Abstract
Neutropenic conditions are prevalent in leukemia patients and are often associated with increased susceptibility to infections. In fact, emergency granulopoiesis (EG), a process regulating neutrophil homeostasis in inflammatory conditions and infections, may occur improperly in leukemic conditions, leading to reduced neutrophil counts. Unfortunately, the mechanisms central to dysfunctional EG remain understudied in both leukemia patients and leukemic mouse models. However, despite no direct studies on EG response in leukemia are reported, recently certain transcription factors (TFs) have been found to function at the crossroads of leukemia and EG. In this review, we present an update on TFs that can potentially govern the fate of EG in leukemia. Transcriptional control of Fanconi DNA repair pathway genes is also highlighted, as well as the newly discovered role of Fanconi proteins in innate immune response and EG. Identifying the TFs regulating EG in leukemia and dissecting their underlying mechanisms may facilitate the discovery of therapeutic drugs for the treatment of neutropenia.
Keywords: emergency granulopoiesis, Fanconi DNA repair pathway, infections, leukemia, neutropenia, neutrophils
Introduction
Bacterial and fungal infections are a leading cause of morbidity and mortality in acute leukemia patients (1, 2). A recent study reported that 71.9% of chronic lymphocytic leukemia (CLL) patients developed infections with a mortality rate of 37.5% (3). Abnormal proliferation of myeloid cells occurs and immature leukocytes accumulate in bone marrow in a cancer microenvironment that can also inhibit antigen-specific T cell response (4). Therefore, leukemia patients are particularly at a high risk for infectious complications. Highly intensive chemotherapy results in prolonged neutropenia, rendering the patients extremely susceptible to microbial infections (5). Prolonged periods of neutropenia proportionately increase the risk of severe infections, which can be exacerbated with the relapse of disease (6, 7).
Neutrophils are key mediators of the early inflammatory response at the time of an infection, and reduced neutrophil counts can lead to life-threatening infections (8). Neutrophil homeostasis is differentially regulated during steady state and infectious episodes. During infections or inflammation, circulating neutrophils become significantly elevated in a process called emergency granulopoiesis (EG). This process involves enhanced generation of neutrophils in the bone marrow through increased myeloid progenitor cell proliferation (9). Neutrophil mobilization is also increased in response to elevation in circulatory granulocyte colony stimulating factor (G-CSF) levels resulting in effective clearance of bacterial and fungal pathogens (10).
Execution of EG occurs in four different stages: (i) it commences with an increase in the peripheral neutrophil count due to vascular demargination and releases from the bone marrow mediated by disruption of CXCL12/CXCR4 signaling (11–15). This phase is accompanied by (ii) de novo generation of neutrophils from increased myeloid progenitor cell proliferation (9) and (iii) their accelerated differentiation by S-phase shortening of the cell cycle stabilized by the Fanconi pathway (16) followed by (iv) termination of EG response, which is partly mediated by interferon regulatory factor (IRF8) (17). A recent study demonstrated that, in cancer chemotherapy-induced neutropenia (CCIN), the neutrophils generated during EG response were functionally immature in both humans and mouse models and displayed weak bactericidal activity (18). Studies evaluating the defects in completion of the EG response during leukemic conditions are scarce; however, certain transcription factors (TFs) have been identified, which share an overlapping role in leukemia and innate immunity. The purpose of this review is to characterize the role of these TFs in leukemia and their link to the EG response. Recently discovered role of the Fanconi DNA repair pathway in innate immunity will also be discussed.
Regulation of Emergency Granulopoiesis
Eemergency granulopoiesis is regulated by various endogenous and exogenous factors. Our knowledge of endogenous factors, predominantly transcriptional regulators, has increased significantly over the past decade. Various TFs play a pivotal role in modulating both EG and leukemia development. Recent studies have shown that dysregulation of these TFs leads to perturbed granulopoiesis along with an aggravation of leukemic state. The role of TFs in normal hematopoiesis, EG, and leukemogenesis is discussed in the following section. Table 1 presents a list of the TFs discussed here.
Table 1.
Transcription factors with intersecting roles in EG and leukemia.
| Transcription Factors | Functions |
|---|---|
| HOXA10 | |
| CEBP-β | |
| IRF8 | |
| STAT3 | |
| STAT5 | |
AML, acute myeloid leukemia; EG, emergency granulopoiesis; G-CSF, granulocyte colony stimulating factor; HSC, hematopoietic stem cell.
HOXA10-Role in Immune Cell Development and Leukemia
HOXA10 is a homeodomain-containing TF which is a part of the A cluster on chromosome 7. HOXA10 is considered a master regulator of postnatal hematopoietic development that controls hematopoietic stem cell (HSC) self-renewal, the development of lymphoid and erythroid/megakaryocyte cells, as well as platelet biogenesis (19–21). It is also abundantly expressed in myeloid progenitors, where it influences myelopoiesis (22–24) and in phagocytic cells, where it represses transcription of the genes encoding p67phox (NADPH oxidase subunit) and gp91phox (cytochrome b subunit beta), thereby influencing its effector functions (25). However, overexpression of HOXA10 in murine bone marrow has been shown to induce a myeloproliferative disorder (MPD) involving expansion of the committed myeloid progenitors, which later evolves into acute myeloid leukemia (AML) (26). Leukemias with chromosomal translocations of the mixed lineage leukemia 1 (MLL1) gene are characterized by increased and sustained transcription of a group of HOX genes (including HOXA10), as fusion proteins generated by MLL1 gene translocations lack ubiquitination/degradation domains (27). Mice transplanted with bone marrow expressing an MLL-fusion protein or overexpressing HOXA10 develop AML (26, 28–30). In a recent study, the MLL-ELL fusion protein was found to increase expression of HOXA9 and HOXA10 directly, by interaction with their promoters, and indirectly via fibroblast growth factor 2 (FGF2), β-catenin, and caudal-type homeobox 4 (CDX4) (31).
HOXA10-Role in Emergency Granulopoiesis through HOXA10-TRIAD1 Interaction
Apart from the leukemogenic role of HOXA10, its role in regulating EG has also been elucidated recently. HOXA10−/− mice showed a fatal EG response, which was rescued by re-expression of TRIAD1 (alias ARIH2) (32). TRIAD1, encoded by the gene ARIH2, is a ubiquitin ligase that regulates myelopoiesis by inhibiting proliferation of myeloid cells (32, 33). In one study, hematopoietic deficiency of ARIH2 caused lethal activation of the immune system. Sustained activity of NF-κB TF subunit p65 (RELA) was found in the nucleus of ARIH2-deficient dendritic cells, which caused lethal immunological responses in ARIH2-sufficent mice reconstituted with ARIH2-deficient hematopoietic stem cells (34). ARIH2 has been shown to be a target gene for HOXA10, with the tandem cis elements in the ARIH2 promoter being activated by HOXA10. In vitro stimulation of myeloid progenitor cells with G-CSF showed HOXA10-dependent increase in TRIAD1 expression (33). As G-CSF is the prime mediator of EG, this study implicates EG-driven up-regulation of TRIAD1 by HOXA10 and presents protein ubiquitination/degradation as a novel mechanism of regulating EG response by HOXA10. Increased TRIAD1 expression degrades FGFR1, thereby reducing the levels of FGF2 and terminating the effect of FGF2 on myeloid progenitor expansion and phagocyte effector function. All these processes culminate in termination of EG, with HOXA10 being the prime mediator. Moreover, in the bone marrow of HOXA10−/− mice, TRIAD1 expression was only slightly decreased at steady state but TRIAD1 expression was totally absent during EG, suggesting the specific role of HOXA10 during EG. Transcription of ARIH2 and expression of TRIAD1 during EG was regulated by tyrosine phosphorylation of HOXA10. Thus, this study elucidated the induction of protein degradation via TRIAD1 as a novel immune modulatory mechanism of HOXA10 (32).
Based on these studies, it can be speculated that, during EG in leukemia, overexpression of HOXA10 leads to sustained activation of TRIAD1, which favors a suppressed EG state, thus identifying one factor that may cause neutropenia in leukemia patients and make them more susceptible to infections (Figure 1). On the other hand, higher leukemia transformations have been reported in severe congenital neutropenia (SCN) patients who required higher G-CSF doses (35), indicating that there are common TFs that mediate leukemogenesis and granulopoiesis.
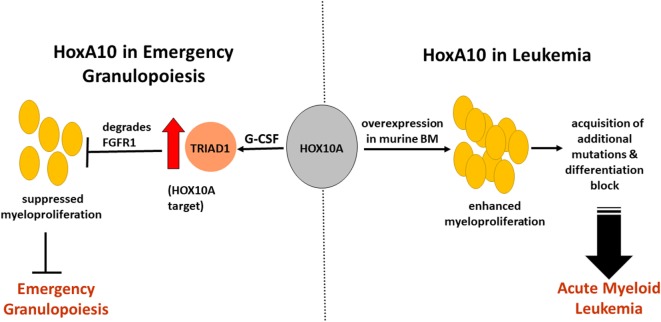
Figure 1.
Impact of HOXA10 on acute myeloid leukemia (AML) development and emergency granulopoiesis (EG) through regulation of myelopoiesis. HOXA10 overexpression in murine bone marrow leads to enhanced myeloproliferation, which progresses into AML through acquisition of additional mutations and differentiation block. HOXA10 effects EG by regulating the gene ARIH2, which encodes TRIAD1, a ubiquitin ligase. In vitro stimulation of myeloid progenitors with granulocyte colony stimulating factor (G-CSF) showed HOXA10-dependent increase in TRIAD1 expression. Increased TRIAD1 expression degrades fibroblast growth factor receptor-1 (FGFR1), reduces the effect of FGF2, and terminates the effect of FGF2 on myeloid progenitor expansion. As G-CSF is the main mediator of EG, these processes result in termination of EG with HOXA10 being the prime regulator. Suppressed EG response during infections in leukemic conditions may serve as one of the causes of neutropenia.
CCAAT/Enhancer Binding Protein Beta—Role in Immune Cell Development and Leukemia
CCAAT/Enhancer Binding Protein beta (C/EBP-β) is a basic leucine zipper (bZIP) domain-containing TF that plays an important role in regulating immune and inflammatory responses (36–39). Both leukemia suppressor and pro-oncogenic roles of C/EBP-β have been reported. C/EBP-β was shown to suppress the leukemogenic potential of 32D-BCR-ABL cells by inducing granulocytic differentiation and by inhibiting cell proliferation. Low C/EBP-β expression is observed in the blast crisis stage of chronic myelogenous leukemia (CML) and is inversely correlated with BCR-ABL tyrosine kinase levels, suggesting that there may be therapeutic potential in restoring its activity in CML-BC (40). In acute promyelocytic leukemia (APL), treatment with all-trans retinoic acid (ATRA) reverses promyelocytic leukemia-retinoic acid receptor α (PML-RARα)-mediated differentiation block at the promyelocyte stage resulting in mature neutrophil-like cells (41, 42). C/EBP-β is upregulated in the presence of PML-RARα during ATRA treatment and promotes the proliferation and differentiation of APL cells, thereby showing a potential anti-cancer role (42).
C/EBP-β exists as several isoforms due to alternative translation initiation: full-length C/EBP-β liver activating protein* (LAP*), a slightly shorter isoform of LAP that lacks the first 21 amino acids and a short isoform of liver inhibitory protein (LIP). LAP* and LAP are trans-activators, whereas LIP is a transcriptional repressor. The relative abundance of LIP and LAP C/EBP-β isoforms mediated through the regulation of translation initiation is important in determining cell fate by controlling proliferation and differentiation (43). In contrast to the leukemia suppressive effect of C/EBP-β, its LIP isoform was shown to promote leukemogenesis in a mouse bone marrow transplantation system by collaborating with Ecotropic viral integration site 1 (Evi1) which is one of the master regulators of AML development. However, experiments performed on human whole BM cells from AML patients revealed that Evi1 closely correlated with both LAP* and LIP expression (44).
CCAAT/Enhancer Binding Protein Beta—Role in Emergency Granulopoiesis
Hirai et al. showed that C/EBP-α is required for steady-state granulopoiesis whereas C/EBP-β is essential for EG (45–47). Only C/EBP-β expression (and not that of other C/EBPs) was upregulated in GMPs after cytokine treatment (45). Using C/EBP β−/− bone marrow cells, it was found that C/EBP-β is involved in cytokine (G-CSF, GM-CSF, IL-3, and IL-6)-induced myeloid proliferation, suggesting that C/EBP-β is required to couple proliferation and differentiation of granulocytes under stress or emergency situations, thereby producing more mature granulocytes (45). The specific role of LAP*, LAP, and LIP isoforms of C/EBP-β in EG is not clear; however, exploring this may help target the specific isoform for both anti-leukemic and anti-neutropenic effects.
IRF8 (Also known as Interferon Consensus Sequence Binding Protein)—Role in Immune Cell Development and Leukemia
The critical role of IRF8 in innate immune response and oncogenesis has been described extensively (48–51). In response to pattern-recognition receptors (PRR) activation, IRF8 induces the expression of proinflammatory cytokines through TLR9-MyD88-dependent signaling (52–55). IRF8 inhibits cell growth and promotes apoptosis in myeloid cells and drives their differentiation toward macrophages while inhibiting neutrophil production (56, 57). IRF8 plays an important role in myeloid cell development, as has been demonstrated by a systemic expansion of neutrophils followed by a fatal blast crisis, resembling human CML in IRF8−/− mice (50). These mice also exhibit increased progenitor cell numbers with enhanced responsiveness to granulocyte-macrophage colony stimulating factor (GM-CSF) and G-CSF. In contrast, responsiveness to macrophage colony stimulating factor (M-CSF) was reduced in IRF8−/− progenitors, implying a role for IRF8 in driving toward the macrophage lineage (57, 58). A tumor suppressor role has been described for IRF8, as very low or absent IRF8 mRNA was found in the peripheral blood of the majority of human myeloid leukemias. Sorted B-cells derived from CML patients also showed the absence of IRF8 mRNA (59). Moreover, high IRF8 mRNA levels were found only in those CML patients who were classified as “good” cytogenetic responders to interferon-α therapy and not in the poor responders (60). One of the target genes of IRF8 is Fas-associated phosphatase 1 (FAP1; the PTPN13 gene), which shares a reciprocal expression profile with IRF8 at all clinical stages of CML. Impaired IRF8 expression in BCR−ABL + myeloid progenitor cells contributed to FAP1-dependent Fas resistance. As Fas resistance contributes to persistence and expansion of CML leukemic stem cells (LSCs), it led to imatinib resistance in BCR − ABL + GMPs through the reduction of IRF8 expression and increased FAP1 expression (61).
IRF8 also represses the Growth Arrest Specific 2 gene (GAS2), which encodes a calpain inhibitor that is involved in cell proliferation and survival. GAS2 expression in BCR-ABL+ cells stabilizes β-catenin, which is a calpain substrate (62). In addition to β-catenin, calpain has other substrates that are involved in the pathogenesis of CML including signal transducer and activator of transcription 5 (STAT5) (63). In a recent study, BCR-ABL-induced SHP2-dependent dephosphorylation of IRF8 was found to impair repression of GAS2, leading to decreased calpain activity and thereby an increase in its substrate protein STAT5, which in turn represses IRF8 promoter. This novel feedback mechanism involving calpain enhances leukemogenesis by increasing STAT5 and repressing IRF8. Hence, therapeutic upregulation of IRF8 can reduce persistent LSCs during treatment of CML with BCRABL-targeted tyrosine kinase inhibitors (TKIs) (64). In another study, induction of IRF8 and repression of β-catenin was found upon arachidonate 15-lipoxygenase (15-LO) inhibition by PD146176 in K562 cells, implicating another mechanism where IRF8 may be involved in eradicating CML LSCs (65). In addition to the role of IRF8 in myeloid leukemia, recently its role in suppressing acute lymphoblastic leukemia has been described. Mice deficient for both PU.1 and IRF8 developed pre-B-ALL at high frequency by reducing the expression of known tumor suppressors, including SPI-B, IKAROS, and BLNK (66).
IRF8 as a Regulatory Component of Emergency Granulopoiesis
In addition to its leukemia suppressor role, IRF8 plays an important regulatory role in innate immune response (51). Some of the target genes of IRF8 include FANCC and FANCF, encoding Fanconi C and F, respectively, which contribute to the Fanconi DNA repair pathway activation during infectious challenge (16, 67). IRF8 activates FANCF cis element in differentiating myeloid cells, thereby protecting them from genotoxic stress associated with differentiation (67). IRF8 was also found to bind and activate a cis element in the proximal FANCC promoter. Re-expression of FANCC rescued DNA repair in IRF8-deficient myeloid cells. Furthermore, IRF8 activates FANCC in murine myeloid progenitor cells upon stimulation with IL-1β and G-CSF, cytokines that are essential for EG (16). As rapid expansion due to S-phase shortening of granulocyte/monocyte progenitor (GMP) populations occurs during the cell cycle, IRF8 contributes to genomic stability during EG through the Fanconi pathway (16, 67) (Figure 2).
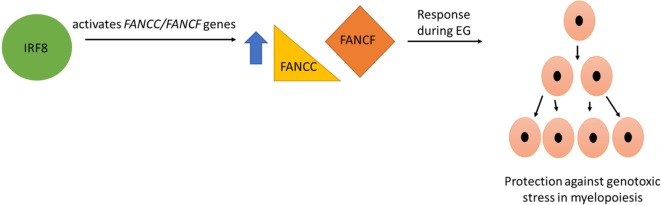
Figure 2.
IRF8 plays an important role in myeloid cell development during emergency granulopoiesis (EG) via regulating Fanconi DNA repair proteins. Some of the target genes of IRF8 include FANCC and FANCF, encoding Fanconi C and Fanconi F, respectively. These Fanconi proteins contribute to Fanconi DNA repair pathway activation during EG and protect cells from the genotoxic stress of myelopoiesis during rapid proliferation phase.
In addition to this, a more specific role of IRF8 in the termination of EG response was recently described. In IRF8−/− mice, sustained granulocyte production was found in response to EG via increased expression of FAP1 and GAS2 in bone marrow myeloid progenitor cells, which leads to decreased FAS sensitivity and increased β-catenin activity in these cells. This implies that IRF8 mediates termination of the EG response by increasing FAS-induced apoptosis and decreasing β-catenin activity in these cells, thereby limiting granulocyte proliferation (17). However, repeated episodes of EG did not increase granulocytes in IRF8−/− mice; instead, an accumulation of myeloid blasts was found leading to AML development (Figure 3). This effect is mediated through reduced expression of IRF8 target proteins FANCC and FANCF with increased sensitivity to DNA damage in bone marrow myeloid progenitors (17).
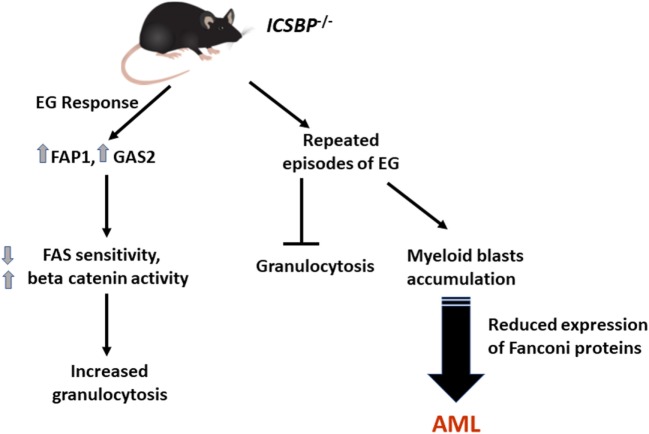
Figure 3.
Repeated episodes of emergency granulopoiesis (EG) lead to leukemia development in IRF8−/− (ICSBP−/−) mice. A sustained expansion of neutrophils was observed in IRF8−/− mice in response to EG through increased expression of FAP1 and GAS2 in bone marrow myeloid progenitors. However, repeated episodes of EG did not increase granulocytes in IRF8−/− mice, but accumulation of myeloid blasts was found, leading to acute myeloid leukemia (AML) development. Reduced expression of FANCC and FANCF, which are IRF8 target proteins, contribute by increasing sensitivity to DNA damage in bone marrow myeloid progenitors.
From these observations, it can be inferred that there may be a dual impact of reduced IRF8 on neutrophil production in leukemic conditions during an EG response. Reduced IRF8 would prolong the termination step of EG (by increasing FAP1 and GAS2 expression), thereby causing granulocytosis with functionally incompetent cells (17). Simultaneously, it will also promote apoptosis of myeloid progenitors during the rapid proliferation step via decreased expression of FANCC and F that rescue collapsed and stalled replication forks during DNA replication. This leads to reduced neutrophil count or enhanced leukemogenesis due to accumulation of mutations. Therefore, gene regulation of Fanconi DNA repair proteins by IRF8 presents an important link between innate immune response and the leukemia suppressor role of IRF8.
STAT Proteins
STAT3—Role in Immune Cell Development and Leukemia
STAT3 is one of the most important TFs phosphorylated by Janus kinases after G-CSF-induced dimerization of G-CSFR (68, 69). G-CSF and IL-6 are potent activators of STAT3 in hematopoietic progenitor cells and neutrophils (69, 70). Constitutively active STAT3 has been reported in human AML cell lines and in pretreatment bone marrow samples of AML patients, where it was associated with shorter disease-free survival (71, 72). Inhibiting the G-CSF-induced STAT3 phosphorylation by a small-molecule STAT3 inhibitor C188-9 induced apoptosis in AML cell lines and primary pediatric AML samples (73). Another STAT3 inhibitor, BP-5-087, in combination with imatinib, reduced the survival of primary tyrosine kinase inhibitor (TKI)-resistant stem and progenitor cells, as activation of pSTAT3 (at amino acid position Y705) is important for BCR-ABL1 kinase-independent TKI resistance (74).
STAT3 interacts directly with c-Jun, as has been reported using the yeast two-hybrid system (75). STAT3, in cooperation with c-JUN and c-FOS, activates the IL-6 response element (IRE) (76). Elevated c-JUN expression has been linked to AML, where it inactivates C/EBP-α via leucine zipper domain interaction. This interaction and C/EBP-α inactivation are necessary to induce proliferation in AML (77). The role of c-JUN during EG has not been explored yet; however, its interaction with STAT3 and IRE activation point toward its involvement as a positive regulator of EG. c-JUN may act as an additional TF that lies at the intersection of EG and leukemia and may enhance the development of leukemia along with skewing of the EG response.
STAT3—Role in Emergency Granulopoiesis
In response to G-CSF, STAT3 accelerates granulocyte cell-cycle progression (G1–S phase transition) and terminal maturation by regulating C/EBP-β, which is an important TF in the EG response. STAT3 and C/EBP-β co-regulate c-MYC through direct interaction with its promoter and displacement of C/EBP-α during demand-driven granulopoiesis (78). A recent study has shown the involvement of STAT3 and C/EBP-β in the activation of FANCC transcription which contributes to DNA repair during EG (79).
STAT3 Regulates Suppressor of Cytokine Signaling-3 (SOCS3) Expression
Studies done in STAT3-deficient mice show neutrophilia and hyper-responsiveness of myeloid cells to G-CSF. This was related to reduced SOCS3 expression, thereby showing the role of STAT3 in inducing SOCS3, which in turn acts as a negative regulator of proliferative signals from G-CSF signaling required during EG (80). SOCS3 deficiency significantly increases STAT3 activation in response to in vivo administration of G-CSF which leads to toxic effects (81). Similarly, in another study on SOCS3-deficient hematopoietic progenitor cells, STAT3 and C/EBP-β activation was increased in response to G-CSF and IL-6 (82). As mentioned above, STAT3 induces C/EBP family TFs and interacts with them to augment the effect of G-CSF signaling (83). SOCS3 is recognized as a negative regulator of G-CSF signaling and EG in myeloid cells (81, 84). Constitutive STAT3 phosphorylation was shown to constitutively activate SOCS3 expression in AML blasts isolated from patients (85). An inefficient EG response during infectious episodes can be expected from constitutively increased expression of SOCS3 under such leukemic conditions. Another study on STAT3-deficient mice showed a failure to accumulate immature granulocytes in the bone marrow after G-CSF exposure, thereby leading to an increase in the ratio of mature to immature neutrophils. However, immature granulocytes are needed during EG for increased neutrophil production. The study characterized impairment in acute neutrophil mobilization which is independent of SOCS3, indicating diverse signaling pathways in response to G-CSF (86).
Recently, activating somatic STAT3 mutations located in the Src homology 2 (SH2) domain have been described in T-cell large granular lymphocytic leukemia with a high frequency of 40% (31 of 77 patients). These patients presented more often with neutropenia than patients without these mutations (87). As discussed earlier, this could be due to the induction of SOCS3 by activated STAT3, which negatively regulates G-CSF signaling (81). Based on these observations, constitutive activation of STAT3 might contribute to failure of the EG response in leukemic patients resulting in neutropenia and increased susceptibility to infections.
Targeting STAT3 can provide highly specific approach to treat failed EG responses in leukemia. Evidently, there is a growing list of STAT3 inhibitors under clinical evaluation which rely on direct or indirect targeting mechanisms (88). Most common STAT3 targeting approaches include inhibition of tyrosine kinases that phosphorylate and upregulate STAT3 activity (89). Others include STAT3 SH2 domain (dimerization) inhibitors thereby preventing the formation of functional STAT3 dimers; STAT3 DNA binding domain inhibitors that prevent binding of STAT3 to its DNA promoter, STAT3 gene expression oligonucleotide inhibitors which compete for STAT DNA binding (88), and STAT3 N-terminal domain inhibitors that disrupt protein–protein interactions between STAT3 and other TFs (90). Therefore, in addition to the possible therapeutic potential of STAT3 inhibitors in cancer treatment they can also be considered for their efficacy in treating infectious episodes in leukemia.
STAT5—Role in Immune Cell Development and Leukemia
STAT5 is an important STAT family protein that is activated by G-CSF (91). STAT5a and STAT5b are two forms of STAT5 that are encoded by two distinct but closely related genes (92) and are activated by tyrosine phosphorylation through many factors and cytokines like prolactin, growth hormone (93), erythropoietin (94), thrombopoietin (95), interleukin 3 (IL-3), GM-CSF (96), and interleukin 2 (IL-2) (97). An anti-apoptotic role of STAT5 has been documented during differentiation of myeloid progenitors. In a study by Kieslinger et al. (98), primary chicken myeloblasts expressing dominant-negative alleles of STAT5 were unable to generate mature cells due to increased apoptosis during differentiation. Bone marrow cells from STAT5a/STAT5b-deficient mice showed increased apoptosis during GM-CSF-dependent maturation in vitro. This apoptotic cell death was rescued by ectopic expression of BCL-X, thereby showing an important role of STAT5 during cytokine-dependent differentiation of myeloid progenitors during inflammation (98).
STAT5 signaling can promote oncogenesis (99) and hyperactivated STAT5 has been implicated in various leukemia types such as BCR-ABL-induced CML and AML, and in MPDs, such as chronic myelomonocytic leukemia and polycythemia vera (99–101).
Constitutive STAT5 activation has been demonstrated to be essential in a mouse model of MPD induced by TEL-JAK2 fusion protein. TEL-JAK2 fusion protein-mediated constitutive STAT5 activation is essential in a mouse model of MPD (102). Constitutively active STAT5 mutant in CD34–c-Kit+Sca-1+ lineage marker− (CD34–KSL) HSCs induced fatal MPD in a mouse model, implying the crucial role of STAT5 in self-renewal of HSCs during MPD development (99). These studies show that STAT5 is involved in both neutropenic conditions and development of hematologic malignancies. Therapeutic approaches to target caSTAT5 are being studied, like the small molecule bromodomain inhibitor JQ1, which reduces STAT5 function in leukemia and lymphoma cells with caSTAT5 (103).
STAT5-Possible Regulatory Role in Emergency Granulopoiesis
STAT5-null (STAT5A and STAT5B) HSCs in mice show an impaired repopulation potential and disrupt multilineage hematolymphoid development in the bone marrow, including a reduction in neutrophil progenitors and mature neutrophils (104). However, constitutively active STAT5 (caSTAT5a) and not wild-type STAT5a is associated with inhibition of lymphoid enhancer-binding factor 1 (LEF-1) in CD34 + cells of congenital neutropenia (CN) patients. LEF-1 positively regulates G-CSF triggered granulopoiesis by promoting proliferation and differentiation of granulocyte precursors (105). caSTAT5a inhibits LEF-1-dependent autoregulation of LEF-1 gene promoter by binding to the LEF-1 protein, recruiting Nemo-like kinase and the E3 ubiquitin-ligase NARF to LEF-1. This leads to LEF-1 ubiquitination and a reduction in LEF-1 protein levels, severely affecting neutrophil production. Interestingly, sustained elevation of phospho-STAT5 in CD34 + cells was observed in the CN patients compared to healthy controls, which was correlated to the development of AML (100).
Emerging Role of Fanconi Anemia (FA) DNA Repair Pathway Interconnects Leukemia and Innate Immune Response
To date, 19 genes belonging to FA complementation groups are known (A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P, Q, R, S, T) (106, 107). The FA pathway is required to repair DNA interstrand crosslinks (ICLs) which involves nucleotide excision repair (NER), translesion synthesis (TLS), and homologous recombination (HR) (108). ICLs affect DNA replication and transcription through stalling of replication forks and preventing strand separation. Therefore, unrepaired ICLs lead to DNA breakage and chromosomal rearrangements resulting in cellular apoptosis or accumulation of mutations (109). In this regard, FA pathway plays an important role in genome maintenance by repairing DNA damage during replication stress responses, especially during S phase of the cell cycle (110). Hypersensitivity to DNA damage agents that induce ICL is observed in cells deficient in any component of FA pathway (111, 112). Cells undergo G2/M arrest and chromosomal breakage on treatment with mitomycin C (MMC) or diepoxybutane (DEB) (113, 114). In humans, germline inactivation of any FA gene predisposes them to increased sensitivity to ICLs, thereby resulting in bone marrow failure (BMF) and cancer development (115–117).
The FA pathway is also important to maintain hematopoietic stem and progenitor cells (HSPC) population. In FA patients, p53/p21 activation and G0/G1 cell cycle arrest occurs in HSPC leading to BMF in FA, whereas p53 deficiency rescued the HSPC defects in human FA cells. Therefore, HSPC instability in FA patients increases their chances to develop AML (118). K-RAS or c-MYC induced oncogenic stress caused a short-lived response in mice deficient for the FA core complex components FANCA or FANCC. Downregulation of Protein Arginine Methyltransferase 5 (PRMT5) led to compromised K-rasG12D-induced arginine methylation of p53 in FANCA deficient cells, thereby demonstrating an arginine methylation-dependent FA-p53 interaction, as forced expression of PRMT5 in FANCA−/− HSPCs prolonged oncogenic response and delayed leukemia development in irradiated recipient mice (119). In another recent study, FANCC deficient aging mice developed hematologic malignancies that precede genomic instability and hematopoietic chromosomal instability or aneuploidy (120). This is further supported by the observation that AML displays an acquired decrease in expression of Fanconi proteins. Role of FANCF in leukemia suppression has also been shown. CHRF-288 (an AML cell line) exhibits a cellular FA phenotype due to lack of FANCF expression, which is corrected by a FANCF-expressing plasmid. FANCF is localized in a hot-spot region for somatic hypermethylation (11p15); therefore, gene silencing due to hypermethylation of the promoter region of the FANCF gene explains the absence of FANCF protein in CHRF-288 cell line (121). FANCF is also an IRF8 target gene that provides genomic stability to myeloid cells from DNA cross-link damage during proliferation and differentiation stages (67). Mitomycin C-induced DNA damage was increased in IRF8 deficient primary murine bone marrow cells, which was rescued by FANCF overexpression (17). Together, these findings strongly suggest a functional cross-talk between cell proliferation and DNA repair pathways.
Until now, the major roles of Fanconi pathway have been shown in maintenance and proliferation of HSPC (122); tumor suppression (123–127); stabilizing the replication fork during S phase; and DNA repair processes to protect against unwanted mutations (108, 128–130). However, its role in innate immune response has emerged recently. As mentioned previously, FANCC is an IRF target gene and IRF8−/− mice succumbed to infectious challenge with failed leukocytosis response (16, 50, 131). This observation implies that EG will be impaired in response to infectious or inflammatory challenge. As Fanconi proteins protect cells from genotoxic stress of myelopoiesis during rapid proliferation phase, their potential role in EG response is plausible. FANCC-deficient mice showed an abnormal response to EG and developed progressive neutropenia and anemia which resulted from excess apoptosis of bone marrow HSCs and myeloid progenitors. These effects led to failed EG episodes which contributed to BMF and suggest that FANCC expression and Fanconi pathway is enhanced during infectious episodes and is an essential element of EG response. Upon treatment with an essential EG cytokine IL-1β, FANCC, FANCJ, and RAD51 were enriched in the chromatin fraction of murine myeloid progenitor cells, signifying their increased expression and diverse DNA repair processes initiated during EG. Moreover, treatment with an IL-1R antagonist (anakinra) in alum-treated FANCC-knockout mice ameliorated BMF (16).
These observations again point toward the overlapping role of FANCC protein in leukemia suppression and completing a successful EG response. Lack of this protein resulted in enhanced susceptibility to AML development and failed EG in response to infectious challenge thereby leading to neutropenia (16).
Conclusion
From the studies reviewed here, it can be inferred that the alterations in the expression of TFs which promote leukemia also cause an improper EG execution leading to neutropenia. Neutropenic conditions will result in increased susceptibility to infections. That is why severe sepsis is observed in cancer patients at a much higher rate than in non-cancer patients (132). So, a counter question can also arise: do neutropenic conditions lead to leukemia? An example can be found in myelodysplastic syndromes where the neutropenic patients are at a higher risk for leukemia. The findings, albeit very few, point to a role of TFs in the pathogenesis of these diseases and it will be of interest to study if failed EG episodes predisposes a person toward leukemia. Identifying TFs that can revert the disease phenotype or selectively treat neutropenia in conjunction with drugs used for leukemia is another highly promising strategy that may be tested. Therefore, targeting intersecting TFs can be of therapeutic value to treat lymphoid and myeloid leukemia and associated disorders including neutropenia.
Author Contributions
SH designed and drafted the manuscript. SH and ARN wrote the manuscript. AR and SH finalized the figures and table. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that there is no conflict of interest, be it financial, commercial, or other.
Abbreviations
EG, emergency granulopoiesis; TFs, transcription factors; HSC, hematopoietic stem cells; AML, acute myeloid leukemia; MLL1, mixed lineage leukemia 1; FGF2, fibroblast growth factor 2; IRF8, interferon regulatory factor; C/EBP-β, CCAAT/enhancer binding protein beta; ICSBP, interferon consensus sequence binding protein; GAS2, growth arrest specific 2; LSCs, leukemic stem cells; FAP1, Fas-associated phosphatase 1; FANCC, Fanconi C; FANCF, Fanconi F; SOCS3, suppressor of cytokine signaling-3; caSTAT5, constitutively active STAT5; MPD, myeloproliferative disorder; BMF, bone marrow failure; FA, Fanconi anemia.
References
- 1.Alexander S, Nieder M, Zerr DM, Fisher BT, Dvorak CC, Sung L. Prevention of bacterial infection in pediatric oncology: what do we know, what can we learn? Pediatr Blood Cancer (2012) 59(1):16–20. 10.1002/pbc.23416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt VR, Viola GM, Ferrajoli A. Invasive fungal infections in acute leukemia. Ther Adv Hematol (2011) 2(4):231–47. 10.1177/2040620711410098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams AM, Baran AM, Meacham PJ, Feldman MM, Valencia HE, Newsom-Stewart C, et al. Analysis of the risk of infection in patients with chronic lymphocytic leukemia in the era of novel therapies. Leuk Lymphoma (2017) 59(3):625–32. 10.1080/10428194.2017.1347931 [DOI] [PubMed] [Google Scholar]
- 4.Almand B, Clark JI, Nikitina E, van Beynen J, English NR, Knight SC, et al. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol (2001) 166(1):678–89. 10.4049/jimmunol.166.1.678 [DOI] [PubMed] [Google Scholar]
- 5.Young LS. Management of infections in leukemia and lymphoma. In: Rubin RH, Young LS, Van Furth R, editors. Clinical Approach to Infection in the Compromised Host. Boston, MA: Springer US; (2002). p. 497–526. [Google Scholar]
- 6.Bodey GP. The changing face of febrile neutropenia-from monotherapy to moulds to mucositis. Fever and neutropenia: the early years. J Antimicrob Chemother (2009) 63(Suppl 1):i3–13. 10.1093/jac/dkp074 [DOI] [PubMed] [Google Scholar]
- 7.Rekha C, Morgan H, Stephen S. Sepsis - an ongoing and significant challenge. In: Azevedo L, editor. Infections in Leukemia. Rijeka: InTech; (2012). 420 p. [Google Scholar]
- 8.Wirths S, Bugl S, Kopp HG. Neutrophil homeostasis and its regulation by danger signaling. Blood (2014) 123(23):3563–6. 10.1182/blood-2013-11-516260 [DOI] [PubMed] [Google Scholar]
- 9.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol (2014) 14(5):302–14. 10.1038/nri3660 [DOI] [PubMed] [Google Scholar]
- 10.Lieschke GJ, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood (1994) 84(6):1737–46. [PubMed] [Google Scholar]
- 11.Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity (2002) 17(4):413–23. 10.1016/S1074-7613(02)00424-7 [DOI] [PubMed] [Google Scholar]
- 12.Semerad CL, Christopher MJ, Liu F, Short B, Simmons PJ, Winkler I, et al. G-CSF potently inhibits osteoblast activity and CXCL12 mRNA expression in the bone marrow. Blood (2005) 106(9):3020–7. 10.1182/blood-2004-01-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levesque JP, Hendy J, Takamatsu Y, Simmons PJ, Bendall LJ. Disruption of the CXCR4/CXCL12 chemotactic interaction during hematopoietic stem cell mobilization induced by GCSF or cyclophosphamide. J Clin Invest (2003) 111(2):187–96. 10.1172/JCI15994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HK, De La Luz Sierra M, Williams CK, Gulino AV, Tosato G. G-CSF down-regulation of CXCR4 expression identified as a mechanism for mobilization of myeloid cells. Blood (2006) 108(3):812–20. 10.1182/blood-2005-10-4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eash KJ, Means JM, White DW, Link DC. CXCR4 is a key regulator of neutrophil release from the bone marrow under basal and stress granulopoiesis conditions. Blood (2009) 113(19):4711–9. 10.1182/blood-2008-09-177287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu L, Huang W, Hjort E, Eklund EA. Increased Fanconi C expression contributes to the emergency granulopoiesis response. J Clin Invest (2013) 123(9):3952–66. 10.1172/JCI69032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu L, Huang W, Hjort EE, Bei L, Platanias LC, Eklund EA. The interferon consensus sequence binding protein (Icsbp/Irf8) is required for termination of emergency granulopoiesis. J Biol Chem (2016) 291(8):4107–20. 10.1074/jbc.M115.681361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Qi X, Sun W, Abdel-Azim H, Lou S, Zhu H, et al. Am80-GCSF synergizes myeloid expansion and differentiation to generate functional neutrophils that reduce neutropenia-associated infection and mortality. EMBO Mol Med (2016) 8(11):1340–59. 10.15252/emmm.201606434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnusson M, Brun AC, Miyake N, Larsson J, Ehinger M, Bjornsson JM, et al. HOXA10 is a critical regulator for hematopoietic stem cells and erythroid/megakaryocyte development. Blood (2007) 109(9):3687–96. 10.1182/blood-2006-10-054676 [DOI] [PubMed] [Google Scholar]
- 20.Buske C, Feuring-Buske M, Antonchuk J, Rosten P, Hogge DE, Eaves CJ, et al. Overexpression of HOXA10 perturbs human lymphomyelopoiesis in vitro and in vivo. Blood (2001) 97(8):2286–92. 10.1182/blood.V97.8.2286 [DOI] [PubMed] [Google Scholar]
- 21.Konieczna IM, DeLuca TA, Eklund EA, Miller WM. Hoxa10 null animals exhibit reduced platelet biogenesis. Br J Haematol (2016) 173(2):303–13. 10.1111/bjh.13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eklund EA, Goldenberg I, Lu Y, Andrejic J, Kakar R. SHP1 protein-tyrosine phosphatase regulates HoxA10 DNA binding and transcriptional repression activity in undifferentiated myeloid cells. J Biol Chem (2002) 277(39):36878–88. 10.1074/jbc.M203917200 [DOI] [PubMed] [Google Scholar]
- 23.Eklund EA, Jalava A, Kakar R. Tyrosine phosphorylation of HoxA10 decreases DNA binding and transcriptional repression during interferon gamma-induced differentiation of myeloid leukemia cell lines. J Biol Chem (2000) 275(26):20117–26. 10.1074/jbc.M907915199 [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Goldenberg I, Bei L, Andrejic J, Eklund EA. HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with histone deacetylase 2. J Biol Chem (2003) 278(48):47792–802. 10.1074/jbc.M305885200 [DOI] [PubMed] [Google Scholar]
- 25.Lindsey S, Zhu C, Lu YF, Eklund EA. HoxA10 represses transcription of the gene encoding p67phox in phagocytic cells. J Immunol (2005) 175(8):5269–79. 10.4049/jimmunol.175.8.5269 [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Lindsey S, Konieczna I, Bei L, Horvath E, Huang W, et al. Constitutively active SHP2 cooperates with HoxA10 overexpression to induce acute myeloid leukemia. J Biol Chem (2009) 284(4):2549–67. 10.1074/jbc.M804704200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bei L, Shah C, Wang H, Huang W, Platanias LC, Eklund EA. Regulation of CDX4 gene transcription by HoxA9, HoxA10, the Mll-Ell oncogene and Shp2 during leukemogenesis. Oncogenesis (2014) 3:e135. 10.1038/oncsis.2014.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J (1998) 17(13):3714–25. 10.1093/emboj/17.13.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Luo RT, Mi S, Sun M, Chen P, Bao J, et al. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res (2009) 69(3):1109–16. 10.1158/0008-5472.CAN-08-3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, et al. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol (1997) 17(1):495–505. 10.1128/MCB.17.1.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah CA, Bei L, Wang H, Altman JK, Platanias LC, Eklund EA. Cooperation between AlphavBeta3 integrin and the fibroblast growth factor receptor enhances proliferation of Hox-overexpressing acute myeloid leukemia cells. Oncotarget (2016) 7(34):54782–94. 10.18632/oncotarget.10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Bei L, Shah CA, Hu L, Eklund EA. HoxA10 terminates emergency granulopoiesis by increasing expression of Triad1. J Immunol (2015) 194(11):5375–87. 10.4049/jimmunol.1401909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Bei L, Shah CA, Horvath E, Eklund EA. HoxA10 influences protein ubiquitination by activating transcription of ARIH2, the gene encoding Triad1. J Biol Chem (2011) 286(19):16832–45. 10.1074/jbc.M110.213975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin AE, Ebert G, Ow Y, Preston SP, Toe JG, Cooney JP, et al. ARIH2 is essential for embryogenesis, and its hematopoietic deficiency causes lethal activation of the immune system. Nat Immunol (2013) 14(1):27–33. 10.1038/ni.2478 [DOI] [PubMed] [Google Scholar]
- 35.Mehta HM, Malandra M, Corey SJ. G-CSF and GM-CSF in neutropenia. J Immunol (2015) 195(4):1341–9. 10.4049/jimmunol.1500861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol (2007) 17(7):318–24. 10.1016/j.tcb.2007.07.004 [DOI] [PubMed] [Google Scholar]
- 37.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J (2002) 365(Pt 3):561–75. 10.1042/BJ20020508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cain DW, O’Koren EG, Kan MJ, Womble M, Sempowski GD, Hopper K, et al. Identification of a tissue-specific, C/EBPbeta-dependent pathway of differentiation for murine peritoneal macrophages. J Immunol (2013) 191(9):4665–75. 10.4049/jimmunol.1300581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J (1995) 14(9):1932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerzoni C, Bardini M, Mariani SA, Ferrari-Amorotti G, Neviani P, Panno ML, et al. Inducible activation of CEBPB, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood (2006) 107(10):4080–9. 10.1182/blood-2005-08-3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Pan J, Yao L, Wu L, Zhu J, Wang W, et al. Acute promyelocytic leukemia with a STAT5b-RARalpha fusion transcript defined by array-CGH, FISH, and RT-PCR. Cancer Genet (2012) 205(6):327–31. 10.1016/j.cancergen.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 42.Duprez E, Wagner K, Koch H, Tenen DG. C/EBPbeta: a major PML-RARA-responsive gene in retinoic acid-induced differentiation of APL cells. EMBO J (2003) 22(21):5806–16. 10.1093/emboj/cdg556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev (2000) 14(15):1920–32. 10.1101/gad.14.15.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe-Okochi N, Yoshimi A, Sato T, Ikeda T, Kumano K, Taoka K, et al. The shortest isoform of C/EBPbeta, liver inhibitory protein (LIP), collaborates with Evi1 to induce AML in a mouse BMT model. Blood (2013) 121(20):4142–55. 10.1182/blood-2011-07-368654 [DOI] [PubMed] [Google Scholar]
- 45.Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol (2006) 7(7):732–9. 10.1038/ni1354 [DOI] [PubMed] [Google Scholar]
- 46.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A (1997) 94(2):569–74. 10.1073/pnas.94.2.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity (2004) 21(6):853–63. 10.1016/j.immuni.2004.11.006 [DOI] [PubMed] [Google Scholar]
- 48.Dror N, Rave-Harel N, Burchert A, Azriel A, Tamura T, Tailor P, et al. Interferon regulatory factor-8 is indispensable for the expression of promyelocytic leukemia and the formation of nuclear bodies in myeloid cells. J Biol Chem (2007) 282(8):5633–40. 10.1074/jbc.M607825200 [DOI] [PubMed] [Google Scholar]
- 49.Burchert A, Cai D, Hofbauer LC, Samuelsson MK, Slater EP, Duyster J, et al. Interferon consensus sequence binding protein (ICSBP; IRF-8) antagonizes BCR/ABL and down-regulates bcl-2. Blood (2004) 103(9):3480–9. 10.1182/blood-2003-08-2970 [DOI] [PubMed] [Google Scholar]
- 50.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell (1996) 87(2):307–17. 10.1016/S0092-8674(00)81348-3 [DOI] [PubMed] [Google Scholar]
- 51.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol (2001) 19:623–55. 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- 52.Zhao J, Kong HJ, Li H, Huang B, Yang M, Zhu C, et al. IRF-8/interferon (IFN) consensus sequence-binding protein is involved in toll-like receptor (TLR) signaling and contributes to the cross-talk between TLR and IFN-gamma signaling pathways. J Biol Chem (2006) 281(15):10073–80. 10.1074/jbc.M507788200 [DOI] [PubMed] [Google Scholar]
- 53.Tsujimura H, Tamura T, Gongora C, Aliberti J, Reis e Sousa C, Sher A, et al. ICSBP/IRF-8 retrovirus transduction rescues dendritic cell development in vitro. Blood (2003) 101(3):961–9. 10.1182/blood-2002-05-1327 [DOI] [PubMed] [Google Scholar]
- 54.Masumi A, Tamaoki S, Wang IM, Ozato K, Komuro K. IRF-8/ICSBP and IRF-1 cooperatively stimulate mouse IL-12 promoter activity in macrophages. FEBS Lett (2002) 531(2):348–53. 10.1016/S0014-5793(02)03556-1 [DOI] [PubMed] [Google Scholar]
- 55.Tsujimura H, Tamura T, Kong HJ, Nishiyama A, Ishii KJ, Klinman DM, et al. Toll-like receptor 9 signaling activates NF-kappaB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J Immunol (2004) 172(11):6820–7. 10.4049/jimmunol.172.11.6820 [DOI] [PubMed] [Google Scholar]
- 56.Yanez A, Ng MY, Hassanzadeh-Kiabi N, Goodridge HS. IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood (2015) 125(9):1452–9. 10.1182/blood-2014-09-600833 [DOI] [PubMed] [Google Scholar]
- 57.Tsujimura H, Nagamura-Inoue T, Tamura T, Ozato K. IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J Immunol (2002) 169(3):1261–9. 10.4049/jimmunol.169.3.1261 [DOI] [PubMed] [Google Scholar]
- 58.Kurotaki D, Tamura T. Transcriptional and epigenetic regulation of innate immune cell development by the transcription factor, interferon regulatory factor-8. J Interferon Cytokine Res (2016) 36(7):433–41. 10.1089/jir.2015.0138 [DOI] [PubMed] [Google Scholar]
- 59.Schmidt M, Nagel S, Proba J, Thiede C, Ritter M, Waring JF, et al. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood (1998) 91(1):22–9. [PubMed] [Google Scholar]
- 60.Schmidt M, Hochhaus A, Nitsche A, Hehlmann R, Neubauer A. Expression of nuclear transcription factor interferon consensus sequence binding protein in chronic myeloid leukemia correlates with pretreatment risk features and cytogenetic response to interferon-alpha. Blood (2001) 97(11):3648–50. 10.1182/blood.V97.11.3648 [DOI] [PubMed] [Google Scholar]
- 61.Huang W, Bei L, Eklund EA. Fas-associated phosphatase 1 mediates Fas resistance in myeloid progenitor cells expressing the Bcr-abl oncogene. Leuk Lymphoma (2013) 54(3):619–30. 10.3109/10428194.2012.720979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W, Zhou W, Saberwal G, Konieczna I, Horvath E, Katsoulidis E, et al. Interferon consensus sequence binding protein (ICSBP) decreases beta-catenin activity in myeloid cells by repressing GAS2 transcription. Mol Cell Biol (2010) 30(19):4575–94. 10.1128/MCB.01595-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oda A, Wakao H, Fujita H. Calpain is a signal transducer and activator of transcription (STAT) 3 and STAT5 protease. Blood (2002) 99(5):1850–2. 10.1182/blood.V99.5.1850 [DOI] [PubMed] [Google Scholar]
- 64.Hjort EE, Huang W, Hu L, Eklund EA. Bcr-abl regulates Stat5 through Shp2, the interferon consensus sequence binding protein (Icsbp/Irf8), growth arrest specific 2 (Gas2) and calpain. Oncotarget (2016) 7(47):77635–50. 10.18632/oncotarget.12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y, Peng C, Abraham SA, Shan Y, Guo Z, Desouza N, et al. Arachidonate 15-lipoxygenase is required for chronic myeloid leukemia stem cell survival. J Clin Invest (2014) 124(9):3847–62. 10.1172/JCI66129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pang SH, Minnich M, Gangatirkar P, Zheng Z, Ebert A, Song G, et al. PU.1 cooperates with IRF4 and IRF8 to suppress pre-B-cell leukemia. Leukemia (2016) 30(6):1375–87. 10.1038/leu.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saberwal G, Horvath E, Hu L, Zhu C, Hjort E, Eklund EA. The interferon consensus sequence binding protein (ICSBP/IRF8) activates transcription of the FANCF gene during myeloid differentiation. J Biol Chem (2009) 284(48):33242–54. 10.1074/jbc.M109.010231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akbarzadeh S, Ward AC, McPhee DO, Alexander WS, Lieschke GJ, Layton JE. Tyrosine residues of the granulocyte colony-stimulating factor receptor transmit proliferation and differentiation signals in murine bone marrow cells. Blood (2002) 99(3):879–87. 10.1182/blood.V99.3.879 [DOI] [PubMed] [Google Scholar]
- 69.Tian SS, Lamb P, Seidel HM, Stein RB, Rosen J. Rapid activation of the STAT3 transcription factor by granulocyte colony-stimulating factor. Blood (1994) 84(6):1760–4. [PubMed] [Google Scholar]
- 70.Zhong Z, Wen Z, Darnell JE, Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science (1994) 264(5155):95–8. 10.1126/science.8140422 [DOI] [PubMed] [Google Scholar]
- 71.Spiekermann K, Biethahn S, Wilde S, Hiddemann W, Alves F. Constitutive activation of STAT transcription factors in acute myelogenous leukemia. Eur J Haematol (2001) 67(2):63–71. 10.1034/j.1600-0609.2001.t01-1-00385.x [DOI] [PubMed] [Google Scholar]
- 72.Benekli M, Xia Z, Donohue KA, Ford LA, Pixley LA, Baer MR, et al. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood (2002) 99(1):252–7. 10.1182/blood.V99.1.252 [DOI] [PubMed] [Google Scholar]
- 73.Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB, Tweardy DJ. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood (2011) 117(21):5701–9. 10.1182/blood-2010-04-280123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eiring AM, Kraft IL, Page BD, O’Hare T, Gunning PT, Deininger MW. STAT3 as a mediator of BCR-ABL1-independent resistance in chronic myeloid leukemia. Leuk Suppl (2014) 3(Suppl 1):S5–6. 10.1038/leusup.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X, Wrzeszczynska MH, Horvath CM, Darnell JE, Jr. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol Cell Biol (1999) 19(10):7138–46. 10.1128/MCB.19.10.7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schuringa JJ, Timmer H, Luttickhuizen D, Vellenga E, Kruijer W. c-Jun and c-Fos cooperate with STAT3 in IL-6-induced transactivation of the IL-6 respone element (IRE). Cytokine (2001) 14(2):78–87. 10.1006/cyto.2001.0856 [DOI] [PubMed] [Google Scholar]
- 77.Rangatia J, Vangala RK, Singh SM, Peer Zada AA, Elsasser A, Kohlmann A, et al. Elevated c-Jun expression in acute myeloid leukemias inhibits C/EBPalpha DNA binding via leucine zipper domain interaction. Oncogene (2003) 22(30):4760–4. 10.1038/sj.onc.1206664 [DOI] [PubMed] [Google Scholar]
- 78.Zhang H, Nguyen-Jackson H, Panopoulos AD, Li HS, Murray PJ, Watowich SS. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood (2010) 116(14):2462–71. 10.1182/blood-2009-12-259630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shah CA, Broglie L, Hu L, Bei L, Huang W, Dressler DB, et al. Stat3 and CCAAT enhancer-binding protein beta (C/ebpbeta) activate Fanconi C gene transcription during emergency granulopoiesis. J Biol Chem (2018). 10.1074/jbc.RA117.000528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee CK, Raz R, Gimeno R, Gertner R, Wistinghausen B, Takeshita K, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity (2002) 17(1):63–72. 10.1016/S1074-7613(02)00336-9 [DOI] [PubMed] [Google Scholar]
- 81.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity (2004) 20(2):153–65. 10.1016/S1074-7613(04)00022-6 [DOI] [PubMed] [Google Scholar]
- 82.Croker BA, Mielke LA, Wormald S, Metcalf D, Kiu H, Alexander WS, et al. Socs3 maintains the specificity of biological responses to cytokine signals during granulocyte and macrophage differentiation. Exp Hematol (2008) 36(7):786–98. 10.1016/j.exphem.2008.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Numata A, Shimoda K, Kamezaki K, Haro T, Kakumitsu H, Shide K, et al. Signal transducers and activators of transcription 3 augments the transcriptional activity of CCAAT/enhancer-binding protein alpha in granulocyte colony-stimulating factor signaling pathway. J Biol Chem (2005) 280(13):12621–9. 10.1074/jbc.M408442200 [DOI] [PubMed] [Google Scholar]
- 84.Kimura A, Kinjyo I, Matsumura Y, Mori H, Mashima R, Harada M, et al. SOCS3 is a physiological negative regulator for granulopoiesis and granulocyte colony-stimulating factor receptor signaling. J Biol Chem (2004) 279(8):6905–10. 10.1074/jbc.C300496200 [DOI] [PubMed] [Google Scholar]
- 85.Schuringa JJ, Wierenga AT, Kruijer W, Vellenga E. Constitutive Stat3, Tyr705, and Ser727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood (2000) 95(12):3765–70. [PubMed] [Google Scholar]
- 86.Panopoulos AD, Zhang L, Snow JW, Jones DM, Smith AM, El Kasmi KC, et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood (2006) 108(12):3682–90. 10.1182/blood-2006-02-003012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med (2012) 366(20):1905–13. 10.1056/NEJMoa1114885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs (2009) 18(1):45–56. 10.1517/13543780802565791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Su F, Ren F, Rong Y, Wang Y, Geng Y, Wang Y, et al. Protein tyrosine phosphatase Meg2 dephosphorylates signal transducer and activator of transcription 3 and suppresses tumor growth in breast cancer. Breast Cancer Res (2012) 14(2):R38. 10.1186/bcr3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Furtek SL, Backos DS, Matheson CJ, Reigan P. Strategies and approaches of targeting STAT3 for cancer treatment. ACS Chem Biol (2016) 11(2):308–18. 10.1021/acschembio.5b00945 [DOI] [PubMed] [Google Scholar]
- 91.Coffer PJ, Koenderman L, de Groot RP. The role of STATs in myeloid differentiation and leukemia. Oncogene (2000) 19(21):2511–22. 10.1038/sj.onc.1203479 [DOI] [PubMed] [Google Scholar]
- 92.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell (1998) 93(5):841–50. 10.1016/S0092-8674(00)81444-0 [DOI] [PubMed] [Google Scholar]
- 93.Gouilleux F, Pallard C, Dusanter-Fourt I, Wakao H, Haldosen LA, Norstedt G, et al. Prolactin, growth hormone, erythropoietin and granulocyte-macrophage colony stimulating factor induce MGF-Stat5 DNA binding activity. EMBO J (1995) 14(9):2005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Quelle FW, Wang D, Nosaka T, Thierfelder WE, Stravopodis D, Weinstein Y, et al. Erythropoietin induces activation of Stat5 through association with specific tyrosines on the receptor that are not required for a mitogenic response. Mol Cell Biol (1996) 16(4):1622–31. 10.1128/MCB.16.4.1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pallard C, Gouilleux F, Benit L, Cocault L, Souyri M, Levy D, et al. Thrombopoietin activates a STAT5-like factor in hematopoietic cells. EMBO J (1995) 14(12):2847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mui AL, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J (1996) 15(10):2425–33. [PMC free article] [PubMed] [Google Scholar]
- 97.Hou J, Schindler U, Henzel WJ, Wong SC, McKnight SL. Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity (1995) 2(4):321–9. 10.1016/1074-7613(95)90140-X [DOI] [PubMed] [Google Scholar]
- 98.Kieslinger M, Woldman I, Moriggl R, Hofmann J, Marine JC, Ihle JN, et al. Antiapoptotic activity of Stat5 required during terminal stages of myeloid differentiation. Genes Dev (2000) 14(2):232–44. 10.1101/gad.14.2.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kato Y, Iwama A, Tadokoro Y, Shimoda K, Minoguchi M, Akira S, et al. Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J Exp Med (2005) 202(1):169–79. 10.1084/jem.20042541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gupta K, Kuznetsova I, Klimenkova O, Klimiankou M, Meyer J, Moore MA, et al. Bortezomib inhibits STAT5-dependent degradation of LEF-1, inducing granulocytic differentiation in congenital neutropenia CD34(+) cells. Blood (2014) 123(16):2550–61. 10.1182/blood-2012-09-456889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Levy DE, Darnell JE, Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol (2002) 3(9):651–62. 10.1038/nrm909 [DOI] [PubMed] [Google Scholar]
- 102.Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cell (2000) 6(3):693–704. 10.1016/S1097-2765(00)00067-8 [DOI] [PubMed] [Google Scholar]
- 103.Liu S, Walker SR, Nelson EA, Cerulli R, Xiang M, Toniolo PA, et al. Targeting STAT5 in hematologic malignancies through inhibition of the bromodomain and extra-terminal (BET) bromodomain protein BRD2. Mol Cancer Ther (2014) 13(5):1194–205. 10.1158/1535-7163.MCT-13-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Snow JW, Abraham N, Ma MC, Abbey NW, Herndier B, Goldsmith MA. STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood (2002) 99(1):95–101. 10.1182/blood.V99.1.95 [DOI] [PubMed] [Google Scholar]
- 105.Skokowa J, Cario G, Uenalan M, Schambach A, Germeshausen M, Battmer K, et al. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat Med (2006) 12(10):1191–7. 10.1038/nm1474 [DOI] [PubMed] [Google Scholar]
- 106.Michl J, Zimmer J, Tarsounas M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J (2016) 35(9):909–23. 10.15252/embj.201693860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) (2014) 19:135–42. 10.1016/j.dnarep.2014.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science (2002) 297(5581):606–9. 10.1126/science.1073834 [DOI] [PubMed] [Google Scholar]
- 109.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer (2007) 7(8):573–84. 10.1038/nrc2167 [DOI] [PubMed] [Google Scholar]
- 110.Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. DNA interstrand cross-link repair requires replication-fork convergence. Nat Struct Mol Biol (2015) 22(3):242–7. 10.1038/nsmb.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Higgs MR, Reynolds JJ, Winczura A, Blackford AN, Borel V, Miller ES, et al. BOD1L is required to suppress deleterious resection of stressed replication forks. Mol Cell (2015) 59(3):462–77. 10.1016/j.molcel.2015.06.007 [DOI] [PubMed] [Google Scholar]
- 112.Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science (2011) 333(6038):84–7. 10.1126/science.1204258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kennedy RD, D’Andrea AD. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev (2005) 19(24):2925–40. 10.1101/gad.1370505 [DOI] [PubMed] [Google Scholar]
- 114.Bagby GC, Jr. Genetic basis of Fanconi anemia. Curr Opin Hematol (2003) 10(1):68–76. 10.1097/00062752-200301000-00011 [DOI] [PubMed] [Google Scholar]
- 115.Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood (2003) 101(3):822–6. 10.1182/blood-2002-05-1498 [DOI] [PubMed] [Google Scholar]
- 116.Quentin S, Cuccuini W, Ceccaldi R, Nibourel O, Pondarre C, Pages MP, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood (2011) 117(15):e161–70. 10.1182/blood-2010-09-308726 [DOI] [PubMed] [Google Scholar]
- 117.Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood (2013) 122(18):3206–9. 10.1182/blood-2013-06-507962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, Regairaz M, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell (2012) 11(1):36–49. 10.1016/j.stem.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Du W, Amarachintha S, Erden O, Wilson A, Pang Q. The Fanconi anemia pathway controls oncogenic response in hematopoietic stem and progenitor cells by regulating PRMT5-mediated p53 arginine methylation. Oncotarget (2016) 7(37):60005–20. 10.18632/oncotarget.11088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cerabona D, Sun Z, Nalepa G. Leukemia and chromosomal instability in aged Fancc−/− mice. Exp Hematol (2016) 44(5):352–7. 10.1016/j.exphem.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tischkowitz M, Ameziane N, Waisfisz Q, De Winter JP, Harris R, Taniguchi T, et al. Bi-allelic silencing of the Fanconi anaemia gene FANCF in acute myeloid leukaemia. Br J Haematol (2003) 123(3):469–71. 10.1046/j.1365-2141.2003.04640.x [DOI] [PubMed] [Google Scholar]
- 122.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature (2013) 493(7432):356–63. 10.1038/nature11863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet (2006) 38(11):1239–41. 10.1038/ng1902 [DOI] [PubMed] [Google Scholar]
- 124.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet (2007) 39(2):165–7. 10.1038/ng1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rafnar T, Gudbjartsson DF, Sulem P, Jonasdottir A, Sigurdsson A, Jonasdottir A, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet (2011) 43(11):1104–7. 10.1038/ng.955 [DOI] [PubMed] [Google Scholar]
- 126.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet (2010) 42(5):410–4. 10.1038/ng.569 [DOI] [PubMed] [Google Scholar]
- 127.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer (2011) 11(7):467–80. 10.1038/nrc3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell (2012) 22(1):106–16. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell (2011) 145(4):529–42. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet (2005) 14(5):693–701. 10.1093/hmg/ddi065 [DOI] [PubMed] [Google Scholar]
- 131.Konieczna I, Horvath E, Wang H, Lindsey S, Saberwal G, Bei L, et al. Constitutive activation of SHP2 in mice cooperates with ICSBP deficiency to accelerate progression to acute myeloid leukemia. J Clin Invest (2008) 118(3):853–67. 10.1172/JCI33742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Williams MD, Braun LA, Cooper LM, Johnston J, Weiss RV, Qualy RL, et al. Hospitalized cancer patients with severe sepsis: analysis of incidence, mortality, and associated costs of care. Crit Care (2004) 8(5):R291–8. 10.1186/cc2893 [DOI] [PMC free article] [PubMed] [Google Scholar]