Abstract
Background
Very little is known about influenza viruses circulating in the Democratic Republic of Congo (DRC). We aim to characterize genetically and antigenically Influenza A(H3N2) and A(H1N1)pdm09 viruses circulating in the country.
Methods
From August to December 2014, specimens were collected from patients with influenza like-illness (ILI) or severe acute respiratory infection (SARI) in various surveillance sites. Specimens were tested using real time reverse transcription polymerase chain reaction (RT-PCR) method for the detection of influenza viruses. Positive influenza samples with a cycle threshold (Ct) <30 were genetically and antigenically characterized.
Results
32 samples tested were found positive to influenza A with Ct <30. At CDC Atlanta, 28 out of 32 samples (88%) were tested positive for influenza A virus, including 26 seasonal influenza A viruses subtype H3N2 and 2 pandemic influenza A viruses subtype H1N1pdm 2009. The majority of influenza A(H3N2) viruses were antigenically related to the A/Switzerland/9715293/2013 vaccine virus, while two influenza A(H1N1)pdm09 isolates were antigenically characterized as A/California/07/2009-like. All A(H3N2) and A(H1N1)pdm09 virus isolates characterized were sensitive to oseltamivir and zanamivir.
Conclusion
Two genetically distinct influenza subtypes were co-circulating in the DRCongo. Effective measures against influenza have been suggested.
Keywords: Molecular, Analysis, Influenza viruses, DRCongo
1. Introduction
Influenza is a contagious respiratory disease caused by the influenza virus, a member of the Orthomyxoviridae family of RNA viruses which have a segmented genome. There are three types of influenza viruses (A, B and C), originally distinguished on the basis of their internal nucleoprotein and matrix proteins. Influenza virus A and B cause annual, recurrent outbreaks in tropical and sub-tropical countries leading to high morbidity and mortality [1], especially in children and the elderly. It is estimated that one million deaths each year are due virus infection [2]. According to the World Health Organization (WHO), about20%of all deaths among children aged <5 years are attributable to influenza-like illnesses (ILI) [3], and the majority of those deaths occur in Africa and Southeast Asia [3, 4].
Seasonal influenza viruses are a population of virus variants, mainly existing as a result of antigenic drift, the accumulation of point mutations in the hemagglutinin (HA) and neuraminidase (NA) surface glycoprotein due to the lack of virus RNA polymerase complex pro of reading activity [5]. The HA is known to be the main antigenic determinant of the host’s neutralizing immune response [6,7,8] and is under selective pressure for antigenic shift, the process by which new virus variants emerge from genetic reassortment among influenza viruses infecting human and animal populations. Antigenic shift results in virus evasion of the preexisting immunity of the host [9]. In addition, genetic mechanisms like nucleic acid substitution, gene insertion and deletion have been reported to contribute with influenza virus variability [9, 10].
Previous studies have shown that influenza is a major contributor of respiratory illness in tropical and subtropical regions worldwide, with more complex patterns of influenza seasonality in Africa and southern Asia [11, 12, and 13]. Tropical countries closer to the equator have year-round influenza activity without a distinct seasonal peak [14, 15, 16, 17]. However, South Africa, Zambia [18], and Senegal [19] seem to have well-defined patterns of seasonality similar to other countries with temperate climates [17]. Furthermore, Madagascar described a yearly circulation of influenza with two distinct peaks, one that corresponds to the northern hemisphere winter (January–March) and a second that corresponds to the southern hemisphere winter (June–October) [18]. Unfortunately, the limited data available from sub-Saharan Africa makes it difficult to comprehensively map and define the seasonality of influenza in the region. The seasonality of influenza coupled with virus genetic and antigenic variation present challenges to the selection of potential virus candidates to be used in the formulation of influenza vaccines. Timely and geographically comprehensive virologic surveillance involving genetic and antigenic analysis of circulating strains is critical to maintaining the effectiveness of the influenza vaccine [20].
Although some countries, such as Madagascar and Senegal, have led national influenza surveillance for many years [21, 22], several countries in the African continent began conducting influenza surveillance recently. In DRCongo, the influenza surveillance system based on laboratory analysis was introduced in 2008 and reported that influenza is present year-round associated with some peaks related to the rainy season [23]. Despite the increased interest on improving influenza surveillance in Africa, the available genetic and epidemiological data are still insufficient to understand disease dynamics in the region. Due to the limited knowledge on influenza virus dynamics in Africa and to contribute in the improvement of the DRC surveillance system, we sought to identify and characterize for the first time the influenza viruses circulating in the Democratic Republic of Congo during the period from August to December of 2014.
2. Materials and Methods
2.1. Ethical Considerations
Specimens and data were collected as part of national public health sentinel surveillance systems, which is considered a non research activity. Influenza sentinel surveillance protocols were adapted from World Health Organization (WHO) guidelines with support from the national influenza program at the DRCongo Ministry of health. This protocol was implemented as part of routine public health surveillance by the Ministry of Health and was therefore considered a service and not subject to human subjects review. Before collecting each specimen, physicians explained the purpose of the surveillance system. After that, patients could refuse to participate. Oral consent was documented in the patient’s study records. Specimens and data were processed anonymously.
2.2. Study area and Sample collection
Nasal, throat and nasopharyngeal swab samples from all enrolled patients presenting with severe acute respiratory infectious (SARI) or influenza-like-illness (ILI) were collected from various surveillance sites selected in DRC and sent to the Influenza National Reference Laboratory at INRB using 3-mL cryovials containing Viral Transport Medium (Copan diagnostics-Murrietaca-USA).
Throat and nasopharyngeal swab specimens were collected from the same patient; both swabs were placed in the same cryovial. The specimens were kept refrigerated at 4°C, until they were packaged using a triple packaging system at the sentinel site. The specimens were then sent in refrigerated cool boxes to the INRB within 72 hours after packaging, where they were divided into 3 aliquots. One aliquot was used for molecular analysis; two others were stored at −80°C for further analysis and safeguard.
2.3. Laboratory analysis
• RNA extraction
Ribonucleic acid (RNA) extractions were performed at INRB, after dividing each sample in three cryovials containing 1 ml aliquots. One aliquot was immediately used for RNA extraction, while the other two aliquots were stored at −80°C for future use. Nucleic acid was extracted from 140 μl of aliquot, using the QIAamp® Viral RNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Purified RNA was stored at −80°C in aliquots.
• Amplification by Reverse Transcription Polymerase Chain Reaction (RT-PCR)
The RT-PCR technique is used as a rapid and sensitive method for the detection of influenza viruses in clinical and virus isolate samples. All samples were screened for influenza A and B viruses by one step real- time reverse transcription polymerase chain reaction. The influenza A one–step real-time RT-PCR was performed using primers for matrix gene (forward: 5′-GACCRATCCTGTCACCTCTGAC-3′; reverse: 5′-AGGGCA TTYTGGACAAAKCGTCTA-3′; probe: 5′-FAM- TGCAGTCCTCGCTCACTGGGCACG-MGB- 3′) provided by the CDC in Atlanta, GA and according to the provided CDC protocol. Type A and B influenza matrix genes were amplified.
Reaction mixtures (25μL) were prepared using the Ag- Path-ID™ One-Step RT-PCR Kit (Ambion, USA) and contained final concentrations: 12.5 μl of 2X RT-PCR Buffer, 0.25 μl of 40 μM reverse primer, 0.25 μl of 40 μM forward primers, 0.25 μl of 10 μM of specific probe, 1μl of 25X RT–PCR enzyme (Taq DNA polymerase and reverse transcriptase) and 5 μl of nucleic acid. Amplification was performed on ABI7500 Fast Real-time PCR System (Applied Biosystems, USA) with the following cycling conditions: 40°C for 10 min, 95°C for 10 min and 95°C for 15s followed by 55°C for 60s (45 cycles). Each specimen was also tested for the human ribonuclease P gene (constitutive gene) as an internal control. Screening for influenza B was simultaneously done by Flu–B matrix gene specific primers and probe. The positive controls for each virus were made of RNA extracts from non infectious material for use with the CDC RT-PCR method for detection and characterization of human seasonal influenza viruses. Water was used as a negative control.
Subsequently, influenza A positive samples were tested to identify the influenza subtypes A(H3N2) or A(H1N1)pdm09 using a single one step real-time RT-PCR with six pairs of primers and probes for the specific detection of influenza A subtypes. The specific HA genes for each influenza A subtype were amplified: seasonal A(H1N1) and A(H3N2), pandemic A(H1N1)pdm09 and avian A(H5N1) and A(H7N9) according to CDC protocol [24]. Influenza isolates with a Cycle threshold (Ct <30) were frozen and shipped in dry ice to the CDC in Atlanta for genetic and antigenic characterization.
• Inoculation into Cell Culture
Among all 32 samples sent to the CDC Atlanta, 23 specimens contained sufficient volume to be inoculated into Madin Darby Canine Kidney (MDCK) or MDCK SIAT-1 cell culture.
• Hemagglutination and Neuraminidase Inhibition Assays
The antigenic analysis using the hemagglutination inhibition (HI) assay was performed on all virus isolates having sufficient hemagglutination (HA) titer. Viruses were tested by HI with post-infection ferret antisera to the vaccine strain as well as representative reference viruses from circulating genetic groups. Influenza A(H3N2) viruses were tested with 0.75% guinea pig red blood cells (RBCs) in the presence of 20 μM oseltamivir and A(H1N1)pdm09 viruses were tested with 0.5% turkey RBCs.
The neuraminidase inhibition assay was performed to assess the virus susceptibility to the NA inhibitors oseltamivir and zanamivir, as previously described [25].
2.4. Sequencing and Genetic Characterization
In collaboration with the INRB, selected influenza A(H3N2) and A(H1N1)pdm09 virus isolates from DRCongo were sent to CDC in Atlanta for sequencing and genetic characterization. The influenza genome was amplified with SuperScript III One-Step RT-PCR with Platinum Taq High Fidelity (Invitrogen) using the Uni/Inf primer set as previously described [26]. Indexed paired-end libraries were generated from the amplicons by utilizing Nextera XT Sample Preparation Kit (Illumina, San Diego, CA) according to manufacturer’s protocol. Libraries were sequenced on the Illumina MiSeqwith a MiSeq v2 300 cycle kit. Reads were assembled and consensus sequences were generated using Iterative Refinement Meta-Assembler (IRMA) [27].
Phylogenetic analyses were done to genetically characterize influenza virus isolates collected at the DRC and to compare them with the vaccine strains recommended by WHO during the 2014-2016 influenza seasons. First, reference tree data sets for A(H1N1)pdm09 and A(H3N2) viruses were created by selecting reference viruses from WHO vaccine selection meeting reports since 2012 and their HA nucleotide sequences were down loaded from the Global Initiative on Sharing Avian Influenza (GISAID) database [28], as shown in the Supplementary Table 1. In addition, GISAID was searched to download the HA gene sequences of A(H1N1)pdm09 and A(H3N2)virus isolates collected in Africa from August to December, 2014. Virus HA sequences from the DRC and other African countries were aligned against the A(H3N2) or A(H1N1)pdm09 WHO reference data sets using MUSCLE [29] and trimmed to the beginning of the mature HA protein gene sequence using BioEdit v7.2.5 [30].
The TREESUB phylogenetic program [31] was used to estimate maximum likelihood phylogenetic trees using RAxML and PAML, followed by branch annotation of amino acid substitutions. Briefly, stop codons were trimmed from the aligned sequences in BioEdit and the vaccine virus was placed at the top of the alignment to be used as the tree out group and reference sequence. The general time reversible + Γ(GTR+GAMMA) nucleotide substitution model was selected in RAxMLv.7.3.0f or tree inference [32]. Ancestral codon substitutions for each gene were estimated using base ml, as implemented in PAML [33], using the ML trees inferred. Non synonymous substitutions were then transcribed on to the consensus gene phylogenies and visualized in FigTree v1.4.2 [34]. Nucleotide sequences from DRCongo used in this study were entered into the GISAID EpiFlu database and accessions are provided in the Supplementary Table 2.
3. Results
From August to December 2014, 806 samples were collected in DRCongo from which 61 were positive for influenza A (Table 1, Figure 1). During this reporting period, no influenza B specimens were detected (Table 1). Only 32 of the 61 influenza A positive samples from different surveillance sites in DRC had a Ct < 30 and these samples were selected and shipped to CDC in Atlanta for antigenic and genetic characterization. Results from this study showed that among the 32 samples tested: 26 were influenza A(H3N2), 2 were influenza A(H1N1)pdm09, 2 had insufficient volume for testing and 2 samples were negative for influenza by RT-PCR, as shown in Table 2.
Table 1.
DRCongo samples analyzed for influenza by RT-PCR, August to December 2014
| Month | Number of Negative Samples | Number of Positive Samples | Total | A(H1N1)pdm09 | A(H3N2) | A/unsubtypable |
|---|---|---|---|---|---|---|
| August | 118 | 1 | 119 | 1 | ||
| September | 160 | 0 | 160 | |||
| October | 153 | 1 | 154 | 1 | ||
| November | 190 | 6 | 196 | 2 | 4 | |
| December | 124 | 53 | 177 | 40 | 13 | |
|
| ||||||
| 745 | 61 | 806 | 3 | 45 | 13 | |
Figure 1.
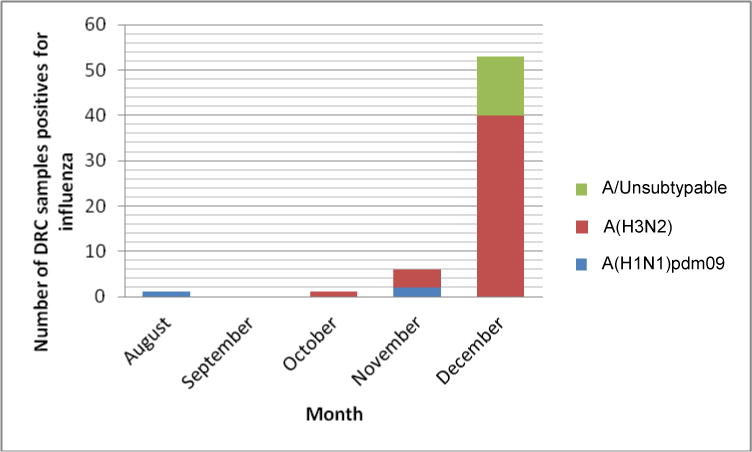
Number of DRCongo isolates positive for influenza and characterized at the CDC Atlanta. Thirty-two DRC samples which were positive for influenza A and had Ct <30 were selected for antigenic and genetic characterization.
Table 2.
Samples from DRCongo characterized at the CDC Atlanta
| Influenza Subtype | Number of sample | Characterization |
|---|---|---|
|
| ||
| A(H1N1)pdm09 | 2 | A/California/07/2009-likeA(H1N1)pdm09 |
| 1 | Insufficient volume for testing | |
|
| ||
| A(H3N2) | 1 | A/Texas/50/2012-likeA(H3N2)GP |
| 3 | A/Texas/50/2012-likeA(H3N2) | |
| 12 | Influenza A(H3N2) | |
| 10 | Influenza A(H3) | |
| 1 | Insufficient volume for testing | |
|
| ||
| Negative | 2 | Negative by PCR for influenza |
• A(H3N2) influenza virus
The majority of A(H3N2) influenza viruses did not have sufficient HA titer to be antigenically characterized by HI. Either full HA sequencing or limited pyrosequencing was done on viruses with insufficient HA titer for HI testing for genetic characterization only. Limited pyrosequencing was also done on A(H3N2) viruses not recovered in cell culture. Among the four A(H3N2) viruses tested by HI, one was antigenically similar to the A/Texas/50/2012 vaccine virus, while three were poorly inhibited by ferret antisera raised against A/Texas/50/2012 (Table 2). The majority of influenza A(H3N2) viruses tested from the DRC were antigenically related to the A/Switzerland/9715293/2013 vaccine virus. All A(H3N2) viruses tested from DRC were sensitive to oseltamivir and zanamivir.
The A(H3N2) phylogenetic tree was divided into 7 genetic groups based on shared amino acid (AA) changes when compared to the previous vaccine strain A/Perth/16/2009 (data not shown). Genetic analysis shows that genetic group 3 (V223I) was further divided into subgroups 3A (N144D, N145S, D487N), 3B (N145S, A198S, N312S, D487N) and 3C (A198S, N312S). Viruses from group 3C (S45N, T48I) diverged into genetic subgroups 3C.1, 3C.2 (Q33R, N145S, N278K, D489N) and 3C.3 (Q33R, T128A, R142G, N145S, N278K). Figure 2 shows the A(H3N2) phylogenetic tree since the divergence of groups 3C.2 and 3C.3 with A/Texas/50/2012 vaccine virus as outgroup. The amino acid differences against A/Texas/50/2012 reference virus are shown in Table 3. Group 3C.2 was divided into two genetic subgroups: 3C.2a (L3I, N144S, F159Y, K160T, N225D, Q311H) and 3C.2b (with AA changes found in group 3C.2a, except for F159Y). Group 3C.3 was split into subgroups 3C.3a (A138S, F159S, N225D, K326R) and 3C.3b (E62K, K83R, N122D, L157S, R261Q, V347K). Sequencing and phylogenetic analysis of A(H3N2) isolates from DRC confirmed that these viruses belonged to genetic group 3C.2a (n=6) and 3C.3 (n=2), as depicted in Table 3 and Figure 2.
Figure 2.
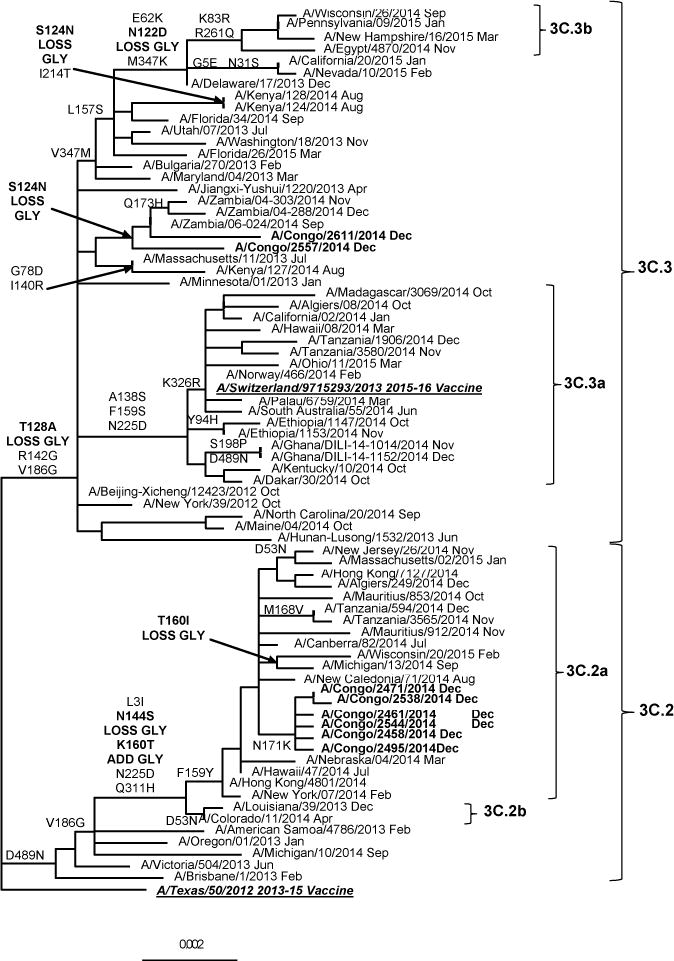
Representative hemagglutinin ML phylogenetic tree of A(H3N2) viruses collected since 2013. The A/Texas/50/2012 (underlined, bold) was used as reference in TREESUB to estimate ML phylogenetic trees via RAxML and PAML, and for automated annotation of amino acid substitutions in the nexus format. The consensus HA tree and the transcribed amino acid substitutions were visualized in FigTree. The scale bar represents the average number of nucleotide substitutions per site. Genetic groups 3C.2 and 3C.3, with the respective subgroups are represented in the tree. DRCongo isolates (bold) grouped with 3C.2a and 3C.3 reference viruses. Other African isolates clustered with the A/Switzerland/9715293/2013 (underlined, bold) vaccine virus from genetic group 3C.3a.
Table 3.
Representative H3 amino acid changes and genetic characterization of DRC isolates compared to A/Texas/50/2012 vaccine virus
| HA Group | Strain Name | Amino Acid Annotation and Numbering after Signal Peptidea | Number of AA Differences |
|||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 38 | 53 | 56 | 62 | 78 | 83 | 94 | 122 | 124 | 128 | 138 | 140 | 142 | 144 | 145 | 157 | 159 | 160 | 168 | 171 | 173 | 186 | 188 | 192 | 198 | 199 | 214 | 219 | 225 | 261 | 262 | 311 | 326 | 347 | 402 | 489 | 510 | 529 | |||
| 3C.1 | A/Texas/50/2012 (2013-15 Vaccine) | L | P | N | D | H | E | G | K | Y | N | S | N | A | I | R | N | N | L | F | K | M | N | Q | V | D | I | P | S | I | F | N | R | S | Q | K | V | V | D | G | V | |
| 3C.2a | A/Congo/2471/2014Dec | I | P/T | T | S | S | Y | T | K | G | S | S | D | H | N | 14 | ||||||||||||||||||||||||||
| A/Congo/2538/2014Dec | I | T | S | S | Y | K | G | S | S | D | H | N | 12 | |||||||||||||||||||||||||||||
| A/Congo/2461/2014Dec | I | T | S | S | Y | T | K | G | S | S | D | H | I | N | 14 | |||||||||||||||||||||||||||
| A/Congo/2544/2014Dec | I | T | S | S | Y | T | K | G | E | S | S | D | H | N | 14 | |||||||||||||||||||||||||||
| A/Congo/2458/2014Dec | I | T | S | S | Y | T | K | G | S | S | D | H | N | 13 | ||||||||||||||||||||||||||||
| A/Congo/2495/2014Dec | I | T | S | S | Y | T | K | G | S | S | D | H | N | 13 | ||||||||||||||||||||||||||||
| A/Algiers/249/2014Dec | I | T | S | S | Y | T | G | S | S | D | H | N | 12 | |||||||||||||||||||||||||||||
| A/Mauritius/853/2014Oct | I | T | S | S | Y | T | G | S | S | D | H | N | I | 13 | ||||||||||||||||||||||||||||
| A/Tanzania/594/2014Dec | I | T | S | S | Y | T | V | G | S | S | D | H | N | 13 | ||||||||||||||||||||||||||||
| A/Tanzania/3565/2014Nov | I | T | S | S | Y | T | V | G | S | S | D | H | N | 13 | ||||||||||||||||||||||||||||
| A/Mauritius/912/2014Nov | T | S | S | Y | T | G | S | S | D | Q | H | N | 12 | |||||||||||||||||||||||||||||
| 3C.3 | A/Congo/2611/2014Dec | S | N | A | G | S | G | S | S | E | 9 | |||||||||||||||||||||||||||||||
| A/Congo/2557/2014Dec | I | N | N | A | G | S | G | V | S | S | 10 | |||||||||||||||||||||||||||||||
| A/Zambia/04-303/2014Nov | N | A | G | S | H | G | S | S | 8 | |||||||||||||||||||||||||||||||||
| A/Zambia/04-288/2014Dec | N | A | G | S | H | G | S | S | 8 | |||||||||||||||||||||||||||||||||
| A/Zambia/06-024/2014Sep | N | A | G | S | G | S | S | 7 | ||||||||||||||||||||||||||||||||||
| A/Kenya/127/2014Aug | D | A | R | G | S | G | S | I/L | S | 9 | ||||||||||||||||||||||||||||||||
| 3C.3a | A/Switzerland/9715293/2013 (2015 Vaccine) | A | S | G | S | S | G | S | S | D | R | 10 | ||||||||||||||||||||||||||||||
| A/Madagascar/3069/2014Oct | H/Y | A | S | G | S | S | G | S | S | D | R | 11 | ||||||||||||||||||||||||||||||
| A/Algiers/08/2014Oct | A | S | G | S | S | G | S | D | R | 9 | ||||||||||||||||||||||||||||||||
| A/Tanzania/1906/2014Dec | H/Y | E/K | H | A | S | G | S | S | G | S | S | D | R | 13 | ||||||||||||||||||||||||||||
| A/Tanzania/3580/2014Nov | A | S | G | S | S | G | S | S | D | N | R | 11 | ||||||||||||||||||||||||||||||
| A/Ethiopia/1147/2014Oct | H | A | S | G | S | S | G | S | S | D | 10 | |||||||||||||||||||||||||||||||
| A/Ethiopia/1153/2014Nov | H | A | S | G | S | S | G | S | S | D | 10 | |||||||||||||||||||||||||||||||
| A/Ghana/DILI-14-1014/2014Nov | A | S | G | S | S | G | S | D | N | 9 | ||||||||||||||||||||||||||||||||
| A/Ghana/DILI-14-1152/2014Dec | A | S | G | S | S | G | S | D | N | 9 | ||||||||||||||||||||||||||||||||
| A/Dakar/30/2014Oct | A | S | G | S | S | G | S | S | D | 9 | ||||||||||||||||||||||||||||||||
| 3C.3b | A/Egypt/4870/2014Nov | K | R | D | A | G | S | S | G | S | S | Q | K | 12 | ||||||||||||||||||||||||||||
Only amino acid differences found in virus isolates from Africa are shown
• A(H1N1)pdm09 influenza viruses
Two influenza A(H1N1)pdm09 isolates from the DRCongo were antigenically characterized as A/California/07/2009-like, the recommended A(H1N1)pdm09 component of the northern and southern hemisphere influenza vaccine formulations since 2009. The A(H1N1)pdm09 phylogenetic tree was divided into 9 major genetic groups (data not shown). The majority of the A(H1N1)pdm09 viruses isolated worldwide in 2014 belonged to genetic group 6. Genetic group 6 (D97N, S185T, S203T, E374K, S451N) had diverged into subgroups 6A (H138R, V249L), 6B (K163Q, A256T, K283E, E499K) and 6C (V2341, K283E, E499K). The A(H1N1)pdm09 DRC isolates (n=2) were classified as group 6B viruses and are represented in Table 4. All A(H1N1)pdm09 viruses tested from DRC were sensitive to oseltamivir and zanamivir.
Table 4.
Representative H1pdm09 amino acid changes and genetic characterization of DRC isolates compared to A/California/07/2009 vaccine virus
| HA Group | Strain Name | Amino Acid Annotation and Numbering after Signal Peptidea | Total Number of AA Differences |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30 | 48 | 83 | 84 | 97 | 116 | 163 | 183 | 185 | 186 | 195 | 203 | 222 | 223 | 234 | 256 | 257 | 283 | 321 | 361 | 374 | 379 | 401 | 451 | 496 | 499 | |||
| A/California/07/2009Vaccine | V | A | P | S | D | I | K | S | S | A | A | S | D/G | Q/R | V | A | M | K | I | Y | E | V | E | S | N | E | ||
| 6B | A/Congo/2278/2014Nov | S | N | N | Q | T | V | T | D | Q | T | E | V | K | N | T | K | 16 | ||||||||||
| A/Congo/2275/2014Nov | F | N | Q | T | T | D | Q | T | E | V | K | D | K | 13 | ||||||||||||||
| A/Ethiopia/63/2014Nov | S | N | Q | T | T | D | Q | T | E | A/V | K | N | K | 14 | ||||||||||||||
| A/Ethiopia/66/2014Nov | P | S | N | Q | T | T | D | Q | T | E | V | F/Y | K | N | K | 15 | ||||||||||||
| A/Ethiopia/1149/2014Oct | P | S | N | Q | T | T | D | Q | T | E | V | K | N | K | 14 | |||||||||||||
| A/Egypt/4372/2014Oct | S | N | Q | P/S | T | T | D | Q | T | E | V | K | N | K | 14 | |||||||||||||
| A/Egypt/4758/2014Oct | S | N | Q | T | T | Q | T | E | V | K | A | N | K | 13 | ||||||||||||||
| A/South Africa/3626/2013Jun | S | N | Q | T | T | D | Q | T | E | V | K | N | K | 15 | ||||||||||||||
| 6C | A/Ghana/FS-14-1022/2014Nov | S | N | M | T | T | T | D | Q | I | T | E | V | K | N | K | 15 | |||||||||||
| A/Ghana/FS-14-1025/2014Nov | A | S | N | M | T | T | T | D | Q | I | E | V | K | N | K | 15 | ||||||||||||
| A/Ghana/FS-14-1069/2014Dec | A | S | N | M | T | T | T | D | Q | I | V | E | V | K | N | K | 16 | |||||||||||
| A/Ghana/Dili-0567/2014Jun | A | S | N | M | T | T | T | D | Q | I | V | E | V | K | N | K | 16 | |||||||||||
Only amino acid differences found in virus isolates from Africa are shown
4. Discussion
The main objective of this study was to identify and characterize influenza viruses circulating in the Democratic Republic of Congo in 2014. To the best of our knowledge, this kind of work has never been done in the DRCongo, while other African countries like Madagascar, Morocco and Egypt, have worked on the improvement of their surveillance system capacity to understand the molecular epidemiology and disease burden caused by circulating influenza viruses [35].
Our findings showed that the influenza activity in DRCongo increased during the month of November of 2014, reaching its peak on December the same year. Of note is that influenza activity was concomitant with the September - April rainy season in DRC, as previously described for the 2009-2011 seasons [36]. Similar results were found in Senegal (Dakar), Côte d’Ivoire (Abidjan), and Niger (Niamey) probably because of lower temperatures, as described elsewhere for Niger in 2010 [36]. This same pattern has also been observed in the north eastern part of tropical Brazil (Fortaleza) [37].
A total of 32 virus samples collected at the DRC from August to December of 2014 were shipped to the CDC in Atlanta, from which 20 were genetically characterized by either full genome sequencing or limited pyrosequencing. Limited pyrosequencing was done in A(H3N2) virus isolates which had insufficient HA titer for HI testing or when viruses were not recovered in cell culture.
Sixteen of the A(H3N2) viruses isolated at the DRC belonged to genetic group3C.2a and two isolates to group 3C.3. Four of these isolates were antigenically similar to the A/Switzerland/9715293/2013 virus, the recommended A(H3N2) vaccine component during the 2015-2016 influenza season. In 2013, the majority of A(H3N2) viruses tested was antigenically similar to the A/Texas/50/2012 vaccine virus. After the WHO northern hemisphere vaccine selection meeting in February 2014, antigenic drift was detected in viruses from genetic group 3C, giving rise to subgroups 3C.2a and 3C. 3a. Ferret antisera raised against A/Texas/50/2012 vaccine virus showed a reduction in HA titer, with the concomitant worldwide increase of 3C.2a and 3C.3a groups from 2014 to date. Despite the genetic diversity, the majority of 3C.2a and 3C.3a viruses were antigenically similar to the new A/Switzerland/9715293/2013 vaccine virus.
Other A(H3N2) HA sequences from African countries, including Algiers, Dakar, Madagascar, Ethiopia, Ghana and Tanzania, available from the GISAID Epiflu database, were classified as 3C.3a, and one isolate from Egypt was found to cluster with other 3C.3b viruses (figure 2). No DRC A(H3N2) isolates from genetic groups 3C.3a and 3C.3b were collected in 2014. However, similarly to the DRC, 3C.3 viruses were also found in Zambia and Kenya after GISAID EpiFlu database searching. Based on the close proximity of East African countries with 3C.3a viruses in circulation, we expect the further expansion of the 3C.3a group to the DRC.
Two A(H1N1)pdm09 isolates from the DRC were classified under genetic group 6B and were antigenically indistinguishable to the A/California/07/2009 virus, which remains the WHO A(H1N1)pdm09 influenza vaccine component since 2009. The A(H1N1)pdm09 HA sequences from Ethiopia and Egypt, also available from the GISAID Epiflu database, clustered with other 6B virus sequences as well. Interestingly, Ghana was the only African country found to have viruses from genetic group 6C in circulation since the summer of 2014 and the reason why further expansion was not observed at the time remains to be elucidated.
Ferret antisera raised against the A/California/07/2009 vaccine virus showed reduced HA titers with a small number of isolates during the 2014-2015 influenza season. These A(H1N1)pdm09 low-reactors often had amino acid changes in HA positions 153-157 and may have acquired these changes through cell culture, which is consistent with results obtained since May 2009. African isolates were not found to have amino acid changes in HA positions 153-157, confirming that no antigenic change has taken place, as established by the antigenic analysis data.
5. Conclusion
Influenza A(H3N2) and A(H1N1)pdm09 viruses have continued to circulate worldwide since their emergence and are expanding at high levels in most countries. Molecular surveillance year-round, especially in sub and tropical areas of Africa, is critical for improving vaccine strategies for the prevention and control of influenza illness. It is also important in contributing to the understanding of genetic variations and in the early detection of antigenic drift.
Supplementary Material
Figure 3.
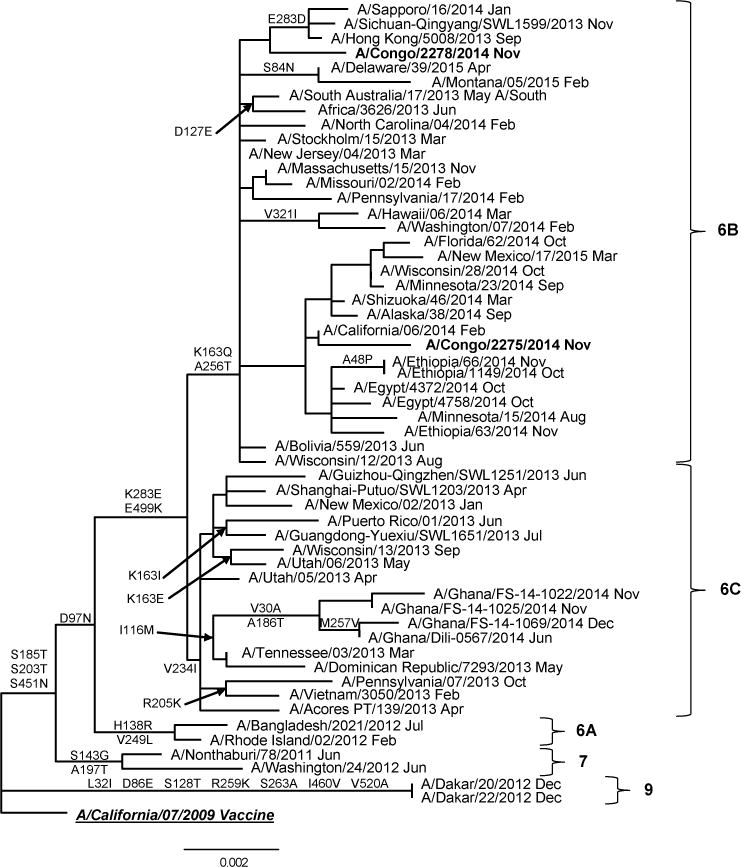
Representative hemagglutinin ML phylogenetic tree of A(H1N1)pdm09 viruses collected since 2009. The A/California/07/2009 (underlined, bold) was used as reference in TREESUB to estimate ML phylogenetic trees via RAxML and PAML, and for automated annotation of amino acid substitutions in the nexus format. The consensus HA tree and the transcribed amino acid substitutions were visualized in FigTree. The scale bar represents the average number of nucleotide substitutions per site. Genetic groups 6, 7 and 9 are depicted in the tree. DRCongo isolates (bold) clustered with viruses from genetic subgroup 6B.
References
- 1.Li CK, Choi BC, Wong TW. Influenza related deaths and hospitalizations in Hong Kong:a sub tropical area. Public Health. 2006;120(6):517–524. doi: 10.1016/j.puhe.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Pan American Health Organization. Technical Advisory group(TAG) Meeting on Vaccine Preventable-Diseases, 2004; Finalreport of the XVITAG meeting on vaccine preventable-diseases of the Pan American Health Organization; Mexico City, Mexico. November 3–5, 2004; Available at http://www.paho.org/English/AD/FCH/IM/TAG16_FinalReport_2004.pdf. [Google Scholar]
- 3.World Health Organization. Influenza-associated burden of disease in low and middle income countries – Examples from Kenya, Bangladesh and China. WHO Peer-reviewed literature. 2012 Available at http://www.who.int/influenza/surveillance_monitoring/updates/2012_05_10_update_GIP_peer_reviewed/en/
- 4.Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis. 2002;2:25–32. doi: 10.1016/s1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 5.Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature. 1982;296(5853):115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 6.Laver WG, Air GM, Webster RG. Mechanism of antigenic drift in influenza virus. Amino acid sequence changes in an antigenically active region of Hong Kong (H3N2) influenza virus hemagglutinin. J Mol Biol. 1981;145(2):339–361. doi: 10.1016/0022-2836(81)90209-6. [DOI] [PubMed] [Google Scholar]
- 7.Sleigh MJ, Both GW, Underwood PA, et al. Antigenic drift in the hemagglutinin of the Hong Kong influenza subtype: correlation of amino acid changes with alterations in viral antigenicity. J Virol. 1981;37(3):845–853. doi: 10.1128/jvi.37.3.845-853.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.G W, Sleigh MJ, Cox NJ, Kendal AP. Antigenic drift in influenza virus H3 hemagglutininfrom 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol. 1983;48(1):52–60. doi: 10.1128/jvi.48.1.52-60.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin YP, Gregory V, Bennett M, Hay A. Recent changes among human influenza viruses. Virus Res. 2004;103(1–2):47–52. doi: 10.1016/j.virusres.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Buonagurio DA, Nakada S, Parvin JD, Krystal M, Palese P, Fitch WM. Evolution of human influenza A viruses over 50years: rapid, uniform rate of change in NS gene. Science. 1986;232(4753):980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- 11.Shek LP, Lee BW. Epidemiology and seasonality of respiratory tract virus infections in the tropics. Paediatr Respir Rev. 2003;4(2):105–111. doi: 10.1016/s1526-0542(03)00024-1. [DOI] [PubMed] [Google Scholar]
- 12.Viboud C, Alonso WJ, Simonsen L. Influenza in tropical regions. PLoS Med. 2006;3(4):e89. doi: 10.1371/journal.pmed.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirve S, Newman LP, Paget J, Azziz-Baumgartner E, Fitzner J, Bha N, et al. Influenza Seasonality in the Tropics and Subtropics -When to Vaccinate? PLoS One. 2016;11(4):e0153003. doi: 10.1371/journal.pone.0153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beckett CG, Kosasih H, Ma’roef C, Listiyaningsih E, Elyazar IR, Wuryadi S, et al. Influenza surveillance in Indonesia: 1999–2003. Clin Infect Dis. 2004;39:443–449. doi: 10.1086/422314. [DOI] [PubMed] [Google Scholar]
- 15.Gachara G, Ngeranwa J, Magana JM, Simwa JM, Wango PW, Lifumo SM, et al. Influenza virus strains in Nairobi, Kenya. J Clin Virol. 2006;35(1):117–118. doi: 10.1016/j.jcv.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Hampson AW. Epidemiological data on influenza in Asian countries. Vaccine. 1999;17(Suppl 1):S19–S23. doi: 10.1016/s0264-410x(99)00100-0. [DOI] [PubMed] [Google Scholar]
- 17.Saha S, Chadha M, AlMamun A, Rahman M, Strum-Ramirez K, Chittaganpitch M, et al. Influenza seasonality and vaccination timing in the tropical and subtropical areas of southern and south-eastern Asia. Bull World Health Organ. 2014;92(5):318–333. doi: 10.2471/BLT.13.124412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gessner BD, Shindo N, Briand S. Seasonal influenza epidemiology in sub-Saharan Africa: a systematic review. Lancet Infect Dis. 2011;11(3):223–235. doi: 10.1016/S1473-. [DOI] [PubMed] [Google Scholar]
- 19.Mbayame Ndiaye N, Annick D, Ndiaye Kader, Sagna Monique, Gregory Victoria, Goudiaby Deborah, Hay Alan, Diop Ousmane M. Sentinel Surveillance for Influenza in Senegal, 1996–2009. J Infect Dis. 2012;206(Suppl 1):S129–135. doi: 10.1093/infdis/jis576. [DOI] [PubMed] [Google Scholar]
- 20.Cox NJ, Bai ZS, Kendal AP. Laboratory-based surveillance of influenza A(H1N1) and A(H3N2) viruses in 1980–81: antigenic and genomic analyses. Bull World Health Organ. 1983;61(1):143–152. [PMC free article] [PubMed] [Google Scholar]
- 21.Rabarijaona LP, Rakotondrarija NT, Rousset D, Soares JL, Mauclere P. Influenza epidemiologic and virologic surveillance in Antananarivo from 1995 to 2002. Arch Inst Pasteur Madagascar. 2003;69(1–2):20–26. [PubMed] [Google Scholar]
- 22.Razanajatovo NH, Richard V, Hoffmann J, et al. Viral etiology of influenza-like illnesses in Antananarivo, Madagascar, July2008 to June 2009. PLoS One. 2011;6(3):e17579. doi: 10.1371/journal.pone.0017579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muyembe Tamfum JJ, Nkwembe E, BiShamamba SK, Bankoshi F, Ilunga BK, Katz KA, et al. Sentinel surveillance for influenza-like illness, severe acute respiratory illness, and laboratory-confirmed influenza in Kinshasa, Democratic Republic of Congo, 2009–2011. J Infect Dis. 2012;206(Suppl 1):S36–40. doi: 10.1093/infdis/jis537. [DOI] [PubMed] [Google Scholar]
- 24.Enami K, Qiao Y, Fukuda R, Enami M. An influenza virus Temperature-sensitive mutant defective in the nuclear-cytoplasmic transport of the negative-sense viral RNAs. Virology. 1993;194(2):822–827. doi: 10.1006/viro.1993.1324. [DOI] [PubMed] [Google Scholar]
- 25.Okomo-Adhiambo M, Mishin VP, Sleeman K, Saguar E, Guevara H, Reisdorf E, et al. Standardizing the influenza neuraminidase inhibition assay among United States public health laboratories conducting virological surveillance. Antiviral Res. 2016;128:28–35. doi: 10.1016/j.antiviral.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Zhou B, Wentworth D. Influenza A virus molecular virology techniques. Methods Mol Biol. 2012;865:175–192. doi: 10.1007/978-1-61779-621-0_11. [DOI] [PubMed] [Google Scholar]
- 27.Shepard S, et al. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics. 2016(17):708. doi: 10.1186/s12864-016-3030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.www.gisaid.org
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high through put. Nucleic Acid Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 31.https://github.com/tamuri/treesub
- 32.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z. PAML4: phylogenetic analysis by maximum likelihood. Mol Bio Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 34.http://tree.bio.ed.ac.uk/software/figtree
- 35.World Health Organization. Report of the1st Africa Flu Alliance Meeting, Marrakesh, Morocco. 2010 Jun 3–4; Available at http://www.who.int/influenza/resources/documents/2010_06_3_afa_mtg_marrakesh_mor.
- 36.Jusot JF, Adamou L, Collard JM. Influenza transmission during a one year period (2009–2010) in a Sahelian city: low temperature plays a major role. Influenza Other Respir Viruses. 2012;6(2):87–89. doi: 10.1111/j.1750-2659.2011.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moura FE, Perdigao AC, Siqueira MM. Seasonality of influenza in the tropics: a distinct pattern in northeastern Brazil. Am J Trop Med Hyg. 2009;81(1):180–183. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


