Abstract
Background
Inhibition of Protein Kinase C alpha (PKCα) enhances contractility and cardioprotection in animal models, but effects in humans are unknown. Genotypes at rs9912468 strongly associate with PRKCA expression in the left ventricle, enabling genetic approaches to measure effects of reduced PKCα in human populations.
Methods and Results
We analyzed the cis expression quantitative trait locus (eQTL) for PRKCA marked by rs9912468 using 313 left ventricular specimens from European Ancestry patients. The forward strand minor allele (G) at rs9912468 associated with reduced PKCα transcript abundance (1.7-fold reduction in minor allele homozygotes, P =1×10−41). This association was cardiac-specific in eQTL datasets that span 16 human tissues. Cardiac epigenomic data revealed a predicted enhancer in complete (R2=1.0) linkage disequilibrium with rs9912468 within intron 2 of PRKCA. We cloned this region and used reporter constructs to verify cardiac-specific enhancer activity in vitro in cardiac and non-cardiac cells and in vivo in zebrafish. The PRKCA enhancer contains two common genetic variants and 4 haplotypes; the haplotype correlated with the rs9912468 PKCα lowering allele (G) showed lowest activity. In contrast to previous reports in animal models, the PKCα lowering allele associated with adverse left ventricular remodeling (higher mass, larger diastolic dimension), reduced fractional shortening, and higher risk of dilated cardiomyopathy in human populations.
Conclusions
These findings support PKCα as regulator of the human heart, but suggest that PKCα inhibition may adversely affect the left ventricle depending on timing and duration. Pharmacologic studies in human subjects are required to discern potential benefits and harms of PKCα inhibitors as an approach to treat heart disease.
Keywords: Protein Kinase C alpha, Heart Failure, Genetics
Journal Subject Terms: heart failure, translational studies, genetics, gene expression and regulation
Introduction
The protein kinase C (PKC) serine-threonine kinases function downstream of multiple membrane-associated signal transduction pathways.1, 2 Approximately 10 different isozymes comprise the PKC family, which are broadly classified by their activation characteristics. The conventional PKC isozymes (α, βI, βII, and γ) are Ca2+- and lipid-activated, while the novel isozymes (ε, θ, η, and δ) and atypical isozymes (ζ and γ) are Ca2+- independent but activated by distinct lipids.1, 2 PKCα is the predominant isozyme expressed in the mouse, human, and rabbit heart3, 4 and is a well-established regulator of myocardial function.
A number of reports have associated PKCα activation or an increase in PKCα expression with hypertrophy, cardiomyopathy, ischemic injury or neuroendocrine stimulation.1, 2 Conversely, PKCα deficiency enhances cardiac contractility and protects against heart failure in multiple model systems, including thoracic aortic constriction,5 Csrp3 deletion,5 and infarction.6, 7 Pharmacologic inhibition of PKCα with the mixed PKCα/β inhibitor ruboxistaurin enhances contractility and cardioprotection in small and large animal models.7–9 The wealth of evidence establishing PKCα as a regulator of cardiac function and injury suggests PKCα inhibition as a strategy to treat heart disease.10 PKCα is elevated in the failing human myocardium,11, 12 yet there is no experience assessing the impact of PKCα inhibition on human myocardial function.
Numerous therapeutic strategies discovered in animal models have failed to translate to effective therapeutics in clinical practice. There are many potential explanations, including species differences and effects of exposures and comorbid conditions on disease progression in patients. Genetic studies have been proposed as an alternative approach to discover targets of higher therapeutic relevance to human disease.13 A well-known recent example is discovery of loss-of-function mutations in PCSK9 associated with extremely low levels of LDL-C.14 This genetic finding inspired development of antibody-based PCSK9 inhibitors now in clinical use to dramatically lower LDL.15 Discovery of loss-of-function mutations in ANGPLT3 provides a similar example that has inspired new therapeutics.16, 17
In previous work, we18 and others19 have discovered a locus marked by the common variant rs9912468 that potently associates with PKCα transcript abundance in the human left ventricle. This variant resides within an intronic region of the PRKCA gene encoding PKCα and therefore marks a cis expression quantitative trait locus (eQTL) for PRKCA. Here we leverage this genetic finding to ask whether an inherited tendency to have low ventricular PKCα is cardioprotective in human subjects. Unlike findings in animal models, variants that reduce left ventricular PKCα associate with adverse cardiac remodeling and cardiomyopathy risk. These findings support PKCα as regulator of the human heart, but raise the possibility that PKCα inhibition may incur cardiac risk depending on timing and duration.
Methods
Human Myocardium
The data, analytic methods, and study materials have been made available to other researchers for purposes of reproducing the results or replicating the procedure20. The Myocardial Applied Genomics Network (MAGNet; www.med.upenn.edu/magnet), collects and banks human cardiac tissue for genomic research. All subjects or next of kin provided written informed consent for tissue donation and analyses, and relevant institutional review boards approved all study protocols. Left ventricular free-wall tissue was harvested at the time of cardiac surgery from subjects with heart failure undergoing transplantation and from unused donor hearts with apparently normal function. In each case the heart was perfused with cold cardioplegia prior to cardiectomy to arrest contraction and prevent ischemic damage, and tissue specimens were frozen in liquid nitrogen. DNA was extracted using the Gentra Puregene Tissue Kit (Qiagen, CA) and total RNA was extracted using the miRNeasy Kit (Qiagen). DNA concentration and quality was assessed using gels and PicoGreen. RNA concentration and quality was assessed using the NanoVue Plus™ spectrophotometer (GE Healthcare) and the Agilent 2100 RNA NanoChip. The final dataset comprised 313 genetically inferred Caucasians.
Fine mapping and annotation of the PRKCA eQTL
DNA samples were genotyped using the Affymetrix Genome-Wide SNP Array 6.0. We excluded samples with cryptic relatedness and limited the samples to genetically inferred European-descent by multi-dimensional scaling of all genotypes. We eliminated SNPs with call rate < 95%, minor allele frequency <15%, or departure from Hardy-Weinberg equilibrium (P < 10−6). A minor allele frequency cutoff of 15% rather than the typical 5% used in large cohort GWAS was used to ensure that adequate numbers of minor allele homozygotes would be available in our sample of 313 subjects. All alleles are encoded on the forward strand. To enable finer genotype coverage, imputation with SNP genotypes in the 1000Genomes Project was performed and filtered using a quality threshold of 0.5 and a minor allele frequency threshold of 0.15. RNA was hybridized with Affymetrix Genechip ST1.1 arrays using manufacturer instructions. CEL files were normalized with robust multiarray analysis (RMA)21 and expression values were further adjusted for potential batch effects using ComBat.22 The final dataset comprised 313 genetic Caucasians and 15,390 RMA expression levels.
Genome-wide associations between transcript abundance and genotypes were tested using an additive genetic model adjusting for age, sex, study site, and patient group (explanted hearts or unused donors) implemented in the R statistical programing language. For each significant locus, the association was retested after conditioning against the strongest eSNP at that locus to determine a set of minimal eSNPs that mark independent eQTLs. We have previously reported a genome-wide significant cis eQTL for PRKCA marked by the common variant rs9912468.18 In this report we use the same dataset to focus on associations between all available genotypes in linkage disequilibrium (R2 ≥ 0.8) with rs9912468 and their association with PKCα transcript levels in the left ventricle. Associations between genotypes near rs9912468 and PKCα transcript abundance in multiple human tissues were assessed using the Genome Tissue Expression database (GTEx).23
Predicted regulatory elements in linkage disequilibrium (R2 ≥ 0.8) with rs9912468 were mapped using results from human left ventricular ChIPseq experiments performed by the Roadmap Epigenomics project using ChromHMM (http://compbio.mit.edu/ChromHMM/). ChromHMM is a software package that predicts chromatin regulatory states from the integration of multiple histone modifications assayed by ChIP-seq.24 ChromHMM data from left ventricle were downloaded as a BED file and were tested for overlap with left ventricular eSNP using the intersect function of the BEDTools suite.25 Regional association plots were constructed using the locusZoom web interface (http://http://locuszoom.org/).
In silico prediction of enhancer binding proteins was attempted using snp2TFBS (http://ccg.vital-it.ch/snp2tfbs/), ePOSSUM (http://mutationtaster.charite.de/ePOSSUM/) and motifBreakR (http://simon-coetzee.github.io/motifBreakR/) applied to the HOCOMOCO TFBS database. SNP2TFBS reported no results and motifBreakR show no significant results after multiple test correction was applied. (Supplemental Table).
PKCα protein studies
A subset of the eQTL study cohort was selected comprised of 20 patients with dilated cardiomyopathy who were homozygous for the major (C) or minor (G) rs9912468 allele and who showed high or low PKCα gene expression by microarray. For each, approximately 100 mg of left ventricular tissue was homogenized in 3 mL of triton lysis buffer (150 mM NaCl, 50 mM Tris pH 8.0, 1% Triton X-100, protease and phosphatase inhibitors). Thirty micrograms of protein were loaded on 10% acrylamide gels for western blotting. Immunoblots were performed using a primary antibody directed against PKCα (SantaCruz H-7 sc-8393, dilution 1:500) and a fluorescent goat anti-mouse IgG1 secondary antibody (LI-COR, dilution 1:3000) in combination with an Odyssey CLx Infrared Imaging System (LI-COR). GAPDH (Fitzgerald, 10R-G109A, dilution 1:20000) was used as a loading control. Western blots were quantified using Image Studio software (LI-COR). Protein extraction and sample preparation were processed in a blinded manner until western blot loading.
Generation of reporter plasmids
The PRKCA enhancer region was amplified using PCR from human genomic DNA (F: ATAAAGCTGAGTTGTCGGGC; R: TGTGCACAAACGATACTGTCA) and cloned into a Firefly Luciferase reporter vector (pGL4.23, Promega) upstream of a CMV minimal promoter. A similar plasmid with the enhancer region replaced by a random non-coding genomic segment was constructed for use as a negative control. A reporter vector containing Renilla Luciferase was used as a control for cell number and transfection efficiency. Site-directed mutagenesis was used to construct reporters for the four enhancer haplotypes using a single clone of the PRKCA enhancer region. This was accomplished using the Q5 Site Directed Mutagenesis kit (New England Biolabs) and back-to-back PCR primers (F: GCAGAGGCGGTCCGGAGCGGC; R: CGCCGCGAGTAGGAAACGCGAG) containing the desired substitutions (major and minor alleles at rs9910355 and rs9303504). Sanger sequencing was performed on all plasmids to confirm mutagenesis efficacy.
Cell Culture
HeLa cells were grown under standard conditions (37°C, 5% CO2) in Dulbecco’s Modified Eagle Medium (DMEM) with L-glutamine (4.5 g/L), sodium pyruvate (4.5 g/L), 10% fetal bovine serum (FBS), penicillin (100 units/ml) and streptomycin (100μg/ml) and maintained until 100% confluent. HL-1 cells were grown on gelatin-fibronectin coated plates under standard conditions in Claycomb medium supplemented with 0.1mM norepinephrine, 2mM L-glutamine, 10% FBS, penicillin (100 units/ml) and streptomycin (100 μg/ml) and maintained until 100% confluent. Neonatal rat ventricular myocytes (NRVM) were isolated using a protocol approved by the Institutional Animal Care and Use Committee. Sprague-Dawley day 0–1 pups were euthanized by decapitation; hearts were immediately extracted; the atria and great vessels were removed, and the left ventricular tissue was minced and subjected to a trypsin-based disaggregation procedure. Minced ventricular tissue was rinsed with Hank’s Balanced Salt Solution (HBSS) with 1% penicillin/streptomycin/glutamine and placed in a 50 ml conical tube containing 10 ml of Trypsin and placed in a shaking incubator (200 rpm at 37°C) for 15 min. The supernatant was discarded after incubation and trypsin procedure repeated for an additional 7 times. After the 8th trypsin treatment, the cells were centrifuged at 660 rpm at 4°C for 5 minutes and the supernatant was discarded. The remaining cell pellet was re-suspended in 20 ml of growth media composed of 4:1 ratio of DMEM and Medium-199, 10% horse serum, 5% fetal bovine serum, 1% HEPES, and 1% solution of penicillin, streptomycin, and glutamine and pre-plated for 1–3 hours in an incubator. Harvested cells were collected after centrifugation at 660 rpm for 5 min at room temperature and plated on fibronectin-coated plates. Growth media was replaced 24 hours after harvest.
In vitro enhancer assays
Cells were seeded in 24-well plates with antibiotic-free media and transiently transfected at ~70% confluency with reporter vectors containing the predicted enhancer and co-transfected with the renilla luciferase reporter. Transfection was performed using Lipofectamine 3000 (Invitrogen) according to manufacturer’s instructions and repeated in three independent experiments with 4 biologic replicates per experiment. Luciferase activity was measured using the Dual-Luciferase Reporter (DLR) Assay System (Promega) according to manufacturer’s instructions. 72 hours after transfection, cells were washed with PBS and lysed with 100μl of passive lysis buffer (PLB) for 15 minutes on a shaker. 20μl of the lysate was transferred into a 96-welled plate and 100μl of luciferase assay reagent II (LAR II) was dispensed into each well. The plate was placed in a luminometer (Biotek) and measured for 10 seconds. After the initial measurement, 100μl of Stop & Glo reagent was added into each well to stop the firefly reaction and start the renilla reaction. Measurements were then repeated in 10-second periods in the luminometer. Bioluminescence was collected as a ratio of firefly:renilla (F/R) and then standardized to the negative control for each condition and reported as luciferase activity relative to the negative control. Two-tailed t-tests were performed for all luciferase assays with a P-value cutoff of 0.05 for statistical significance with the exception of the PRKCA enhancer assay showing allelic differences in enhancer activity. For that experiment, a linear regression was performed that modeled for the genotype at each SNP as well as an interaction between the two SNPs. This methodology allowed us to test whether the genotype at each SNP produced an effect on luciferase expression and whether the genotypes at the two SNPs acted together to produce synergistic effects on luciferase expression.
Zebrafish studies
Zebrafish experiments were performed using standard protocols approved by the Institutional Animal Care and Use Committee. For enhancer activity screening, the predicted enhancer region was cloned into a tol2-flanked EGFP reporter vector upstream of a CMV minimal promoter for injection into zebrafish embryos. Zebrafish embryos were microinjected at the single cell stage with the vector (25ng/μl) along with transposase mRNA (10ng/μl). Fluorescent reporter activity was assayed at 72 hours post fertilization. Low magnification micrographs, which detail expression across the entire embryo, were acquired on an Olympus SZX16 fluorescent dissecting microscope. For high-resolution images of cardiac specific expression, embryos were anesthetized in tricaine, and hearts were manually excised. Following 30 minutes of fixation in 4% paraformaldehyde, hearts were counterstained in rhodamine-labeled phalloidin, and mounted using Vectashield. A z-series of micrographs were acquired on a Zeiss 510 confocal microscope using a 63X 1.4 NA oil immersion lens with 4μm section separation. Final images represent a 2D projection of the entire acquisition stack.
For analyzing the effects of PRKCA knockdown on cardiac structure, we performed morpholino-mediated knockdown as described.26 Antisense morpholino oligonucleotides targeting the 3rd exon-intron junction of prkca (5′-GCGCATTTGCATACTTACATCTGTA-3′, Genetools, LLC, USA) were injected into the yolk of single cell zebrafish embryos. At 72 hours post-fertilization, embryos were either collected for analysis of knockdown efficiency or analyzed for cardiac dimensions. For calculations of knockdown efficiency, RNA was collected using the quick-RNA mini kit (Zymo Research, USA) and reverse transcribed using iScript (BioRad Inc, USA). Amplification of prkca was performed using primers spanning from exon 1–5. Percent knockdown was measured using ImageJ. Gross morphology was examined using an Olympus SZX16 fluorescent dissecting microscope. Analysis of cardiac dimensions was performed on excised hearts, which were then stained with rhodamine-labeled phalloidin (Sigma-Aldrich, USA), and imaged on a Zeiss LSM510 confocal microscope at 20X magnification. 2D projections of z-series were generated by averaging all frames, with dimensions measured using ImageJ. Statistical comparisons for longitudinal and transverse dimensions of the ventricle were compared using two-way ANOVA in Graphpad Prism7.
Association between rs9912468 and cardiac structure, function, and failure in human populations
We leveraged extant GWAS data to assess the effect of rs9912468 on cardiac structure and function in community-based cohorts. We began by leveraging GWAS data that has determined the association between common variants and the QRS interval, an electrocardiographic measure of left ventricular mass.27 Four related and clinically applied QRS quantitative traits were assessed: the Sokolow-Lyon,28, 29 Cornell,29–31 12-lead-voltage duration products (12-leadsum),29, 31 and QRS duration.28 Briefly, GWAS summary data were combined from 24 studies with up to 2,766,983 autosomal SNPs. These studies together comprise 60,255 individuals of European ancestry ascertained in North America and Europe, with a maximum sample size of 54,993 for Sokolow-Lyon, 58,862 for Cornell, 48,632 for 12-leadsum, and 60,255 for QRS duration. For each SNP, evidence of association was combined across studies using an inverse-variance fixed-effect meta-analysis carried out by two independent groups.
Next, we leveraged GWAS data from the EchoGen consortium32 to determine the association between rs9912468 echocardiographic measured LV mass, LV end-diastolic dimension, LV wall thickness, and LV fractional shortening. This meta-analysis included up to 7 cohorts with total sample sizes up to 11,314 for our traits of interest. Each cohort ran individual linear regression models and data from all cohorts were then combined using inverse-variance weighted fixed effects meta-analysis after applying genomic controls to each cohort.
For all-cause heart failure in the general population, we obtained association summary statistics for SNPs of interest from the published meta-analysis of 4 cohorts of European ancestry (Atherosclerosis Risk in Communities [ARIC] Study, Cardiovascular Health Study [CHS], Framingham Heart Study [FHS], Rotterdam Study [RS]).33 The analysis included a total of 20,926 participants free of clinical heart failure at baseline, in whom 2,526 incident heart failure events occurred during follow-up. Definitions of heart failure varied between the four contributing study cohorts. ARIC relied on ICD-9 codes collected from hospital discharge summaries and death certificates. For CHS, FHS, and RS, heart failure events were identified by self-report or administrative data and were validated by physician’s review of medical records. CHS and FHS applied their published criteria, and RS applied the European Society of Cardiology criteria. All studies imputed their data to HapMap for genome-wide association analyses using Cox proportional hazards regression and performed study-specific quality control. After applying genomic controls to each cohort, data from the four cohorts were meta-analyzed using inverse variance weighted fixed effects meta-analysis.
For prevalent dilated cardiomyopathy in referral cohorts, we tested the association between rs9912468 and odds of dilated cardiomyopathy (DCM) in a meta-analysis of a new case-control GWAS (MAGNet association study; 631 cases with DCM, 1,596 controls) and a previously published dilated cardiomyopathy GWAS (Meder et al.34; 909 DCM cases, 2,120 controls). For the MAGNet association study, we recruited 631 subjects with dilated cardiomyopathy defined as patients with heart failure and an ejection fraction < 40% in the absence of hypertension, primary valvular disease, or coronary artery disease from the University of Pennsylvania Health System and compared them to controls recruited from the same center who had no history of heart disease. All subjects provided written informed consent. Whole genome SNP genotypes for cases and controls were generated using the Illumina OmniExpressExome Array and were imputed to SNP genotypes in the 1000 Genomes Project using Minimac.35 We compared genotypes between cases and controls using an additive genetic model adjusting for gender and 2 principal components of race using SNPtest. There was minimal genomic inflation (γ= 1.0335). To increase power, we meta-analyzed these findings with those from Meder et al using METAL.36 The effective total sample size was 4,342, and there was no evidence of heterogeneity by study (I2=18.5%, heterogeneity P=0.27).
Results
Fine mapping of the PRKCA eQTL reveals a cardiac-specific enhancer
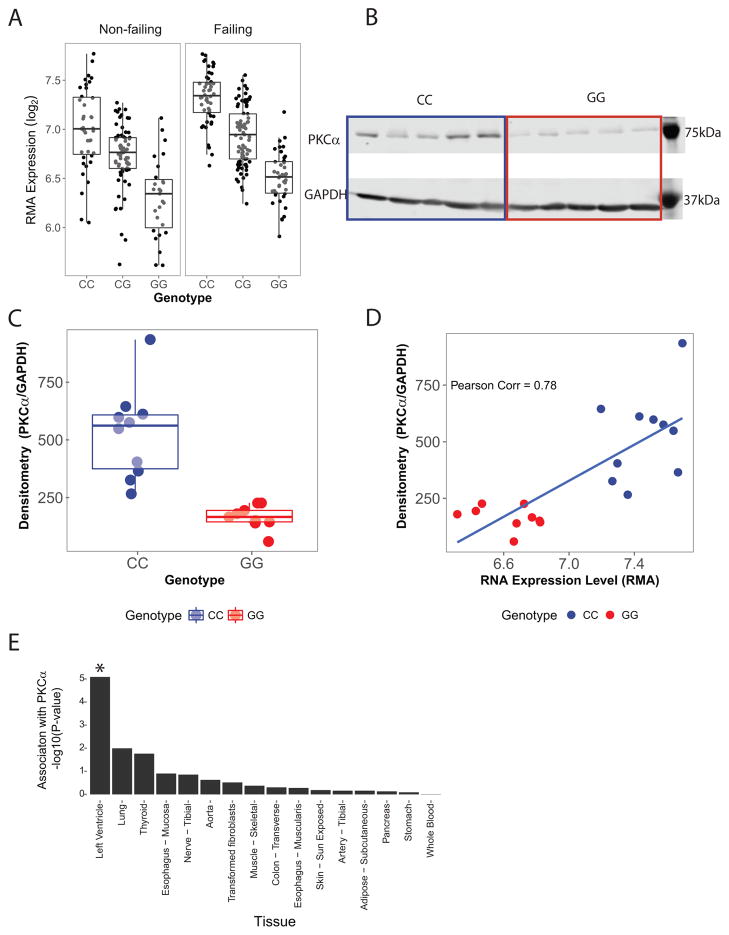
As previously described,18 we performed genome-wide eQTL mapping in 313 human left ventricles from patients of European genetic ancestry (Table 1) and identified a genome-wide significant association between variants at rs9912468 and expression of PRKCA. Minor allele carriers (forward strand G) at rs9912468 showed substantially decreased left ventricular PKCα transcript levels both in the absence and in the presence of heart failure (Figure 1A, P = 1×10−41). No other left ventricular transcripts showed significant association with rs9912468. Western blotting in human LV samples verified that minor allele carriers showed lower PKCα protein abundance (Figures 1B and 1C). Moreover, individual measures of transcript and protein abundance were strongly correlated (Figure 1D), indicating that PKCα transcript abundance is a strong surrogate for PKCα protein abundance in the human LV. To assess the extent to which rs9912468 affects PRKCA expression in tissues other than the heart, we interrogated eQTL data from the Genome Tissue Expression Project (GTEx; Figure 1E).23 Genotypes at rs9912468 were again strongly associated with left ventricular PKCα transcript levels. No other tissue showed statistically significant associations after Bonferroni correction for multiple comparisons.
Table 1.
MAGNet left ventricular eQTL study population (n=313)*
| Unused Cardiac Donors | Heart Failure Explants | |
|---|---|---|
| N | 136 | 177 |
| Age (years) | 49±15 | 56±11 |
| Female Gender | 62 (46) | 33 (18) |
| European Genetic Ancestry | 136 (100) | 136 (100) |
| Left ventricular Ejection Fraction (%) | 50±17 | 16±8 |
| Patient Height (cm) | 170±13 | 174±9 |
| Patient Weight (kg) | 79±25 | 83±15 |
| Heart Weight (g) | 387±132 | 526±124 |
| Normalized Heart Weight (g/m2 BSA) | 203±58 | 268±63 |
| Ischemic Heart Disease | 0 | 94 (53) |
Values are mean ± s.d. or number (%)
Figure 1.
cis eQTL for PRKCA in the human left ventricle. A, Human left ventricular free wall PKCα transcript abundance versus rs9912468 genotype in patients without (n = 136) and with (n =177) heart failure. The minor allele at rs9912468 (forward strand G) is associated with lower PKCα transcript levels (joint effects p = 1×10−41). B, Western blot comparing left ventricular PKCα protein levels in rs9912468 major allele homozygotes (CC, blue) and minor allele homozygotes (GG, red) in a subset of the patients in A. C, Quantitative analysis using densitometry (n=20) demonstrated reduced PKCα protein in minor allele homozygotes (P < 0.001) D, PKCα transcript and protein levels assessed in the same subjects showed strong correlation, indicating that PKCα transcript level is an accurate surrogate for protein level in the human left ventricle. E, Association between rs9912468 genotype and PKCα transcript levels in multiple human tissues based on GTEx data (*Bonferroni P < 0.05). Associations are strongest in left ventricle.
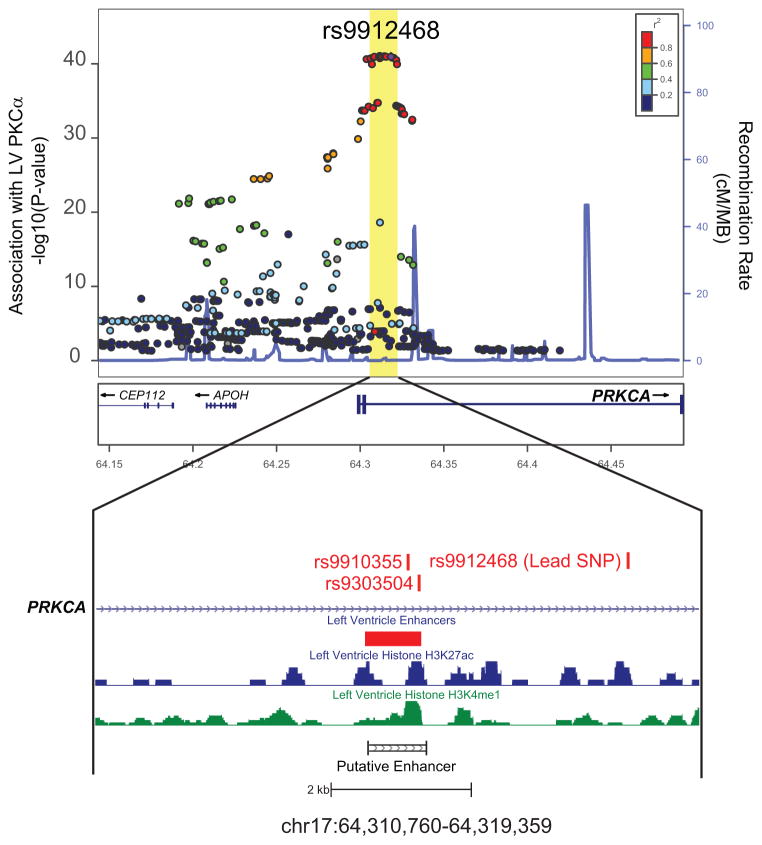
To understand the locus marked by rs9912468 in detail, we constructed a fine map using 1000Genomes imputation to test associations between left ventricular PRKCA expression and genotypes near rs9912468. This revealed a ~20kb block containing multiple variants that strongly associated with PRKCA expression and were in strong linkage disequilibrium (R2 ≥ 0.8) with the sentinel rs9912468 variant. We denote this 20 kb region as the PRKCA eQTL (Figure 2). Roadmap epigenomic histone marks (H3K4me1 and H3K27ac) from human LV ChIPSeq experiments and predicted enhancer regions from ChromHMM revealed that this region contains a single putative enhancer. Two common SNPs (rs9910355 and rs9303504) associated with PRKCA expression physically reside within this enhancer and are in perfect linkage disequilibrium with rs9912468 (Figure 2).
Figure 2. Fine map of the PRKCA cis eQTL in the human left ventricle reveals an enhancer region.
LocusZoom plot of associations between genotypes surrounding rs9912468 and left ventricular PKCα transcript levels were assessed in n = 313 patients. The rs9912468 variant marks a single linkage disequilibrium block within intron 2 of PRKCA that contains variants strongly associated PKCα transcript levels. Within this block is a single putative enhancer based on ChromHMM analysis of human epigenomic data, including results from ChIP-seq experiments assessing enhancer marks H3K27ac (blue) and H3K4me1 (green) in human left ventricular tissue. We cloned this region for functional study (putative enhancer).
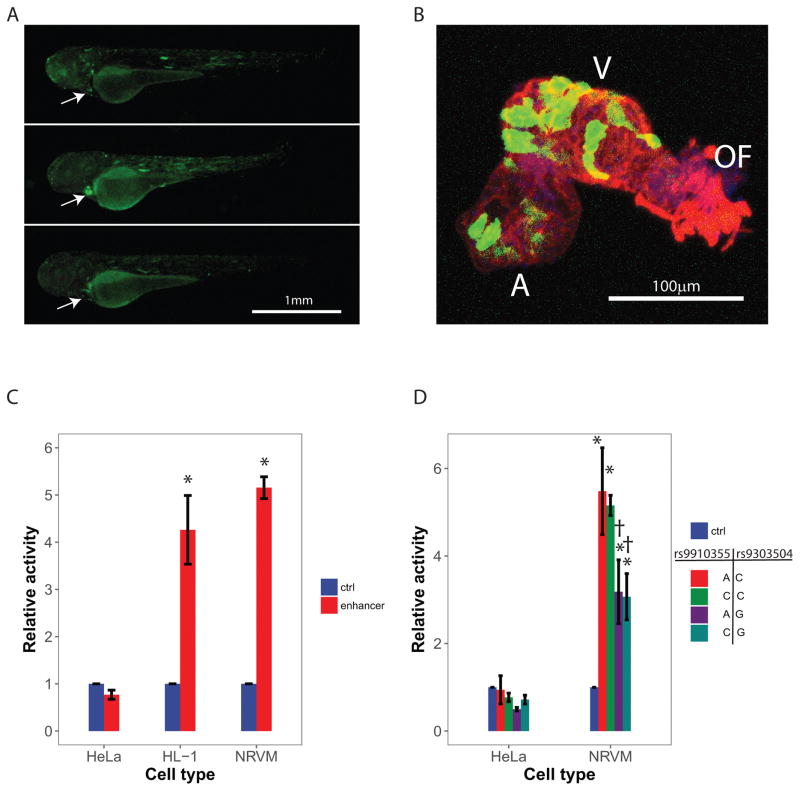
To test enhancer function and specificity in vivo, eGFP constructs containing the cloned enhancer region were microinjected into zebrafish embryos at the single cell stage. Survey of eGFP expression post-injection at 72 hpf revealed marked eGFP expression in cardiac tissue (Figure 3A). Expression of eGFP was also observed to a lesser degree in skeletal muscle cells. Closer interrogation of cardiac eGFP expression within the isolated zebrafish heart (Figure 3B) suggested preferential expression in the ventricle relative to the atrium. To quantify enhancer activity in non-cardiac and in cardiac cells, we cloned the predicted enhancer upstream of firefly luciferase and transfected a non-cardiac cell line (HeLa), the cardiac derived HL-1 myoblast cell line, and freshly isolated neonatal rat ventricular myocytes (NRVM). As shown in Figure 3C, the enhancer region increased luciferase activity ~5-fold in cardiac-derived (HL-1 and NRVM) but not in HeLa cells. Thus, human eQTL data and in vivo and in vitro functional assays all support that the region of interest is a cardiac specific enhancer. Potential binding proteins for this enhancer are unknown and could not be reliably predicted by in silico algorithms.
Figure 3. Cardiac specific PRKCA enhancer activity and identification of a low-activity enhancer haplotype.
A, The cloned human PRKCA enhancer was inserted into an eGFP reporter construct and microinjected into zebrafish embryos. Fluorescent microscopy was used to detail enhancer activity (green). Cardiac specific activity was evident (arrow) in vivo. B, 2-dimensional projections of confocal z-series through excised zebrafish hearts suggest greater enhancer activity (green) in the ventricle (V) when compared to the outflow tract (OF) or atrium (A). The red channel displays the distribution of F-actin with nuclei in blue. C, The cloned human PRKCA enhancer was inserted into a luciferase reporter construct and activity was measured relative to a negative control construct after transfection into different cell types. Enhancer activity was evident after transfection into cardiac-derived cells (the HL-1 cell line and primary neonatal rat ventricular myocytes, NRVM) but not after transfection in non-cardiac cells (HeLa cell line). Error bars represent standard errors for 3 biological replicates for HeLa and HL-1 and 4 biological replicates for NRVM. D, Site-directed mutagenesis was used to create the four enhancer haplotypes observed in humans by altering genotypes at rs9910355 and rs9303504. These were incorporated into luciferase reporter constructs and assessed in vitro. All haplotypes were inactive after transfection into HeLa cells, but were active in NRVMs. Enhancer activity was 40% lower for haplotypes containing the minor allele (G) at rs9303504. These ‘low activity haplotypes’ are in perfect linkage disequilibrium (R2 = 1.0) with the minor allele (G) at the sentinel variant rs9912468 described in Figure 1. Error bars represent standard errors of 4 biologic replicates. *P<0.05 vs. control. †P<0.05 vs. haplotype-AC.
Effect of enhancer haplotypes on PRKCA expression
The variants rs9910355 and rs9303504 described above physically reside within the enhancer and together define four haplotypes. To compare their activity, we cloned the enhancer region, performed site directed mutagenesis to produce all four haplotypes, and compared their ability to drive a luciferase reporter in NRVMs (Figure 3D). Regardless of haplotype, cells transfected with the enhancer sequence showed increased luciferase activity when compared to control. We found that haplotypes containing the minor allele, G, at rs9303504 showed a 40% decrease in luciferase activity compared to the major allele, C (P = 0.004), whereas luciferase activity was unaffected by genotype at rs9910355 (P = 0.8). These findings parallel the expected directionality based on the eQTL associations in the human left ventricle. That is, the minor allele, G, at the lead SNP rs9912468 marks a PRKCA enhancer haplotype containing a minor allele at rs9303504 that reduces enhancer activity and left ventricular PRKCA expression.
The rs9912468 minor allele G associates with adverse remodeling, impaired contractile function, and dilated cardiomyopathy
Having established that the minor allele at rs9912468 tags a cardiac specific enhancer with reduced activity and reduced ventricular PRKCA expression, we assessed the impact of PRKCA through expression modulation in the zebrafish embryo and through examination of rs9912468 genotypes in human subjects using extant GWAS data. After knockdown in zebrafish, no effect on ventricular dimensions was observed (Supplemental Figure), although embryonic phenotypes may not be the most appropriate model of adult cardiomyopathy. For human analyses, traits interrogated include measures of left ventricular mass, dilatation, wall thickness, contractile function, all-cause heart failure in the community, and dilated cardiomyopathy in referral cohorts (Table 2). The PKCα lowering allele (G) at rs9912468 associated with increased left ventricular mass measured by electrocardiogram27 using the Cornell voltage criteria (P = 0.0026), 12-lead sum area (P = 4.99 × 10−11), Sokolow-Lyon area (P = 3 × 10−12), and QRS duration (P = 1.65 × 10−11). In the EchoGEN Consortium32, the PKCα lowering allele associated with increased left ventricular internal diastolic dimension (P = 0.0003), thinning of the left ventricular wall (P = 0.0039), and a potential decrease in fractional shortening (P = 0.039). An association with LV mass by echocardiogram was not observed, nor was there an association with incident all-cause heart failure in the community in the CHARGE consortium.37 In a meta-analysis of a new and a previously published34 dilated cardiomyopathy case-control GWAS, the PKCα lowering allele associated with dilated cardiomyopathy (OR 1.19, P= 0.0012). The directionality across all associations is consistent and suggests that individuals inheriting the PKCα lowering allele have higher LV mass, more LV dilation, LV wall thinning, and higher risk of clinically diagnosed dilated cardiomyopathy.
Table 2.
Effect of PKCα lowering allele (rs9912468 forward strand G) on LV structure, function, and disease*
| Trait | Sample Size | Allele frequency | Beta | Odds Ratio or Hazard Ratio (95% CI) | P-value |
|---|---|---|---|---|---|
| LV mass by electrocardiogram voltage | |||||
| Cornell | 52,620 | 0.44 | +9.8 mV | - | 0.0026 |
| Leadsum | 39,939 | 0.42 | +156 mV | - | 5.0 × 10−11 |
| QRS duration | 60,025 | 0.43 | +0.4 mV | - | 1.7 × 10−11 |
| Sokolow | 48,740 | 0.44 | +32 msec | - | 3.0 × 10−12 |
| LV structure and function by echocardiography | |||||
| LV mass index | 11,273 | 0.44 | +0.03 g/m2 | - | 0.95 |
| LV internal diastolic dimension | 11,314 | 0.44 | +0.20 mm | - | 0.0003 |
| LV wall thickness | 11,311 | 0.44 | −0.081 mm | - | 0.0039 |
| LV fractional shortening | 11,167 | 0.44 | −0.15 | - | 0.039 |
| All-cause heart failure in community cohorts | 2,526 incident cases 20,926 total population | 0.43 | - | 1.02 (0.96 – 1.07) | 0.55 |
| Dilated cardiomyopathy in referral cohorts | 1,660 prevalent cases 3,220 controls | 0.43 | - | 1.19 (1.06 – 1.32) | 0.0012 |
Beta indicates average change in parameter per allele copy; odds and hazard ratios indicate average fold-increase in risk per allele copy;
CI, confidence interval; LV, left ventricular
Discussion
This report combines genetic, transcriptomic, and epigenetic data to uncover a cardiac specific enhancer that regulates PRKCA expression in the human left ventricle. Common variants within this enhancer potently affect expression of PRKCA in the human heart, but not in other tissues, and functional studies in vitro and vivo verify that enhancer activity is specific to the heart. Paradoxically, minor allele carriers at rs9912468, which marks the enhancer haplotype that reduces PKCα in the left ventricle, show increases in adverse cardiac remodeling and cardiomyopathy risk. In sum, these data suggest a mechanism whereby minor allele carriers inherit genetically blunted left ventricular PRKCA expression that causes mild adverse effects on the human heart.
Li et al38 recently reported an allelic imbalance study in 51 left ventricular tissue specimens that associates the minor allele at rs9909004 with lower PKCα transcript abundance. Further, they speculate that this allele could have favorable cardiac effects because it associates with a shortening of the electrocardiographic QT interval in a GWAS meta-analysis of cardiac repolarization.18 The rs9909004 variant is in perfect linkage disequilibrium with rs9912468 (R2 = 1.0) and thus marks the same locus studied herein with the same reported effect on PKCα reduction. However, Sotoodehnia et al39 demonstrate that although the minor allele at this locus associates with QT shortening and may reduce the risk of torsade de pointe, the same allele also associates with QRS prolongation, and may promote heart failure through left ventricular dyssynchrony. These discordant effects on QRS and QT intervals are evident for several loci that affect electrocardiographic traits.38 To shed more light on the clinical impact of the PKCα lowering allele, we examined associations with additional cardiac traits (Table 2). Overall, these show a pattern that is not favorable, with increased LV mass by EKG and echo, more ventricular dilatation, and higher risk of clinically diagnosed cardiomyopathy.
These findings appear to contradict previously published animal studies. Whereas studies in rodents and pigs found cardioprotective effects of PKCα inhibition,5–8 our genetic data suggest adverse effects. There are several potential explanations for this paradox. Species differences are an obvious consideration. Alternatively, the discrepancy between our human genetic and previous animal studies may reflect differences in dosage and time course of change in PKCα activity. Whereas pharmacologic inhibition in animal studies occurs in the acute setting with near complete abolishment of PKCα activity, the variant in our study is associated with a life-long and partial decrease in PRKCA expression. It may be reasonable to expect differential cardiac effects of reduced PKCα during the preclinical phase of heart failure development compared to PKCα inhibition in established heart failure. Even among patients with established heart failure, there is tremendous heterogeneity in etiologic factors, severity of disease, treatment approaches, and disease setting (e.g., chronic heart failure versus acutely decompensated heart failure). Given previously observed inotropic effects of PKCα inhibition in animal models,6 PKCα inhibition may have utility in acute settings, but toxicity in the setting of heart failure prevention. Detailed study of the effects of the PRKCA eQTL on human left ventricular PKCα activity and the phosphorylation state of PKCα targets may provide additional insights.
The convergence of work in animal models and in human genetics identifying PRKCA as a modifier of myocardial disease raises its profile as a pharmacologic target. However the path forward for drug development is not obvious. Currently, there is no experience assessing the impact of PKCα inhibition on human myocardial function. The only cardiovascular analysis of PKCα inhibition in humans comes from studies with ruboxistaurin, a mixed PKCα and PKCβ inhibitor. In two separate early phase trials, ruboxistaurin administration in diabetics induced improvements in flow40 and methacholine41 mediated peripheral artery dilatation. Microvascular effects have been assessed in approximately 1400 patients with diabetic retinopathy across a number of clinical trials. Interestingly, aggregate analysis of these trials suggests a reduction in all-cause mortality with ruboxistaurin.42 Assessing therapeutic potential in human heart failure will require carefully designed pharmacologic studies to determine which subgroups might or might not benefit. These studies should consider the pharmacogenomic implications of the PRKCA eQTL in their design.
A final observation of importance is that there are clearly endophenotypes of heart failure that have distinct genetic architectures. The PRKCA eQTL had a consistent influence on heart failure characterized by left ventricular dilatation, reduction in contractile function, and increased risk of dilated cardiomyopathy. But there was no discernible effect on all-cause heart failure in the community, a heterogeneous phenotype that also includes heart failure with preserved ejection fractions and ischemic heart disease. Precise definition of heart failure subtypes should increase our ability to elucidate the genetic architecture of myocardial disease, which likely differs across subtypes.
In conclusion, we uncovered common enhancer variants that reduce PRKCA expression in the human heart and in so doing affect cardiac structure, function, and disease. These findings support PKCα as regulator of the human heart, but raise the possibility that PKCα inhibition may incur cardiac risk depending on timing and duration. Pharmacologic studies in human subjects are required to discern the potential benefits and harms of PKCα inhibitors as an approach to treat heart disease.
Supplementary Material
Clinical Perspective.
Protein kinase C alpha (PKCα) is a serine-threonine kinase that functions downstream of membrane-associated signal transduction pathways to regulate myocardial function. PKCα inhibition enhances contractility and cardioprotection in animal models and has been proposed as a therapeutic strategy for myocardial disease. However, there is no experience assessing the impact of PKCα inhibition on human myocardial function. Our group and others have recently shown that common variants within the gene encoding PKCα (PRKCA) are linked to reduced gene expression and lower PKCα transcript levels in the human left ventricle. We analyzed these variants in detail to understand the mechanisms and clinical impact of inherited reduction in ventricular PKCα. A cardiac-specific enhancer within intron 2 of PRKCA containing variants that impair enhancer activity may explain the reduction in gene expression. In contrast to findings in animal models, individuals who inherit alleles that lower PKCα show a pattern that is not favorable, with increased left ventricular mass, more ventricular dilatation, and higher risk of clinically diagnosed cardiomyopathy. These findings support PKCα as regulator of the human heart, but raise the possibility that PKCα inhibition may increase cardiac risk depending on timing and duration. Pharmacologic studies in human subjects are required to discern the potential benefits and harms of PKCα inhibitors as an approach to treat heart disease.
Acknowledgments
Source of Funding: Supported by grants from the US National Institutes of Health (T32HL007208, TL1TR000138, HL105993, HL088577, HL098283, HL113933, HL092577, HL105780), Doris Duke Charitable Foundation, Burroughs Wellcome Fund, European Union BestAgeing FP7, Dutch Kidney Foundation, EU Project Grant GENECURE, Netherlands Organisation for Health Research and Development, Dutch Inter University Cardiology Institute Netherlands (ICIN), American Heart Association (13EIA14220013), Fondation Leducq (14CVD01), and the George L. Nardi Memorial Research Fund.
Footnotes
Acknowledgements: None.
Disclosures: Dr. Ellinor is principal investigator on a grant from Bayer AG focused on the genetics and therapeutics of atrial fibrillation.
References
- 1.Vlahos CJ, McDowell SA, Clerk A. Kinases as therapeutic targets for heart failure. Nat Rev Drug Discov. 2003;2:99–113. doi: 10.1038/nrd1009. [DOI] [PubMed] [Google Scholar]
- 2.Dorn GW, 2nd, Force T. Protein kinase cascades in the regulation of cardiac hypertrophy. J Clin Invest. 2005;115:527–37. doi: 10.1172/JCI24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hambleton M, Hahn H, Pleger ST, Kuhn MC, Klevitsky R, Carr AN, et al. Pharmacological- and gene therapy-based inhibition of protein kinase Calpha/beta enhances cardiac contractility and attenuates heart failure. Circulation. 2006;114:574–82. doi: 10.1161/CIRCULATIONAHA.105.592550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ping P, Zhang J, Qiu Y, Tang XL, Manchikalapudi S, Cao X, et al. Ischemic preconditioning induces selective translocation of protein kinase C isoforms epsilon and eta in the heart of conscious rabbits without subcellular redistribution of total protein kinase C activity. Circ Res. 1997;81:404–14. doi: 10.1161/01.res.81.3.404. [DOI] [PubMed] [Google Scholar]
- 5.Braz JC, Gregory K, Pathak A, Zhao W, Sahin B, Klevitsky R, et al. PKC-alpha regulates cardiac contractility and propensity toward heart failure. Nat Med. 2004;10:248–54. doi: 10.1038/nm1000. [DOI] [PubMed] [Google Scholar]
- 6.Hambleton M, York A, Sargent MA, Kaiser RA, Lorenz JN, Robbins J, et al. Inducible and myocyte-specific inhibition of PKCalpha enhances cardiac contractility and protects against infarction-induced heart failure. Am J Physiol Heart Circ Physiol. 2007;293:H3768–71. doi: 10.1152/ajpheart.00486.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladage D, Tilemann L, Ishikawa K, Correll RN, Kawase Y, Houser SR, et al. Inhibition of PKCalpha/beta with ruboxistaurin antagonizes heart failure in pigs after myocardial infarction injury. Circ Res. 2011;109:1396–400. doi: 10.1161/CIRCRESAHA.111.255687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyle AJ, Kelly DJ, Zhang Y, Cox AJ, Gow RM, Way K, et al. Inhibition of protein kinase C reduces left ventricular fibrosis and dysfunction following myocardial infarction. J Mol Cell Cardiol. 2005;39:213–21. doi: 10.1016/j.yjmcc.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Chen X, Macdonnell SM, Kranias EG, Lorenz JN, Leitges M, et al. Protein kinase C{alpha}, but not PKC{beta} or PKC{gamma}, regulates contractility and heart failure susceptibility: implications for ruboxistaurin as a novel therapeutic approach. Circ Res. 2009;105:194–200. doi: 10.1161/CIRCRESAHA.109.195313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Molkentin JD. Protein kinase Calpha as a heart failure therapeutic target. J Mol Cell Cardiol. 2011;51:474–8. doi: 10.1016/j.yjmcc.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowling N, Walsh RA, Song G, Estridge T, Sandusky GE, Fouts RL, et al. Increased protein kinase C activity and expression of Ca2+-sensitive isoforms in the failing human heart. Circulation. 1999;99:384–91. doi: 10.1161/01.cir.99.3.384. [DOI] [PubMed] [Google Scholar]
- 12.Simonis G, Briem SK, Schoen SP, Bock M, Marquetant R, Strasser RH. Protein kinase C in the human heart: differential regulation of the isoforms in aortic stenosis or dilated cardiomyopathy. Mol Cell Biochem. 2007;305:103–11. doi: 10.1007/s11010-007-9533-3. [DOI] [PubMed] [Google Scholar]
- 13.Rader DJ. New therapies for coronary artery disease: genetics provides a blueprint. Sci Transl Med. 2014;6:239ps4. doi: 10.1126/scitranslmed.3008535. [DOI] [PubMed] [Google Scholar]
- 14.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 15.Ridker PM, Revkin J, Amarenco P, Brunell R, Curto M, Civeira F, et al. Cardiovascular Efficacy and Safety of Bococizumab in High-Risk Patients. N Engl J Med. 2017;376:1527–1539. doi: 10.1056/NEJMoa1701488. [DOI] [PubMed] [Google Scholar]
- 16.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–7. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dewey FE, Gusarova V, Dunbar RL, O’Dushlaine C, Schurmann C, Gottesman O, et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N Engl J Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46:826–36. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koopmann TT, Adriaens ME, Moerland PD, Marsman RF, Westerveld ML, Lal S, et al. Genome-wide identification of expression quantitative trait loci (eQTLs) in human heart. PLoS One. 2014;9:e97380. doi: 10.1371/journal.pone.0097380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Morley M, Brandimarto J, Hannenhalli S, Hu Y, Ashley EA, et al. RNA-Seq Identifies Novel Myocardial Gene Expression Signatures of Heart Failure. NCBI GEO Datasets. 2015 doi: 10.1016/j.ygeno.2014.12.002. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57338. [DOI] [PMC free article] [PubMed]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, Normalization, and Summaries of High Density Oligonucleotide Array Probe Level Data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 23.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinner MF, Tucker NR, Lunetta KL, Ozaki K, Smith JG, Trompet S, et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation. 2014;130:1225–35. doi: 10.1161/CIRCULATIONAHA.114.009892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Harst P, van Setten J, Verweij N, Vogler G, Franke L, Maurano MT, et al. 52 Genetic Loci Influencing Myocardial Mass. J Am Coll Cardiol. 2016;68:1435–48. doi: 10.1016/j.jacc.2016.07.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokolow M, Lyon TP. The ventricular complex in right ventricular hypertrophy as obtained by unipolar precordial and limb leads. Am Heart J. 1949;38:273–94. doi: 10.1016/0002-8703(49)91335-6. [DOI] [PubMed] [Google Scholar]
- 29.Carlsson MB, Tragardh E, Engblom H, Hedstrom E, Wagner G, Pahlm O, et al. Left ventricular mass by 12-lead electrocardiogram in healthy subjects: comparison to cardiac magnetic resonance imaging. J Electrocardiol. 2006;39:67–72. doi: 10.1016/j.jelectrocard.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Casale PN, Devereux RB, Kligfield P, Eisenberg RR, Miller DH, Chaudhary BS, et al. Electrocardiographic detection of left ventricular hypertrophy: development and prospective validation of improved criteria. J Am Coll Cardiol. 1985;6:572–80. doi: 10.1016/s0735-1097(85)80115-7. [DOI] [PubMed] [Google Scholar]
- 31.Molloy TJ, Okin PM, Devereux RB, Kligfield P. Electrocardiographic detection of left ventricular hypertrophy by the simple QRS voltage-duration product. J Am Coll Cardiol. 1992;20:1180–6. doi: 10.1016/0735-1097(92)90376-x. [DOI] [PubMed] [Google Scholar]
- 32.Vasan RS, Glazer NL, Felix JF, Lieb W, Wild PS, Felix SB, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, Loehr LR, et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2010;3:256–66. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meder B, Ruhle F, Weis T, Homuth G, Keller A, Franke J, et al. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J. 2014;35:1069–77. doi: 10.1093/eurheartj/eht251. [DOI] [PubMed] [Google Scholar]
- 35.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, Loehr LR, et al. The Association of Genome-Wide Variation with the Risk of Incident Heart Failure in Adults of European and African Ancestry: A Prospective Meta-Analysis from the CHARGE Consortium. Circ Cardiovasc Genet. 2010;3:256–66. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Zhang L, Binkley PF, Sadee W, Wang D. Regulatory Variants Modulate Protein Kinase C alpha (PRKCA) Gene Expression in Human Heart. Pharm Res. 2017;34:1648–1657. doi: 10.1007/s11095-017-2102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sotoodehnia N, Isaacs A, de Bakker PI, Dorr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–76. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehta NN, Sheetz M, Price K, Comiskey L, Amrutia S, Iqbal N, et al. Selective PKC beta inhibition with ruboxistaurin and endothelial function in type-2 diabetes mellitus. Cardiovasc Drugs Ther. 2009;23:17–24. doi: 10.1007/s10557-008-6144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beckman JA, Goldfine AB, Gordon MB, Garrett LA, Creager MA. Inhibition of protein kinase Cbeta prevents impaired endothelium-dependent vasodilation caused by hyperglycemia in humans. Circ Res. 2002;90:107–11. doi: 10.1161/hh0102.102359. [DOI] [PubMed] [Google Scholar]
- 42.Javey G, Schwartz SG, Flynn HW, Aiello LP, Sheetz MJ. Ruboxistaurin: Review of safety and efficacy in the treatment of diabetic reinopathy. Clin Med Insights Ther. 2010;2:625–631. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.