556-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.World Health Organization. Dengue guidelines for diagnosis, treatment, prevention and control: new edition. 2009. [PubMed]
- 133.Simmons CP, Farrar JJ, van Vinh Chau N, Wills B. Dengue. N Engl J Med. 2012;366:1423–32. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 134.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL. et al. The global distribution and burden of dengue. Nature. 2013;496:504–7. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim WR. The burden of hepatitis C in the United States. Hepatology. 2002;36:S30–4. doi: 10.1053/jhep.2002.36791. [DOI] [PubMed] [Google Scholar]
- 136.Deuffic-Burban S, Poynard T, Sulkowski MS, Wong JB. Estimating the future health burden of chronic hepatitis C and human immunodeficiency virus infections in the United States. J Viral Hepat. 2007;14:107–15. doi: 10.1111/j.1365-2893.2006.00785.x. [DOI] [PubMed] [Google Scholar]
- 137.Shuhart MC, Bronner MP, Gretch DR, Thomassen LV, Wartelle CF, Tateyama H. et al. Histological and clinical outcome after liver transplantation for hepatitis C. Hepatology. 1997;26:1646–52. doi: 10.1002/hep.510260638. [DOI] [PubMed] [Google Scholar]
- 138.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales Jr FL. et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 139.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH. et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405–16. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 140.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC. et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368:1878–87. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 141.Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH. et al. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med. 2014;370:1993–2001. doi: 10.1056/NEJMoa1316145. [DOI] [PubMed] [Google Scholar]
- 142.Jacobson IM, Dore GJ, Foster GR, Fried MW, Radu M, Rafalsky VV. et al. Simeprevir with pegylated interferon alfa 2a plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-1): a phase 3, randomised, double-blind, placebo-controlled trial. The Lancet. 2014;384:403–13. doi: 10.1016/S0140-6736(14)60494-3. [DOI] [PubMed] [Google Scholar]
- 143.Ghany MG, Gara N. Quest for a cure for hepatitis C virus: the end is in sight. The Lancet. 2014;384:381–3. doi: 10.1016/S0140-6736(14)60807-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mason WS GW, Taylor JM, Kann M, Mizokami T, et al. Family Hepadnaviridae. In: King AMQ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses (Ninth Report of the International Committee on Taxonomy of Viruses) Amsterdam; 2011. pp. 445–55. [Google Scholar]
- 145.Ganem D, Prince AM. Hepatitis B virus infection-natural history and clinical consequences. N Engl J Med. 2004;350:1118–29. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 146.Pan CQ, Zhang JX. Natural History and Clinical Consequences of Hepatitis B Virus Infection. Int J Med Sci. 2005;2:36–40. doi: 10.7150/ijms.2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.World Health Organization. Hepatitis B fact sheet No. 204, 2000. Geneva, Switzerland: World Health Organization; 2007. [Google Scholar]
- 148.Thomas HC. Best practice in the treatment of chronic hepatitis B: a summary of the European Viral Hepatitis Educational Initiative (EVHEI) J Hepatol. 2007;47:588–97. doi: 10.1016/j.jhep.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 149.Sarri G, Westby M, Bermingham S, Hill-Cawthorne G, Thomas H. Diagnosis and management of chronic hepatitis B in children, young people, and adults: summary of NICE guidance. BMJ; 2013. p. 346. [DOI] [PubMed] [Google Scholar]
- 150.Meng XJ, Anderson D, Arankalle VA, Emerson SU, Harrison TJ, Jameel S, In: King AMQ, Carstens EB, Lefkowitz EJ, ed. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. London; 2012. pp. 1021–8. [Google Scholar]
- 151.Harrison TJ. Hepatitis E virus - an update. Liver. 1999;19:171–6. doi: 10.1111/j.1478-3231.1999.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 152.Liu HH CX, Yang Y, Liu MG, Wang YF. Array-based nano-amplification technique was applied in detection of hepatitis E virus. J Biochem Mol Biol. 2006;39:247–52. doi: 10.5483/bmbrep.2006.39.3.247. [DOI] [PubMed] [Google Scholar]
- 153.Pellett PE, Eberle R, Ehlers B, Hayward GS, Lacoste V, Minson AC, et al. Family Herpesviridae. In: King AMQ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. California, USA; 2012. pp. 111–22. [Google Scholar]
- 154.Spear PG, Longnecker R. Herpesvirus entry: an update. J Virol. 2003;77:10179–85. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol. 2011;9:382–94. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 156.Gupta R, Warren T, Wald A. Genital herpes. The Lancet. 2008;370:2127–37. doi: 10.1016/S0140-6736(07)61908-4. [DOI] [PubMed] [Google Scholar]
- 157.Roizman B, Whitley RJ. The nine ages of herpes simplex virus. Herpes. 2001;8:23–7. [PubMed] [Google Scholar]
- 158.Looker KJ, Garnett GP, Schmid GP. An estimate of the global prevalence and incidence of herpes simplex virus type 2 infection. Bull World Health Organ. 2008;86:805–12. doi: 10.2471/BLT.07.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bernstein DI, Bellamy AR, Hook EW, Levin MJ, Wald A, Ewell MG. et al. Epidemiology, clinical presentation, and antibody response to primary infection with herpes simplex virus type 1 and type 2 in young women. Clin Infect Dis. 2013;56:344–51. doi: 10.1093/cid/cis891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grinde B. Herpesviruses: latency and reactivation-viral strategies and host response. J Oral Microbiol; 2013. p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Suazo PA, Tognarelli EI, Kalergis AM, González PA. Herpes simplex virus 2 infection: molecular association with HIV and novel microbicides to prevent disease. Med Microbiol Immunol. 2015;204:161–76. doi: 10.1007/s00430-014-0358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhu X-P, Muhammad ZS, Wang J-G, Lin W, Guo S-K, Zhang W. HSV-2 vaccine: current status and insight into factors for developing an efficient vaccine. Viruses. 2014;6:371–90. doi: 10.3390/v6020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Friedman HM. Immune Evasion by Herpes Simplex Virus. 43rd Annual Meeting: Idsa; 2005. [Google Scholar]
- 164.Suazo PA, Ibañez FJ, Retamal-Díaz AR, Paz-Fiblas MV, Bueno SM, Kalergis AM, Evasion of Early Antiviral Responses by Herpes Simplex Viruses. Mediators Inflamm. 2015; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010;20:202–13. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 166.Boeckh M, Geballe AP. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest. 2011;121:1673. doi: 10.1172/JCI45449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kotton CN, Kumar D, Caliendo AM, Åsberg A, Chou S, Snydman DR. et al. International consensus guidelines on the management of cytomegalovirus in solid organ transplantation. Transplantation. 2010;89:779–95. doi: 10.1097/TP.0b013e3181cee42f. [DOI] [PubMed] [Google Scholar]
- 168.Lanari M, Lazzarotto T, Venturi V, Papa I, Gabrielli L, Guerra B. et al. Neonatal cytomegalovirus blood load and risk of sequelae in symptomatic and asymptomatic congenitally infected newborns. Pediatrics. 2006;117:e76–e83. doi: 10.1542/peds.2005-0629. [DOI] [PubMed] [Google Scholar]
- 169.Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pass RF, Zhang C, Evans A, Simpson T, Andrews W, Huang M-L. et al. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med. 2009;360:1191–9. doi: 10.1056/NEJMoa0804749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Plotkin S, Orenstein W, Offit P. Vaccines. 6th ed. Edinburgh, UK: Elsevier; 2013. [Google Scholar]
- 172.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM. et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–9. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 173.Ganem D. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol Mech Dis. 2006;1:273–96. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- 174.Moore P CY. Kaposi's sarcoma-associated herpesvirus. In: Knipe DM, Griffin DE, Lamb RA, Martin MA, Straus SE, editors. Field's virology. 4th ed. Pennsylvania, USA: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 175.Moore PS, Kingsley LA, Holmberg SD, Spira T, Gupta P, Hoover DR. et al. Kaposi's sarcoma-associated herpesvirus infection prior to onset of Kaposi's sarcoma. AIDS. 1996;10:175–80. doi: 10.1097/00002030-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 176.Whitby D, Boshoff C, Hatzioannou T, Weiss R, Schulz T, Howard M. et al. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. The lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 177.McCauly HS, Kaverin NV, Kochs G, Lamb RA, Matrosovich MN, Perez DR, et al. Family Orthomyxoviridae. In: King AMQ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. California, USA: Elsevier Academic Press; 2012. pp. 193–210. [Google Scholar]
- 178.Kaye D, Pringle CR. Avian influenza viruses and their implication for human health. Clin Infect Dis. 2005;40:108–12. doi: 10.1086/427236. [DOI] [PubMed] [Google Scholar]
- 179.Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 180.Lamb RA. Orthomyxoviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields virology. 4th ed. Pennsylvania, USA; London, UK: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 181.Harper SA, Bradley JS, Englund JA, File TM, Gravenstein S, Hayden FG. et al. Seasonal influenza in adults and children—diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1003–32. doi: 10.1086/604670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Crowther Z. Influenza and RSV: know the diagnostic options. MLO Med Lab Obs. 2012;44:36. 8. [PubMed] [Google Scholar]
- 183.World Health Organization. Pandemic H1N1 2009 update 112. 2010.
- 184.Nichol KL. Cost-benefit analysis of a strategy to vaccinate healthy working adults against influenza. Arch Intern Med. 2001;161:749–59. doi: 10.1001/archinte.161.5.749. [DOI] [PubMed] [Google Scholar]
- 185.Nichol KL, Heilly SJ, Greenberg ME, Ehlinger E. Burden of influenza-like illness and effectiveness of influenza vaccination among working adults aged 50-64 years. Clin Infect Dis. 2009;48:292–8. doi: 10.1086/595842. [DOI] [PubMed] [Google Scholar]
- 186.Antiviral drugs for seasonal influenza 2014-2015. JAMA. 2015;313:413–4. doi: 10.1001/jama.2014.18380. [DOI] [PubMed] [Google Scholar]
- 187.DiazGranados CA, Dunning AJ, Kimmel M, Kirby D, Treanor J, Collins A. et al. Efficacy of high-dose versus standard-dose influenza vaccine in older adults. N Engl J Med. 2014;371:635–45. doi: 10.1056/NEJMoa1315727. [DOI] [PubMed] [Google Scholar]
- 188.Hauk L. ACIP Releases Updated Recommendations on Influenza Vaccination to Include the 2014-2015 Season. Am Fam Physician. 2014;90:584–7. [Google Scholar]
- 189.Bernard HU, Burk RD, de Villiers EM, zur Hausen H. Family Papillomaviridae. In: King AMQ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. California, USA; 2012. pp. 235–48. [Google Scholar]
- 190.de Villiers EM. Cross-roads in the classification of papillomaviruses. Virology. 2013;445:2–10. doi: 10.1016/j.virol.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 191.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR. et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30:F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 192.Munoz N, Castellsague X, de Gonzalez AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006;24:1–10. doi: 10.1016/j.vaccine.2006.05.115. [DOI] [PubMed] [Google Scholar]
- 193.Connelly LB, Frazer I. Screening to prevent cervical cancer: Guidelines for the management of asymptomatic women with screen detected abnormalities: Commonwealth of Australia; 2005.
- 194.Franco EL, Villa LL, Sobrinho JP, Prado JM, Rousseau M-C, Désy M. et al. Epidemiology of acquisition and clearance of cervical human papillomavirus infection in women from a high-risk area for cervical cancer. J Infect Dis. 1999;180:1415–23. doi: 10.1086/315086. [DOI] [PubMed] [Google Scholar]
- 195.Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, Winer R. et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: A meta-analysis update. Int J Cancer. 2007;121:621–32. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- 196.Howell-Jones R, Bailey A, Beddows S, Sargent A, de Silva N, Wilson G. et al. Multi-site study of HPV type-specific prevalence in women with cervical cancer, intraepithelial neoplasia and normal cytology, in England. Br J Cancer. 2010;103:209–16. doi: 10.1038/sj.bjc.6605747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L. et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30:F12–F23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 198.Lu B, Kumar A, Castellsagué X, Giuliano AR. Efficacy and safety of prophylactic vaccines against cervical HPV infection and diseases among women: a systematic review & meta-analysis. BMC Infect Dis. 2011;11:13. doi: 10.1186/1471-2334-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Reisinger KS, Block SL, Lazcano-Ponce E, Samakoses R, Esser MT, Erick J. et al. Safety and persistent immunogenicity of a quadrivalent human papillomavirus types 6, 11, 16, 18 L1 virus-like particle vaccine in preadolescents and adolescents: a randomized controlled trial. Pediatr Infect Dis J. 2007;26:201–9. doi: 10.1097/01.inf.0000253970.29190.5a. [DOI] [PubMed] [Google Scholar]
- 200.Stanley M. Prophylactic HPV vaccines: prospects for eliminating ano-genital cancer. Br J Cancer. 2007;96:1320–3. doi: 10.1038/sj.bjc.6603695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Azizah N, Hashim U, Gopinath SC, Nadzirah S. Gold nanoparticle mediated method for spatially resolved deposition of DNA on nano-gapped interdigitated electrodes, and its application to the detection of the human Papillomavirus. Mikrochim Acta. 2016;183:3119–26. [Google Scholar]
- 202.Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, Picornaviridae. California, USA: Elsevier; 2012. [Google Scholar]
- 203.Woo PC, Lau SK, Choi GK, Huang Y, Teng JL, Tsoi H-W. et al. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine picodicistrovirus, in the picornavirus-like superfamily. J Virol. 2012;86:2797–808. doi: 10.1128/JVI.05481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rotbart H. Clinical significance, diagnosis, and treatment of picornavirus infections. In: Semler BL, Wimmer E, editors. Molecular biology of picornaviruses. Washington D.C, USA: ASM Press; 2002. pp. 357–65. [Google Scholar]
- 205.Wasley FS, Bell BP. Hepatitis A Virus. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. 7th ed. Pennsylvania, USA: Elsevier; 2009. [Google Scholar]
- 206.Ferreira C, Vieira S, Kieling C, Silveira T. Hepatitis A acute liver failure: follow-up of paediatric patients in southern Brazil. J Viral Hepat. 2008;15:66–8. doi: 10.1111/j.1365-2893.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- 207.Daniels D, Grytdal S, Wasley A, Centers for Disease C, Prevention. Surveillance for acute viral hepatitis - United States, 2007. MMWR Surveill Summ. 2009;58:1–27. [PubMed] [Google Scholar]
- 208.Wasley A, Fiore A, Bell BP. Hepatitis A in the era of vaccination. Epidemiol Rev. 2006;28:101–11. doi: 10.1093/epirev/mxj012. [DOI] [PubMed] [Google Scholar]
- 209.Attoui MPPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, Chappell JD, et al. Reoviridae. In: King AMQ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. London, UK: Elsevier; 2012. pp. 541–637. [Google Scholar]
- 210.Vogt PK. Historical introduction to the general properties of retroviruses; 1997. [PubMed]
- 211.Balvay L, Lastra ML, Sargueil B, Darlix J-L, Ohlmann T. Translational control of retroviruses. Nat Rev Microbiol. 2007;5:128–40. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.HIV/AIDS JUNPo. Global report: UNAIDS report on the global AIDS epidemic. 2010: UNAIDS; 2010. [Google Scholar]
- 213.Mindel A, Tenant-Flowers M. ABC of AIDS: Natural history and management of early HIV infection. BMJ. 2001;322:1290. doi: 10.1136/bmj.322.7297.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Picker LJ, Hansen SG, Lifson JD. New paradigms for HIV/AIDS vaccine development. Annu Rev Med. 2012;63:95. doi: 10.1146/annurev-med-042010-085643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Richards R, Bönnemann H. Synthetic approaches to metallic nanomaterials. In: Challa K, Hormes J, Leuschner C, editors. Nanofabrication Towards Biomedical Applications: Techniques, Tools, Applications, and Impact. Mörlenbach, Germany: Wiley-VCH Verlag GmbH & Co; 2005. [Google Scholar]
- 216.Wang H, Shen G, Yu R. Aspects of recent development of immunosensors. In: Yu R, editor. Electrochemical Sensors, Biosensors and their Biomedical Applications. Amsterdam, The Netherlands: Elsevier; 2008. pp. 237–260. [Google Scholar]
- 217.Bossuyt RJ, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, Lijmer JG. et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Clin Chem. 2003;49:1–6. doi: 10.1373/49.1.1. [DOI] [PubMed] [Google Scholar]
- 218.Walsh JH, Yalow RS, Berson SA. Radioimmunossay of Australia Antigen. Vox Sang. 1970;19:217–24. doi: 10.1111/j.1423-0410.1970.tb01515.x. [DOI] [PubMed] [Google Scholar]
- 219.Detecting the AIDS virus directly with PCR. Oncol Nurs Forum. 1988;15:408. [PubMed] [Google Scholar]
- 220.Hanaee H, Ghourchian H, Ziaee AA. Nanoparticle-based electrochemical detection of hepatitis B virus using stripping chronopotentiometry. Anal Biochem. 2007;370:195–200. doi: 10.1016/j.ab.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 221.Wan ZX, Wang YF, Li SSC, Duan LL, Zhai JX. Development of array-based technology for detection of HAV using gold-DNA probes. J Biochem Mol Biol. 2005;38:399–406. doi: 10.5483/bmbrep.2005.38.4.399. [DOI] [PubMed] [Google Scholar]