Abstract
Microglia play essential roles in central nervous system (CNS) homeostasis and influence diverse aspects of neuronal function. However, the transcriptional mechanisms that specify human microglia phenotypes are largely unknown. We examined the transcriptomes and epigenetic landscapes of human microglia isolated from surgically resected brain tissue ex vivo and following transition to an in vitro environment. Transfer to a tissue culture environment results in rapid and extensive downregulation of microglia-specific genes that are induced in primitive mouse macrophages following migration into the fetal brain. Substantial subsets of these genes exhibit altered expression in neurodegenerative and behavioral diseases and are associated with non-coding risk variants. These findings reveal an environment-dependent transcriptional network specifying microglia-specific programs of gene expression and facilitate efforts to understand the roles of microglia in human disease.
Microglia are the resident macrophage population of the central nervous system (CNS) that perform functions required for typical development, as well as homeostasis, plasticity, immunity and repair (1–5). Dysregulation of microglia functions contribute to the pathogenesis of neurodegenerative diseases, including Alzheimer’s disease (AD) (6–8), Parkinson’s disease (PD) (9–12) and Huntington’s disease (HD) (13–15). In addition, microglia may play roles in neurodevelopmental and psychiatric diseases such as autism, schizophrenia (Scz) and depression (16, 17). Collectively, these observations provide a compelling incentive to define the mechanisms that control microglia functions and to investigate the potential to alter these functions for therapeutic purposes.
Microglia possess distinctive ontogeny, morphology, gene expression patterns, and functional characteristics and account for 5–20% of all glial cells, depending on brain regions. Studies in mice indicate that microglia are derived from yolk sac progenitor cells that populate the brain early in development (18, 19). Microglia share many traits with other subsets of resident tissue macrophages, including dependence on the Csf1 receptor for differentiation and survival (18, 20), a requirement for PU.1 as an essential lineage-determining transcription factor (LDTF) (21, 22), highly efficient phagocytic and tissue debris clearance abilities (2, 5), and the ability to quickly trigger an inflammatory response following detection of pathogens or tissue damage (23).
Microglia also express hundreds of genes at higher levels than other macrophage subsets (24–26) that may be influenced by factors present in the CNS microenvironment (25, 27), suggesting mechanisms by which the brain environment influences the activity and functions of one of its primary support cells. As microglia participate in the developmental refinement of synaptic networks (28) and elaborate neuromodulatory factors for adult motor learning (29), alterations in the brain microenvironment may lead to pathogenic programs of gene expression in microglia.
Insights into mechanisms that control the development and function of specific cell types can be obtained by analyses of their enhancer repertoires and corresponding epigenetic landscapes (30). Microglial enhancers are primed by relatively simple combinations of LDTFs (31, 32) to become sites of activity of broadly expressed signal-dependent transcription factors (SDTFs). These enhancers thereby integrate cellular signaling pathways with DNA. Within cell-specific enhancers, DNA sequences provide information about the identities of key LDTFs and the activity states of signaling pathways that control the functions of SDTFs (31, 33–35). Furthermore, the locations and sequence content of enhancers provide insights into how non-coding genetic variation may affect gene expression and cellular phenotypes (32, 36–38).
Human microglia transcriptomes
Microglia were isolated from brain tissue resected for treatment of epilepsy, brain tumors or acute ischemic events in 19 individuals. Patient characteristics, medications, and assays performed for each sample are provided in Table S1. For patients with epilepsy, the resected tissue provided did not include the brain area displaying the strongest epileptogenic activity. Although most specimens from individuals with epilepsy exhibited some degree of cortical dysplasia, three samples were determined to be histologically normal. For patients with brain tumors (8), samples were considered to be tumor negative by pathological examination. The two brain samples resected to treat acute ischemic injuries were from individuals who had no prior evidence of brain pathology. As judged by neuropathological diagnostic evaluation, none of the samples displayed evidence of pathological microgliosis (i.e. loss of tiled distribution, process retraction and enlarged cell bodies) despite the occasional presence of reactive cells (Fig. S1).
Tissues were immediately placed on ice in the operating room and processed for microglia isolation within a few hours of surgical resection (25). A microglia-rich cell fraction was isolated by Percoll gradient centrifugation and further purified by FACS, with final gating on live-CD11b+CD45LowCD64+CX3CR1High single cells (Fig. S2A). This strategy excluded cells expressing moderate to high levels of CD45, which were present in variable amounts depending on the specific sample (Fig. S2A). Microglia were then triaged to specific analysis workflows depending on yields and depth of existing data sets (Table S1). For five individuals, RNA-seq was also performed from a portion of the intact cortical brain tissue used to isolate microglia (hereafter referred to as “cortex”).
Isolated microglia exhibited a high degree of viability and RNA integrity. In one case, brain tissue was simultaneously obtained from the occipital and parietal lobes of the same individual, enabling independent isolation of microglia from each region. RNA expression levels for these two samples exhibited a Pearson correlation coefficient of 0.929 (Fig. S3A). In a second case, an individual underwent sequential surgical resections approximately one year apart. The RNA expression levels in microglia obtained from this individual at these two time points exhibited a Pearson correlation coefficient of 0.905 (Fig. S3B). On the basis of these observations, the variance in gene expression noted in these samples likely overestimates the variance in the isolation of microglia and implies a high degree of precision in the RNA-Seq data set. Comparisons of the RNA-seq profiles of any two individuals for genes expressed > 2 log2 transcripts per million (TPM) yielded Pearson correlation coefficients of approximately 0.8, as exemplified by a comparison of microglia isolated from a 1-year-old female patient diagnosed with atypical rhabdoid tumor to microglia isolated from a 7-year-old female patient with epilepsy (Fig. S3C). When all transcripts were considered (i.e., including genes expressed at ≤ 2 log2 TPM), Pearson correlation coefficients were in the range of 0.94 to 0.98 (Fig. S3D), indicating no substantial outliers among samples. RNA-Seq data indicated low to absent levels of neuronal, astrocyte, oligodendrocyte and endothelial-specific genes, confirming insignificant levels of contaminating cells (Fig. S2B, Table S2).
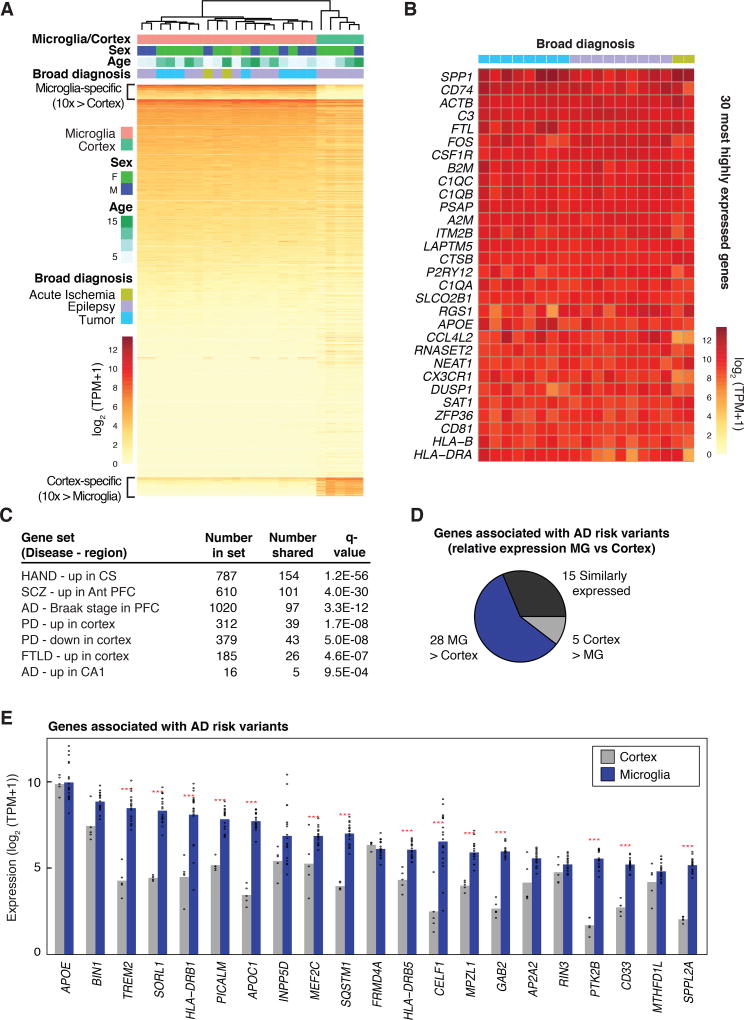
Hierarchical clustering of expressed genes from all microglia samples in comparison to cortex is illustrated in Figure 1A. While there are likely to be alterations in gene expression related to disease, no strong patterns emerged that were related to diagnosis or age (Fig. 1A), perhaps due in part to exclusion of most reactive cells at the time of sorting (Fig. S2A). A small set of genes exhibited significant sex-specific differences in gene expression, and these were mostly related to X- and Y-linked genes (Fig. S4). Relative expression values for the 30 most highly expressed genes across each of the patients are illustrated in Figure 1B, indicating substantial individual variation for some transcripts. This analysis confirmed that human microglia expressed very high levels of numerous genes associated with regulation of microglia ramification and motility, including the fractalkine receptor CX3CR1, purinergic receptor P2RY12, and genes involved in synaptic remodeling, such as complement factors C3, C1QA, C1QB, and C1QC (Table S2).
Figure 1. Human microglia transcriptomes.
(A) Heat map of mRNA expression values determined in microglia isolated from 19 individuals and in cortex from 5 of these individuals. 881 genes exhibiting > 10-fold higher average expression in microglia compared to cortex are indicated at the top of the heat map. (B) Individual variation in gene expression of the 30 most highly expressed genes in human microglia. (C) Enrichment of the 881 microglia-enriched genes upregulated in HIV-associated neurocognitive disorder (HAND) in centrum semiovale (CS) (48), upregulated in schizophrenia (SCZ) in the anterior prefrontal cortex (PFC) (49), positively correlated with Braak stage of AD in PFC (50), upregulated or downregulated in PD in cortex (51), upregulated in frontotemporal lobar degeneration (FTLD) in frontal cortex (52), or upregulated in AD in the CA1 region of the brain (53). (D) Pie chart depicting the relative expression of genes associated with AD risk variants in brain cortex and microglia. (E) Combined bar graphs and dot plots of the TPM expression levels of the 21 most highly expressed genes associated with AD risk variants in microglia and cortex. Red stars indicate significant differential expression (> 2-fold, FDR 0.05).
Nearly all studies of differentially regulated genes in human neurodegenerative or behavioral disorders are based on RNA profiling of intact tissues. As gene expression profiles for individual cell types in the human brain are poorly characterized, it is difficult to assign cell types responsible for disease-related changes. Using a stringent cutoff of 10-fold increased expression relative to cortex tissue at a false discovery rate (FDR) of 0.05, we defined a microglial gene signature in the human brain comprising 881 transcripts (Fig. 1A, Table S2). We then intersected this gene list with 46 publicly available data sets for differentially regulated genes in neurodegenerative and behavioral disorders derived from microarray or RNA-seq analysis of intact tissue (39–53). Two-sided Fisher’s exact tests revealed significant enrichment or depletion of microglia signature genes in 28 of the 46 data sets (Table S3). For example, hundreds of genes in the microglia gene signature exhibited significant overlaps with sets of genes that were positively correlated with Braak stage in AD, were upregulated in frontotemporal lobar degeneration (FTLD) in frontal cortex, were upregulated in HIV-associated neurocognitive disorder (HAND) or were upregulated in Scz (Fig. 1C). Interestingly, different subsets of the microglia signature gene set were upregulated or downregulated in cortex in PD (Fig. 1C).
Characterization of the microglia transcriptome, in conjunction with parallel whole brain cortical gene expression profiling, also enabled evaluation of the relative expression levels of genes associated with AD, PD, multiple sclerosis (MS) and Scz risk alleles (54). Many of these genes were preferentially expressed in microglia, with the relative proportion varying by disorder (Fig. 1D and Fig. S5). With respect to AD, for example, expression for 28 of 48 AD genes was higher in microglia, including TREM2, SORL1, INPP5D, MEF2C and CD33 (Figure 1E). For MS, 48 of 81 genes associated with risk alleles were preferentially expressed in microglia (Fig. S5). In contrast, approximately one third of the genes associated with PD and Scz risk alleles exhibited preferential expression in microglia (Fig. S5). As the risk alleles for these disorders are largely non-overlapping, these findings suggest different roles for microglia in each disorder context.
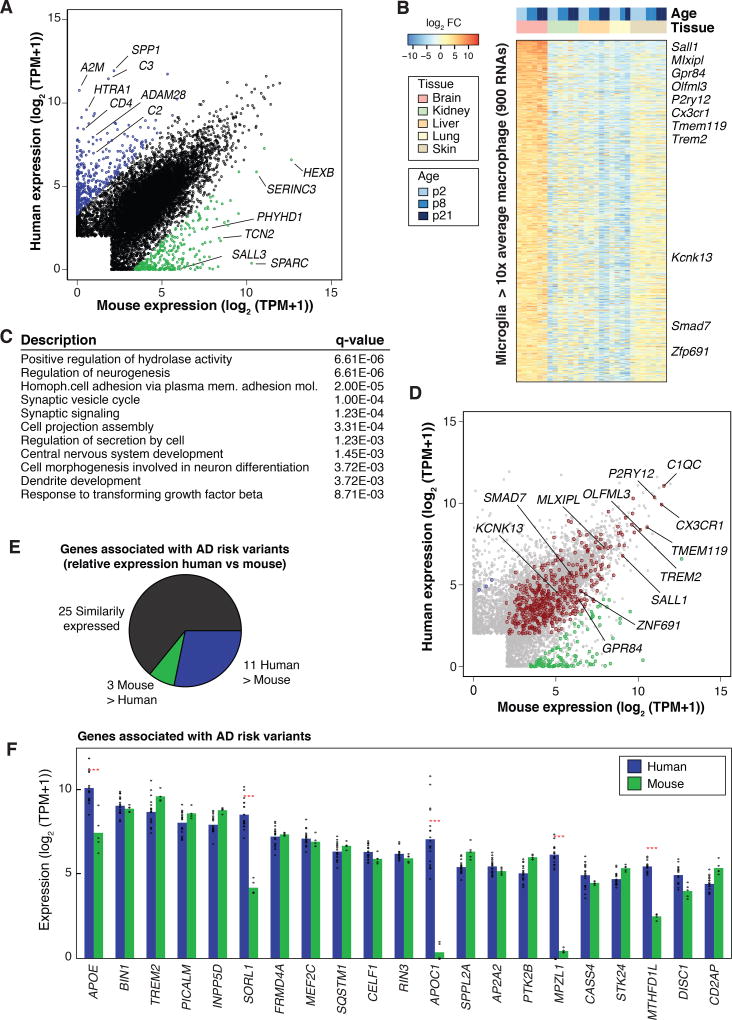
The extent to which findings in mice and humans are similar can vary depending on the system evaluated (55). We compared average transcript levels of human microglia mRNAs with orthologous mRNAs microglia isolated from 7- to 10-week-old male C57BL/6 mice using an identical protocol (25) (Fig. 2A and S6, Table S4 and S5). Overall, the microglia transcriptome comparison between the two species revealed overall similarities in patterns of gene expression (Pearson’s r = 0.806), with the majority of orthologous genes pairs (13,253 of 15,768) expressed within a 4-fold range. Nonetheless, this analysis also revealed genes with species-specific bias in expression magnitude. At a cutoff of 10-fold difference and an FDR of 0.05, 400 orthologous mRNAs were preferentially expressed in human microglia and 293 mRNAs were more highly expressed in mouse microglia (Fig. 2A). Notable examples included higher expression in human microglia of regulators of the complement system (e.g., C2, C3, VSIG4, SERPING1) and genes involved in brain structure development, including SYNDIG1, GLDN, CTTNBP2, and ROBO3. In contrast, genes more highly expressed in mouse microglia included Hexb, Sparc, and Sall3. Of the subset of these genes that also exhibited microglia-specific expression, a similar species-specific pattern of expression was observed in the intact cortical tissue used for microglia isolation, indicating that these differences were not a consequence of the isolation procedure (Fig. S7).
Figure 2. Comparison of human and mouse microglia transcriptomes.
(A) Scatter plot depicting mRNA expression levels of mouse and human genes with one-to-one orthologs, highlighting significantly differentially expressed genes in human (blue) and mouse microglia (green) (> 10-fold, FDR 0.05). (B) Identification of a mouse microglia gene signature consisting of 900 mRNAs expressed > 10-fold higher in microglia (FDR 0.05) compared to the average expression values of macrophages from skin, lung, liver, and kidney. (C) Functional annotations of the microglia gene signature. (D) Scatter plot depicting mRNAs expression levels of human and mouse microglia with orthologs intersected with the microglia gene signature expression profile from (B). Genes with conserved expression are depicted in red, genes more highly expressed in human are in blue, and genes more highly expressed in mice are in green. (E) Pie chart showing relative expression of genes associated with AD risk alleles in human and mouse microglia. (F) Combined bar graphs and dot plots illustrating the expression of the top 21 most abundant genes associated with AD risk variants in human and mouse microglia (average TPM-rank ordered). Red stars indicate significant differential expression (> 2-fold, FDR 0.05).
We compared the levels of mRNA transcripts in C57BL/6 microglia with different resident tissue macrophage populations (26, 56) and observed 900 mRNAs with levels of expression in microglia that were 10-fold greater than the integrated mean of expression levels in the other macrophage subsets at a FDR of 0.05 (Fig. 2B). This definition captures a mouse microglia gene signature set that is larger but mostly inclusive of previously described microglia signature sets (27, 57, 58). Gene ontology analysis of this extended microglia gene signature returned significant functional enrichment for CNS-related categories such as regulation of neurogenesis, synaptic signaling, CNS development, response to transforming growth factor beta (TGF-β), and regulation of neurotransmitter transport (Fig. 2C). RNA-seq data sets are not available for a sufficient number of human resident macrophage populations to generate a corresponding human microglia signature gene list. However, the relative expression levels of the human and mouse orthologous genes corresponding to the mouse microglia signature genes revealed that ~53% (477 of 900) of the genes that were highly specific to microglia in the mouse were expressed at comparable levels in human microglia (Fig. 2D).
Comparing the expression of genes linked to human neurodegenerative and behavioral disorders, of the 39 AD-associated and expressed genes, 11 were higher in human microglia whereas three were increased in mouse microglia (Fig. 2E). Interestingly, of the top 25 most expressed, two genes, APOC1 and MPZL1, were essentially not expressed in C57BL/6 microglia; the expression of a third one, SORL1, was nearly 20 times lower in mouse than human (Fig. 2F). PD-associated genes also displayed similar human vs mouse distributions (Fig. S8). PD-associated genes SNCA, LRRK2, CD46, and CXCL12 were expressed to a greater extent in human microglia than mouse microglia. Corresponding comparisons for all expressed genes linked to AD, PD, MS, and Scz are illustrated in Figure S8. Collectively, these results highlight human vs mouse similarities and differences in the expression profile of core microglia genes.
Enhancer landscape of human microglia
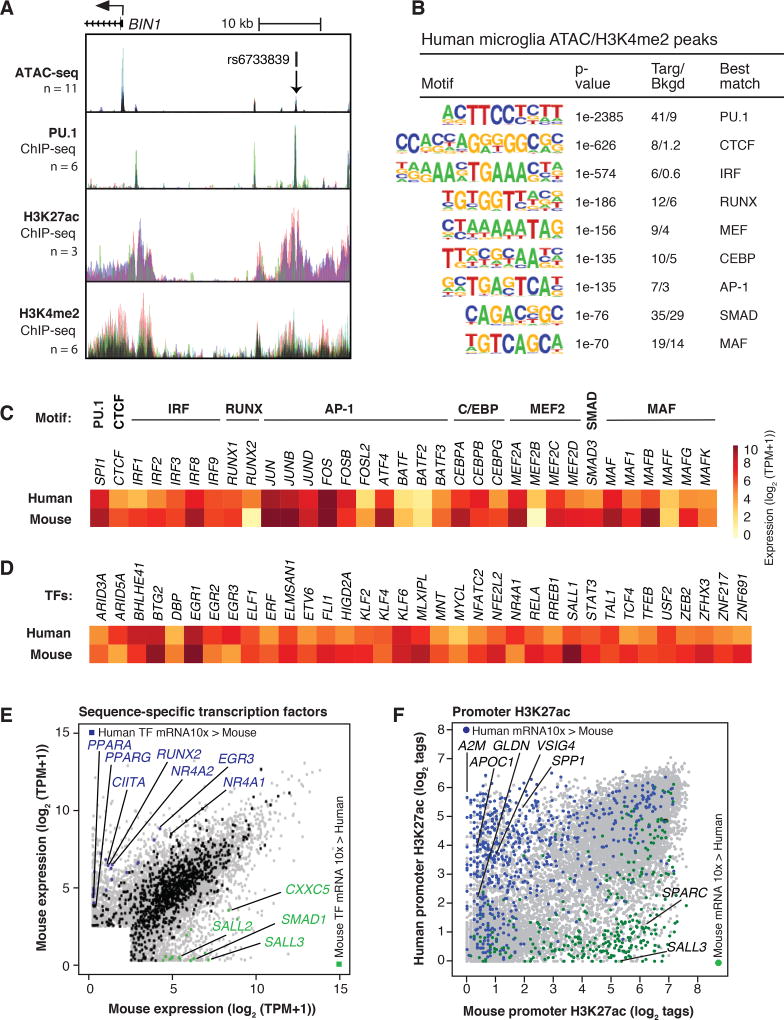
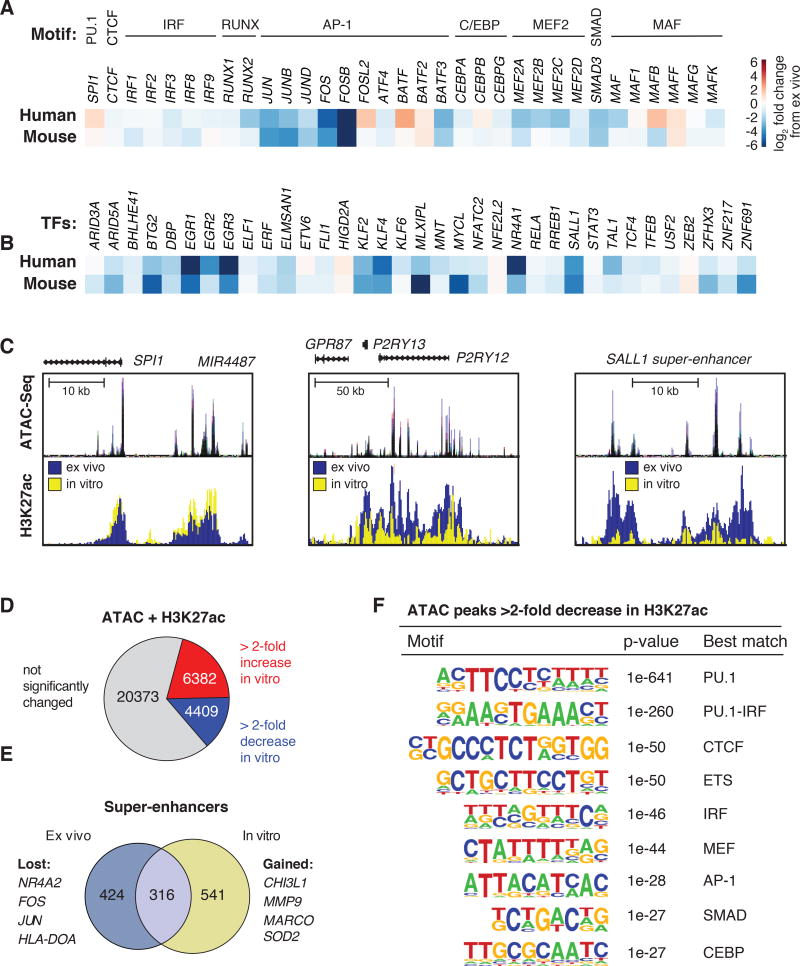
We performed ATAC-seq to define open regions of chromatin and ChIP-seq for H3K4me2 and H3K27ac to define regions of poised/active and active chromatin, respectively, in both human and mouse microglia. Mouse ATAC-seq data were aligned with previous H3K4me2 and H3K27ac data (25). ChIP-seq and ATAC-seq data were highly correlated across human individuals and mouse biological replicates (Fig. S9). A representative UCSC genome browser track for human microglia data in the vicinity of the BIN1 gene is illustrated in Figure 3A, and for CX3CR1 and TREM2 in Figure S10A. rs6733839 is a non-coding risk allele for AD (59) that lies within an enhancer-like region approximately 25 kb upstream of the BIN1 transcriptional start site, suggesting regulatory function. De novo motif analysis of ATAC-seq peaks associated with H3K4me2 was performed to identify the most highly enriched transcription factor recognition motifs associated with poised or active chromatin. These analyses returned very similar motifs for both mouse and human microglia. A dominant signature was observed for PU.1 in both species, with highly significant enrichment also observed for motifs assigned to CTCF, IRF, RUNX, MEF2, C/EBP, AP-1, SMAD and MAF factors (Fig. 3B, Fig. S10B).
Figure 3. Active genomic regulatory regions of human microglia.
(A) ATAC-seq and ChIP-seq data sets for PU.1, H3K27ac and H3K4me2 in the vicinity of the BIN1 gene locus. Each color represents a different individual. rs6733839 is a significant risk allele associated with AD and is located within a region highly bound by PU.1. (B) Motifs enriched at distal ATAC-seq peaks associated with H3K4me2 in human and mouse microglia. (C) Heat map of mRNA expression values of most highly expressed transcription factors recognizing motifs shown in panel B. (D) Heat map of mRNA expression values of transcription factors associated with super-enhancers in human and mouse microglia. (E) Scatter plot depicting mRNA expression values of orthologous genes encoding transcription factors in human and mouse microglia relative to overall transcriptome profiles (gray dots). Orthologous transcription factors similarly expressed are denoted in black, and transcription factors more highly expressed (> 10-fold change, FDR 0.05) in human are colored in blue, and those more elevated in mice are colored in green. (F) Scatter plot of H3K27ac tag counts at human and mouse promoters. Promoters associated with RNA transcripts expressed 10-fold higher in human microglia are colored in blue and promoters associated with RNA transcripts expressed 10-fold higher in mouse microglia are colored in green.
Expression levels of transcription factors recognizing the motifs identified in Figure 3B are illustrated in Figure 3C. SPI1 (encoding PU.1) was the most highly expressed ETS domain transcription factor, and the top motifs were nearly perfect matches for its consensus recognition sequence. The assignment of the second most enriched motif to CTCF was unexpected because CTCF is ubiquitously expressed and is thought to primarily play roles in the organization of broad domains of chromatin architecture. However, the identified motif was nearly a perfect match for the CTCF consensus recognition site, and CTCF was the only transcription factor expressed in microglia known to recognize this motif. In contrast, the remaining motifs were clearly recognized by multiple related members of transcription factor families that were reliably co-expressed in microglia.
We used H3K27ac as a means of identifying super-enhancers (SEs), which are regions of the genome exhibiting a disproportionately high density of active regulatory marks and transcription factor binding (60–62). Common SEs were associated with genes that are known to play essential roles in the development and function of virtually all macrophages whereas mouse microglia-specific SEs were associated with genes known to play restricted or selected roles in this cell type (25). Corresponding analysis of the H3K27acHigh regions in human microglia identified ~700 SEs that were also associated with active regions of the genome overlapping or in close proximity to genes highly expressed in or specific to microglia, including CX3CR1, P2RY12 and TMEM119 (Table S6). The intersection of transcription factor genes associated with SEs in human and mouse microglia captured many of the transcription factors illustrated in Figure 3C. An additional 35 factors were associated with SEs, some of which have been linked to functional roles in microglia, including SALL1, STAT3 and RELA (Fig. 3D). These factors also exhibited similar expression levels in mouse and human microglia.
Evaluation of mRNAs annotated as encoding orthologous transcription factors indicated similar expression levels in human and mouse for the great majority. However, using a 10-fold cutoff at an FDR of 0.05, 20 factors were preferentially expressed in human microglia, including CIITA, RUNX2, EGR3, and NR4A2. In contrast 16 factors were preferentially expressed in mouse microglia, including Sall3 and Smad1 (Fig. 3E).
To establish the relationship between enriched motifs and actual transcription factor binding, we performed ChIP-seq for PU.1 in microglia isolated from six individuals. These experiments yielded a highly consistent pattern of genomic binding (Fig. 3A, Fig. S10A), with PU.1 being localized at 51.2% of ATAC-seq-defined regions of open chromatin associated with H3K4me2 and at 60.9% of regions exhibiting these features plus H3K27ac. De novo motif enrichment analysis identified the PU.1 motif as the top hit, followed by motifs for IRF, AP-1, MEF2, C/EBP and RUNX (Fig.3 B, Fig. S10B). Similar results were observed for Pu.1 binding sites in mouse microglia. The frequency distribution of many of these motifs is consistent with the corresponding transcription factors forming ternary complexes with PU.1 (e.g., the IRF motif, Fig. S10C) or functioning as collaborative binding partners (e.g., CTCF, AP-1 and RUNX motifs, Fig. S10C).
We also investigated the relationships between PU.1 binding, open chromatin, and H3K27ac for genes that are highly differentially expressed between human and mouse microglia. H3K27ac at promoters was strongly correlated with differential gene expression (Fig. 3F). Differential gene expression and H3K27ac were also associated with species-specific PU.1 binding and ATAC-seq signal exemplified by the SALL1, SPARC, VSIG4 and APOC1 loci (Fig. S11). Collectively, these findings provide evidence that the transcriptional mechanisms that control gene expression are largely conserved between human and mouse microglia and that differences in gene expression are primarily driven by species-specific organization of cis regulatory elements at target genes. However, as a small number of transcription factors thought to play important roles in microglia are divergently expressed, such as CIITA and RUNX2 proteins, there is also the potential for significant trans effects at the subsets of genes regulated by these factors.
Alterations of the human microglia transcriptome in vitro
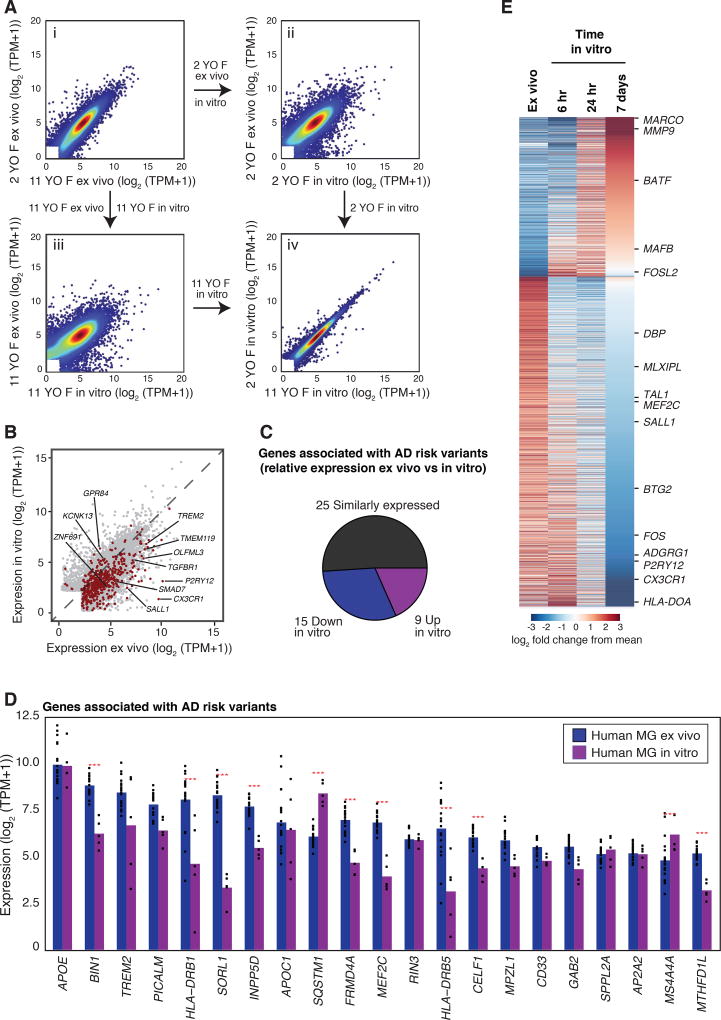
Tissue environment has emerged as an important determinant of microglia identity, but many studies of microglia are performed in vitro. Transcriptomic analysis of mouse microglia transferred to a tissue culture environment for 7 days documented significant changes in expression of hundreds of genes, with a preferential reduction in expression of microglia-specific genes (25). Similar to what was observed with mouse microglia, transfer of human microglia to the in vitro environment resulted in significant changes in the expression of thousands of genes. Responses of human microglia isolated from different individuals to the in vitro environment were concordant (Fig. 4Aiv). For the individuals with both ex vivo and in vitro samples available, gene expression within culture conditions (mean Pearson correlation = 0.95) was more correlated (p=0.008 by permutation test) than between the ex vivo and in vitro samples of the same individual (mean Pearson correlation = 0.89). A paired analysis of samples for which both ex vivo and 7-day in vitro microglia populations were obtained revealed more than 300 genes induced greater than 10-fold and more than 700 genes that were repressed more than 10-fold in vitro (FDR 0.05). At a 2-fold cutoff, 2,263 genes were upregulated in vitro and 3,702 genes were downregulated at a FDR of 0.05. Thirty-three percent of the conserved signature gene set and 31% of the genes associated with AD risk variants exhibited more than 2-fold reduction in expression in vitro (Figures 4B, C, and D). A similar proportion (30%) of genes associated with PD also displayed such reduction in vitro, whereas more limited changes for genes associated with MS and Scz were observed (Fig. S12).
Figure 4. Influence of tissue culture environment on microglia gene expression.
(A) Comparison of gene expression of ex vivo microglia from two individuals (panel i), effect of transition to an in vitro environment in presence of IL-34 for 7 days on microglia from each individual (panels ii and iii), and comparison of in vitro gene expression profiles (panel iv). (B) Scatter plot illustrating effects of culture environment on the conserved microglial gene signature (red) in human microglia. (C) Pie chart of the genes associated with AD risk variants that are expressed in ex vivo or in vitro microglia above a log2 (TPM +1) value of 2. Genes more highly expressed ex vivo are in blue and gene more highly expressed in vitro are in purple. (D) Combined bar graphs and dot plots of the 21 most expressed genes, indicating effects of in vitro culture on expression of genes associated with AD risk alleles. Red stars indicate significant differential expression (> 2-fold, FDR 0.05). (E) Heat map illustrating changes in microglia gene expression 6, 24 h and 7 days after transfer to culture environment.
We performed RNA-seq in human microglia at 6, 24 and 168 h following transfer to tissue culture. Highly dynamic responses were observed beginning at 6 h and lasting throughout the time course (Fig. 4D). Cultured microglia remained positive for Iba1 staining, whereas no expression of markers for astrocytes or neurons was observed, indicating that changes in RNA expression were not due to contaminating cells (Fig. S13A). A total of 1957 mRNAs were induced more than 4-fold by 6 h following plating. Gene ontology analysis of highly induced genes indicated strong enrichment for terms related to inflammation and stress responses (Fig. S13B, C). Remarkably, more than 2,000 genes were downregulated more than 4-fold at 6 h following plating, with some genes exhibiting partial recovery at the 7-day time point (Figs. 4E, S13D). Downregulated genes were strongly enriched for terms related to immune cell function and signaling, as well as immune, blood vessel and brain development (Fig. S13E). Similar observations of gene expression over time were made in mouse microglia cultured for 6 h, 24 h and 168 h (i.e. 7 days; Fig. S14 and Table S4).
An environment-dependent transcriptional network
The perturbation in gene expression resulting from the transition to an in vitro environment provided an opportunity to gain insight into the transcriptional network controlling human microglia identity. While SPI1 levels increased slightly, the majority of the mRNAs encoding transcription factors recognizing enriched motifs in microglia enhancers exhibited significant reductions in vitro. In particular, AP-1 and MEF2 family members, as well as MAF, RUNX, and SMAD3 were highly sensitive to the environmental perturbation (Fig. 5A). Similarly, the majority of transcription factors associated with SEs were also downregulated (Fig. 5B).
Figure 5. Environment-dependent enhancer landscapes.
(A) Heat map illustrating the effect of culture environment on human and mouse microglia mRNA expression of transcription factors recognizing motifs highly enriched ex vivo. (B) Heat map illustrating the effect of culture environment on human and mouse microglia on mRNA expression of transcription factors associated with super-enhancers ex vivo. (C) UCSC browser visualization of regulatory elements near genes SPI1 and P2RY12, and the SALL1 SE in human microglia. Top panels display regions of accessible chromatin in microglia ex vivo, as defined by ATAC-seq. Bottom panels display H3K27ac abundance ex vivo (blue) and in vitro (yellow). (D) Fractions of ATAC-seq + H3K27ac regions in human microglia exhibiting similar, gained or reduced H3K27ac signal after transfer to a culture environment for 7 days. (E) Venn diagram of super-enhancers identified in ex vivo and in vitro human microglia based on H3K27ac signal. (F) Motifs enrichment at distal accessible chromatin regions in ex vivo human microglia defined by ATAC-seq that are associated with 2-fold loss of H3K27ac signals following maintenance in culture environment for 7 days.
We performed ATAC-seq and ChIP-seq for H3K27ac in microglia placed into culture for 7 days (Fig. 5C). Overall, of the 31,164 ATAC-seq peaks associated with at least 16 normalized H3K27ac tags (i.e., high confidence sites), ~14% exhibited a loss of H3K27ac and ~20% a gain of H3K27ac (Fig. 5D), indicating remodeling of the regulatory landscape in vitro. Extending this analysis, 424 of the 740 SEs defined ex vivo were lost and 541 new SEs were observed (Fig. 5E). De novo motif analysis of the ATAC-seq peaks associated with a > 2-fold decrease in H3K27ac returned recognition elements for PU.1, IRF, CTCF, AP-1, SMAD and MEF2 motifs as the most significantly enriched (Fig. 5F), consistent with PU.1 establishing microglia enhancers and reduced expression and/or activity of many of the collaborative and signal-dependent factors recognizing these motifs (Fig. 5A). Notably, loss of the SMAD motif is consistent with reduced TGF-β signaling in the in vitro environment.
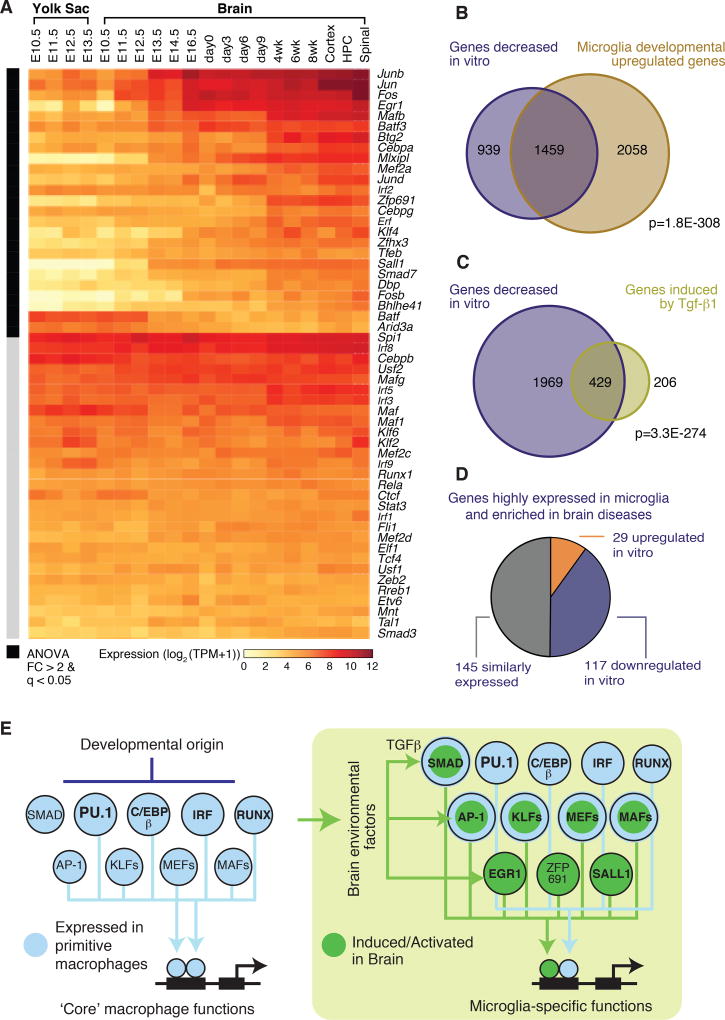
We compared the expression of the mouse orthologues of transcription factors with those present in primitive yolk sac macrophages and their progeny in developing and adult mouse brain (63). A subset of mRNAs encoding these factors was expressed in primitive macrophages and maintained high levels of expression in brain microglia (Fig. 6A). In contrast, an alternative subset of factors that recognize DNA motifs enriched in microglia enhancers and/or were preferentially expressed in adult human and mouse microglia exhibited increases in expression from the embryonic to the adult stage of brain development (Fig. 6A). Notably, a majority of the adult microglia transcription factors exhibiting reduced expression following transfer to the in vitro environment were induced following migration of primitive macrophages into the developing brain, consistent with their induction by local environmental factors.
Figure 6. A network of developmentally programmed and environment-dependent transcription factors establishes microglia identity and function.
(A) Heat map displaying changes in mRNA expression of key microglia transcription as a function of development of the mouse brain. (B) Venn diagram illustrating significant overlap of mouse microglia genes repressed (> 2-fold, FDR 0.05) in culture and genes that increased (> 2-fold, FDR 0.05) in expression during brain development. p-value of the overlap is provided (Fisher’s exact test). (C) Venn diagram illustrating significant overlap of mouse microglia genes repressed (> 2-fold, FDR 0.05) in culture after 7 days and genes maintained/induced (> 2-fold, FDR 0.05) by chronic stimulation with Tgf-β1. p-value of the overlap is provided (Fisher’s exact test). (D) Pie chart of microglia-enriched genes compared to brain cortex overlapping with differentially expressed genes in neurodegenerative or behavioral diseases from Fig. 1C color-coded according to whether they are increased, decreased or unchanged after transfer to a tissue culture environment. (E) Diagram illustrating main features of transcription factor network regulating microglia cell identity and functions.
We compared the set of genes robustly upregulated during brain development in mouse microglia to those consistently downregulated upon transition to the in vitro environment. A highly significant overlap was observed: of 3,517 genes that increased during development, 1,459 intersected with the 2,398 genes “down” in culture (p < 1E-308; Fig. 6B). This finding illustrates that a significant component of the microglia unique signature that emerges during development is downregulated in vitro.
The correlation between genes upregulated in fetal microglia compared to yolk sac macrophages and genes downregulated upon transfer of adult microglia to an in vitro environment provided evidence that the changes observed in vitro result from loss of signals that are part of the brain environment. We cultured mouse microglia in vitro for 7 days and treated them with Tgf-β1 cytokine (Figs. 6C, S15), which is necessary for maintenance of the in vivo mouse microglia phenotype (27, 64). Of the 2,398 genes downregulated > 2-fold in vitro, Tgf-β1 induced expression > 2-fold of 429, including Sall1, Tmem119, and P2ry12. Corresponding studies of TGF-β1 treatment of human microglia in vitro yielded similar results, although variation between individuals limited the number of genes achieving statistical significance following correction for multiple testing. Nonetheless, SALL1 was among the genes exhibiting significant upregulation (Fig. S15B). Because of the relatively modest effects of TGF-β1, we evaluated several other tissue culture conditions to investigate whether any might be more effective in maintaining an ex vivo gene expression signature. Although most treatment conditions produced some effect on gene expression, none resulted in preservation of the ex vivo molecular phenotype (Fig. S16). It has not yet been possible to maintain primary human microglia in a serum-free environment. Recent studies have derived microglia like cells from human induced pluripotent stem cells (iPSCs) in a defined media that more closely resembles cerebrospinal fluid (65). However, these cells exhibited a transcriptomic phenotype that was much more similar to human microglia cultured in vitro than ex vivo microglia (Fig. S17).
Lastly, we investigated whether genes expressed at least 10-fold higher in ex vivo microglia compared to brain cortex tissue and that are associated with differentially regulated genes in neurodegenerative or behavioral disorders (Fig. 1C) were environment-sensitive. Of the non-redundant set of 291 microglia transcripts associated with differentially regulated genes in AD, PD, HAND, SCZ or FTD, 117 were downregulated in vitro whereas 29 were upregulated in vitro (Fig. 6D). These observations provide evidence that differential expression of microglia-enriched genes in these disorders is at least partially due to changes in brain environment.
Discussion
The transcriptomes and epigenomic features of human microglia show that disease status, age, sex, and treatment history had a relatively minor influence on the overall pattern of gene expression. It is thus likely that the composite transcriptomic and epigenomic profiles identified largely capture a representative human microglial gene signature and its underlying network of genomic regulatory elements. We find that mouse and human microglia transcriptomes are largely conserved but they also exhibit large differences in the expression of several hundred genes that appear to be primarily driven by cis-regulatory differences.
Genetic evidence suggests that microglia dysregulation is a core feature of numerous chronic neurodegenerative and psychiatric diseases. To this end, our profiling of the human microglia transcriptome enabled us to estimate a weighted influence of microglia on the development of different brain disorders. In addition, by defining a microglial gene signature in the human brain, it is possible to link changes in gene expression in neurodegenerative and behavioral disorders to alterations in the number and/or function of microglia. The observation that the microglial gene signature overlaps with genes that are both up- and downregulated in PD provides strong evidence that differential gene expression at the whole tissue level reflects changes in microglia gene expression and not simply changes in the microglia population number. This interpretation is further reinforced by the observation that many of the microglia-enriched genes that change in the context of neurodegenerative and behavioral disorders also show significant changes in expression upon transfer to an in vitro environment. Although we also observed substantial individual variation in genes linked to disease risk, more individuals will be required to power analyses linking genetic variation to gene expression.
Examining differences in ex vivo and in vitro microglia, we observe rapid and profound alterations in gene expression by 6 h, characterized by activation of genes associated with acute inflammatory responses and decreased expression of genes associated with diverse immune functions. The incomplete recovery of downregulated genes at 7 days was at least in part due to the absence of brain-specific signals. At present, the spectrum of brain-specific signals that are necessary to maintain microglia phenotypes remains largely unknown, but this finding highlights the limitations of investigating microglia biology in a tissue culture setting. Importantly, current efforts to reprogram various starting cell populations to microglia-like cells largely achieve an in vitro phenotype. It is likely that more complex culture conditions, such as co-culture with neurons and astrocytes in three-dimensional or organoid contexts, will be required to more precisely model the in vivo phenotype. Integration of these findings with recent studies of yolk sac and embryonic microglia (56, 63) suggests a model by which intersecting developmentally determined and environment-dependent transcriptional networks establish microglia identity and function (Fig. 6E). We speculate that regulatory elements are targets of environmental perturbations associated with disease and are also important sites of action of natural genetic variation.
In sum, our work demonstrates that human and mouse microglia display relatively well-conserved transcriptomic and epigenomic phenotypes and it provides a critical resource that will allow for a better understanding of mouse models of human brain disorders, guide reprogramming efforts for generation of human microglia in vitro, and enable a better interpretation of the functions of disease variants associated with disorders of the central nervous system.
Methods
Human tissue
Microglia were isolated from brain tissue (in excess of that needed for pathological diagnosis) resected for treatment of epilepsy, brain tumors, or acute ischemia in 19 individuals. For patients with epilepsy (9), the resected tissue displayed epileptogenic activity. However, the epileptic focus exhibiting the strongest epileptogenic activity was not provided and was separated at the time of resection for pathological analysis. For patients with brain tumors (8), samples consisted of incidentally resected brain tissue, i.e., brain tissue resected to gain access to the tumor and considered to be tumor negative by the neurosurgeon intraoperatively. Two brain samples were resected to treat acute ischemic injuries from individuals who had no prior evidence of brain pathology. Brain tissue was obtained with informed consent under a protocol approved by the UC San Diego and Rady Children’s Hospital Institutional Review Board. Resected brain tissue was immediately placed on ice and transferred to the laboratory for microglia isolation within 3 hours after resection.
Human and mouse microglia isolation
Resected human brain tissues and mouse brains were homogenized by gentle mechanical dissociation, as performed in Gosselin et al. (25). Microglia were then enriched by Percoll gradient, washed, and stained with cell surface markers for final purification by flow cytometry, using a BD Influx cell sorter. Human microglia were defined as live/DAPI− CD11b+CD45LowCD64+CX3CR1High single cells. C57BL/6 mouse microglia were defined as live/DAPI− CD11b+CD45Low single cells.
Primary culture of isolated microglia
Isolated microglia were maintained in culture for 6 h, 24 h, or 7 days. Culture media consisted of DMEM/F12 (Life Technologies, 113300-032) supplemented with 5% FBS (heat inactivated, Omega Scientific, FB-12) and 1× Anti-Anti (Life Technologies, 152240-062). No media changes were performed for the duration of the culture experiments. CSF1R ligand Interleukin-34 (R&D, human 5265-IL-010; mouse 5195-ML-010) was supplemented daily at a concentration of 20 ng/ml. For experiments investigating the effects of TGF-β1 on microglia gene expression in culture, recombinant human or mouse TGF-β1 cytokine (R&D, human 240-B-002; mouse 7666-MB-005) was added daily at a concentration of 10 ng/ml.
RNA-seq library preparation: Isolation and fragmentation of poly(A) RNA and synthesis of cDNA
Isolated microglia were pelleted and put into 150 µl lysis/Oligo d(T) Magnetic Beads binding buffer and stored at −80°C until processing. For human brain cortical RNA, 500 ng of RNA was diluted with proper volume of 2× lysis/ Oligo d(T) Magnetic Beads binding buffer to a final concentration of 1× lysis/Oligo d(T) Magnetic Beads binding buffer. mRNAs were enriched by incubation with Oligo d(T) Magnetic Beads (NEB, S1419S) and then fragmented/eluted by incubation at 94 °C for 9 min. cDNA was then synthesized with Superscript III (first-strand synthesis; Life Technologies kit 18080-044) and then DNA Polymerase I (second-strand synthesis; Enzymatics P7050L)
Chromatin immunoprecipitation
For ChIP-seq experiments, a cross-linking step with paraformaldehyde was performed immediately after staining but before microglia sorting. Chromatin immunoprecipitation for histone modification H3K4me2, H3K27ac and transcription factor PU.1 was performed as described in Gosselin et al. (25).
RNA-seq and ChIP-seq final library preparation and sequencing
Sequencing libraries were prepared from recovered DNA (ChIP) or generated cDNA (RNA) by blunting, A-tailing, and adapter ligation as previously described in Heinz et al. (31). Prior to final PCR amplification, RNA-seq libraries were digested by 30 min of incubation at 37°C with Uracil DNA Glycosylase. RNA-seq and ChIP-seq librairies were single-end sequenced for 51 cycles on an Illumina HiSeq 4000 or NextSeq 500 (Illumina, San Diego, CA) according to manufacturer’s instruction.
Data Analysis
Preprocessing
FASTQ files from sequencing experiments were mapped to either the UCSC genome build hg38 (for human) or mm10 (for mouse). STAR with default parameters was used to map RNA-seq experiments. Bowtie2 with default parameters was used to map ATAC-seq and ChIP-seq experiments. HOMER was used to convert aligned reads into “tag directories” for further analysis.
RNA-seq
The “analyzeRepeats” function of HOMER was used to generate a table of read counts for each experiment using the parameters “-raw -count exons -strand both condenseGenes” and a table of transcripts per million (TPM) values using “-tpm normMatrix 1e7 -count exons -strand both -condenseGenes.” The base-2 logarithm of the TPM values was taken after adding a pseudocount of 1 TPM to each gene. Within-species differential gene expression analysis was performed using the R package limma using the voom normalization. Mapping of orthologous genes between species was done using the one-to-one orthologs from the Ensembl (version 84) Compara database.
ChIP-seq
Tag directories for the ChIP-seq experiments were combined into ex vivo and in vitro pools for each target in both mouse and human. The tag directories for the input DNA were likewise combined. Peaks were then called on the pooled tags with the pooled input DNA as background using HOMER’s “findPeaks” command with the following parameters for the PU.1 ChIP-seq: “-style factor -size 200 -minDist 200” and the following parameters for the H3K27ac and H3K4me2 ChIP-seq: “-style histone -size 500 -minDist 1000 -region.” The “-tbp” parameter was set to the number of replicates in the pool.
Motif enrichment
Peaks were considered overlapping if their coordinates overlapped by at least one base pair. De novo motif enrichment analysis was performed on ATAC-seq and PU.1 ChIP-seq peaks that overlapped either H3K4me2 (ATAC-seq) or H3K27ac (PU.1), using HOMER’s “findMotifsGenome” command with the following parameters: “-size given -len 14,12,10,8 -mask.”
Supplementary Material
Acknowledgments
We thank J. Collier for technical assistance, L. Van Ael for her assistance with manuscript preparation, and M.L. Gage for her assistance with manuscript editing. We also thank D. Malicki for assistance in obtaining patient samples for Iba-1 immunohistological analysis. We thank J. A. Miller from the Allen Institute for Brain Science for assistance with the analysis of brain disease-associated gene sets. These studies were supported by the Larry L. Hillbloom Foundation and NIH grants NS096170, DK063491 and GM085764. DG was supported by a Canadian Institute of Health Research fellowship and a Multiple Sclerosis Society of Canada fellowship. DS was supported by UCSD institutional predoctoral training grant (T32DK007541). IRH was supported by University Medical Center Groningen Institutional postdoctoral traveling grant, the Dutch MS Research Foundation, and the Gemmy and Mibeth Tichelaar Foundation. JCMS was supported by the DFG (SCHL2102/1-1.) FHG is supported by the JPB Foundation, Dolby Family Ventures, The Paul G. Allen Family Foundation, and the Engman Foundation. CKG is supported by the Ben and Wanda Hildyard Chair in Hereditary Diseases. Author contribution: DG, NGC, FHG and CKG conceived the study. NGC coordinated tissue acquisition, clinical chart review and IRB approval. MLL and DGG consented patients, and resected and evaluated brain tissue. DG and JCMS isolated microglia, with assistance from BNJ, MP and AA. DG, JCMS, CO, CF and MPP performed microglia sorting. JCMS isolated monocytes. NGC and MP performed histological analyses. DG performed culture experiments. DG, JCMS, and ES prepared, respectively, microglia, monocyte, and intact brain sequencing libraries. DG and CO analyzed flow cytometry data. DG, DS, IRH and CKG analyzed sequencing data. DG, IHR, DS and CKG wrote the manuscript, with contributions from NGG, RMR and FHG.
References
- 1.Rivest S. Regulation of innate immune responses in the brain. Nat Rev Immunol. 2009;9:429–439. doi: 10.1038/nri2565. [DOI] [PubMed] [Google Scholar]
- 2.Katsumoto A, Lu H, Miranda AS, Ransohoff RM. Ontogeny and functions of central nervous system macrophages. J Immunol. 2014;193:2615–2621. doi: 10.4049/jimmunol.1400716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shemer A, Erny D, Jung S, Prinz M. Microglia Plasticity During Health and Disease: An Immunological Perspective. Trends Immunol. 2015;36:614–624. doi: 10.1016/j.it.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Schafer DP, Stevens B. Microglia Function in Central Nervous System Development and Plasticity. Cold Spring Harb Perspect Biol. 2015;7:a020545. doi: 10.1101/cshperspect.a020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casano AM, Peri F. Microglia: multitasking specialists of the brain. Dev Cell. 2015;32:469–477. doi: 10.1016/j.devcel.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Malm TM, Jay TR, Landreth GE. The evolving biology of microglia in Alzheimer's disease. Neurotherapeutics. 2015;12:81–93. doi: 10.1007/s13311-014-0316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derecki NC, Katzmarski N, Kipnis J, Meyer-Luehmann M. Microglia as a critical player in both developmental and late-life CNS pathologies. Acta Neuropathol. 2014;128:333–345. doi: 10.1007/s00401-014-1321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosher KI, Wyss-Coray T. Microglial dysfunction in brain aging and Alzheimer's disease. Biochem Pharmacol. 2014;88:594–604. doi: 10.1016/j.bcp.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch EC, Vyas S, Hunot S. Neuroinflammation in Parkinson's disease. Parkinsonism Relat Disord. 2012;18(Suppl 1):S210–212. doi: 10.1016/S1353-8020(11)70065-7. [DOI] [PubMed] [Google Scholar]
- 10.Croisier E, Moran LB, Dexter DT, Pearce RK, Graeber MB. Microglial inflammation in the parkinsonian substantia nigra: relationship to alpha-synuclein deposition. J Neuroinflammation. 2005;2:14. doi: 10.1186/1742-2094-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins LM, Toulouse A, Connor TJ, Nolan YM. Contributions of central and systemic inflammation to the pathophysiology of Parkinson's disease. Neuropharmacology. 2012;62:2154–2168. doi: 10.1016/j.neuropharm.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 12.Saijo K, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crotti A, et al. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci. 2014 doi: 10.1038/nn.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Politis M, et al. Microglial activation in regions related to cognitive function predicts disease onset in Huntington's disease: a multimodal imaging study. Hum Brain Mapp. 2011;32:258–270. doi: 10.1002/hbm.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tai YF, et al. Microglial activation in presymptomatic Huntington's disease gene carriers. Brain. 2007;130:1759–1766. doi: 10.1093/brain/awm044. [DOI] [PubMed] [Google Scholar]
- 16.Bilimoria PM, Stevens B. Microglia function during brain development: New insights from animal models. Brain Res. 2014 doi: 10.1016/j.brainres.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 17.Frick LR, Williams K, Pittenger C. Microglial dysregulation in psychiatric disease. Clin Dev Immunol. 2013;2013:608654. doi: 10.1155/2013/608654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 20.Elmore MR, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith AM, et al. The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia. 2013;61:929–942. doi: 10.1002/glia.22486. [DOI] [PubMed] [Google Scholar]
- 22.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 23.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schafer DP, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkhurst CN, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.C. Roadmap Epigenomics et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G. Epigenomics: Roadmap for regulation. Nature. 2015;518:314–316. doi: 10.1038/518314a. [DOI] [PubMed] [Google Scholar]
- 36.Kasowski M, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McVicker G, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilpinen H, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avramopoulos D, Szymanski M, Wang R, Bassett S. Gene expression reveals overlap between normal aging and Alzheimer's disease genes. Neurobiol Aging. 2011;32:2319, e2327–2334. doi: 10.1016/j.neurobiolaging.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blalock EM, et al. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capurro A, Bodea LG, Schaefer P, Luthi-Carter R, Perreau VM. Computational deconvolution of genome wide expression data from Parkinson's and Huntington's disease brain tissues using population-specific expression analysis. Front Neurosci. 2014;8:441. doi: 10.3389/fnins.2014.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colangelo V, et al. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- 43.Liang WS, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set. Physiol Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClintick JN, et al. Ethanol treatment of lymphoblastoid cell lines from alcoholics and non-alcoholics causes many subtle changes in gene expression. Alcohol. 2014;48:603–610. doi: 10.1016/j.alcohol.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra M, et al. Gene expression analysis of frontotemporal lobar degeneration of the motor neuron disease type with ubiquitinated inclusions. Acta Neuropathol. 2007;114:81–94. doi: 10.1007/s00401-007-0240-7. [DOI] [PubMed] [Google Scholar]
- 46.Satoh J, Yamamoto Y, Asahina N, Kitano S, Kino Y. RNA-Seq data mining: downregulation of NeuroD6 serves as a possible biomarker for alzheimer's disease brains. Dis Markers. 2014;2014:123165. doi: 10.1155/2014/123165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster JA, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borjabad A, et al. Significant effects of antiretroviral therapy on global gene expression in brain tissues of patients with HIV-1-associated neurocognitive disorders. PLoS Pathog. 2011;7:e1002213. doi: 10.1371/journal.ppat.1002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maycox PR, et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry. 2009;14:1083–1094. doi: 10.1038/mp.2009.18. [DOI] [PubMed] [Google Scholar]
- 50.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riley BE, et al. Systems-based analyses of brain regions functionally impacted in Parkinson's disease reveals underlying causal mechanisms. PLoS One. 2014;9:e102909. doi: 10.1371/journal.pone.0102909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen-Plotkin AS, et al. Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration. Hum Mol Genet. 2008;17:1349–1362. doi: 10.1093/hmg/ddn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Med. 2013;5:48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burns TC, Li MD, Mehta S, Awad AJ, Morgan AA. Mouse models rarely mimic the transcriptome of human neurodegenerative diseases: A systematic bioinformatics-based critique of preclinical models. Eur J Pharmacol. 2015;759:101–117. doi: 10.1016/j.ejphar.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 56.Mass E, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353 doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiu IM, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature genetics. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matcovitch-Natan O, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 64.Buttgereit A, et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol. 2016 doi: 10.1038/ni.3585. [DOI] [PubMed] [Google Scholar]
- 65.Muffat J, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olah M, et al. An optimized protocol for the acute isolation of human microglia from autopsy brain samples. Glia. 2012;60:96–111. doi: 10.1002/glia.21251. [DOI] [PubMed] [Google Scholar]
- 67.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 71.Tripathi S, et al. Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leinonen R, Sugawara H, Shumway M. C. International Nucleotide Sequence Database, The sequence read archive. Nucleic acids research. 2011;39:D19–21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao S, Guo Y, Sheng Q, Shyr Y. Advanced heat map and clustering analysis using heatmap3. Biomed Res Int. 2014;2014:986048. doi: 10.1155/2014/986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whickam H. Ggplot2 : elegant graphics for data analysis. Springer. 2009 [Google Scholar]
- 75.M. Developers, matplotlib: v1.5.3 [Data set] Zenodo; 2016. [Google Scholar]
- 76.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.