Abstract
Sanger sequencing, the traditional “gold standard” for mutation detection, has been wildly used in genetic testing of pulmonary artery hypertension (PAH). However, with the advent of whole-exome sequencing (WES), few studies have compared the accuracy of WES and Sanger sequencing in routine genetic testing of PAH. PAH individuals were enrolled from Fu Wai Hospital and Shanghai Pulmonary Hospital. WES was used to analyze DNA samples from 120 PAH patients whose bone morphogenetic protein receptor type 2 (BMPR2) mutation statuses had been previously studied using Sanger sequencing. The Sanger sequencing and WES agreement was 98.3% (118/120) with near-perfect agreement (κ coefficient = 0.848). There was no significant difference between the two methods on the McNemar–Bowker test (P > 0.05). Twenty-one BMPR2 mutation carriers and 99 non-carriers were detected by Sanger sequencing. Among the 21 BMPR2 carriers detected by Sanger sequencing, one variant (c.1040 T > A) was not found by WES. Among the 99 BMPR2 non-carriers, WES detected an extra mutation carrier (c.76 + 1 G > C) missed by Sanger sequencing. Both false-positive and false-negative results were highly conserved and were re-analyzed by Sanger sequencing. WES improved the accuracy of Sanger sequencing and detected 1% (1/99) false-positive and 4.8% (1/21) false-negative results of Sanger sequencing. No false-positive and false-negative results of WES were identified in our analysis. WES is non-inferior to Sanger sequencing and may play a more important role in genetic testing of PAH patients in the future.
Keywords: whole-exome sequencing, Sanger sequencing, genetic testing, pulmonary artery hypertension
Pulmonary artery hypertension (PAH) is a fatal disease that is clinically silent in the early stages, while the symptoms develop with progression of the disease. Early diagnosis and timely intervention in PAH may translate into better long-term outcomes.1,2 Pre-symptomatic genetic screening of at-risk populations such as known heritable/familial PAH consequently results in careful and regular clinical follow-up of asymptomatic mutation carriers and facilitates early PAH diagnosis.1
The dominant genetic cause of familial PAH is mutations in the gene of bone morphogenetic protein receptor type 2 (BMPR2) that account for approximately 75% of heritable PAH and up to 25% of presumably idiopathic PAH (IPAH).1 Compared with non-BMPR2 mutation carriers, BMPR2 mutation carriers are diagnosed at a younger age and have worse hemodynamic parameters, less response to acute vasodilators, and fewer benefits from the treatment of calcium channel blockade.2,3
Sanger sequencing is a traditional DNA sequencing technology using chain-terminating inhibitors with capillary array electrophoresis. As is generally affordable and feasible for most laboratories, Sanger sequencing has been widely used in the genetic testing for PAH.4 However, traditional Sanger sequencing only focuses on certain regions of chromosomes and needs much manual operation from DNA synthesis and sequencing to result interpretation.5,6 Obtaining a complete genetic view of disease, requiring high-quality sequencing of large number of genes and genomic regions, is beyond the scope and capacity of Sanger sequencing and highlights the importance of next-generation sequencing (NGS).4
Whole-exome sequencing (WES) utilizes NGS to sequence all protein-coding regions, or exons, of the human genome. WES relies on sequencing by synthesis to obtain nucleic acid sequences from the amplified libraries which provides a much faster, cheaper, and higher throughput alternative to sequencing DNA.7 Meanwhile, recently developed statistical methods have improved disease-related variants calling rate, especially for the single nucleotide polymorphism detection.8 Compared with manual reading electropherograms in Sanger sequencing, WES could automatically accomplish splicing, annotation, interpretation, and result output which avoids the potential artificial errors caused by Sanger sequencing.5,7
Using WES, Austin et al. and Ma et al. identified caveolin 1 (CAV1) and potassium two pore domain channel subfamily K member 3 (KCNK3) as two new candidate genes in patients with BMPR2-negative familial PAH.9,10 Recently, there have been increasing studies reporting the superiority of WES, not only for the high diagnostic yield11 but also for the ability to detect mutations missed by Sanger sequencing, such as in Charcot–Marie–Tooth disease type 2.12
With the advent of NGS, more and more studies have made a comparison of NGS with Sanger sequencing in various diseases.13–15 However, few studies have directly compared the accuracy of WES with Sanger sequencing in PAH. The aim of the present study is to evaluate whether WES can provide more accurate results (defined by fewer false-negative/-positive results) than Sanger sequencing in the routine genetic testing of BMPR2 in PAH patients.
Materials and method
Setting and study participants
The study was conducted between December 2013 and January 2015 at two PAH centers in China. PAH patients, visited in Beijing Fuwai Hospital and Shanghai Pulmonary Hospital, were screened for BMPR2 mutation using Sanger sequencing and WES. PAH was diagnosed by right heart catheterization as defined by mean pulmonary artery pressure ≥25 mmHg, pulmonary capillary wedge pressure ≤ 15 mmHg, and pulmonary vascular resistance > 3 Wood units.16 Heritable PAH was recognized if there was more than one confirmed case in first- to third-degree relatives in the family. IPAH was the diagnosis after exclusion of other disorders known to cause pulmonary hypertension (PH), as summarized in the updated guideline,16 by clinical evaluation and objective tests and absence of a family history of PAH.
This study was approved by the Ethics Committee of Fu Wai and Shanghai Pulmonary Hospital and all participants signed informed consent for genetic analyses before participation.
Molecular methods of Sanger sequencing
Genomic DNA was isolated from peripheral blood leukocytes using proteinase, chloroform extraction, ethanol precipitation, and Tris-buffered EDTA resuspension. Direct screening was performed using the ABI 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA) to detect point mutations in the coding regions and intron/exon boundaries of BMPR2 gene. Fourteen pairs of PCR primers were designed to amplify 13 exons (Table 1). Valid sequencing data were mapped to the reference sequence of the BMPR2 gene (RefSeq accession number NM_001204.5) using the ABI SeqScape software, version 2.5 (Applied Biosystems). Sanger sequencing data were analyzed in a double-blind, back-to-back manner. The mutation nomenclature was referred to current guidelines recommended by the Human Genome Variation Society. The mutation number employed in this report was based on the cDNA sequence, where +1 designates the A of the ATG initiation codon. All participants were further sequenced by WES.
Table 1.
Primers of BMPR2.
| EXON | Primer | Sequence (5’ to 3’) | Base number | Product size |
|---|---|---|---|---|
| EXON1 | F | AGGAAGCACCGAAGCGAAA | 19 | 827 |
| R | TGGAAGTGGGGATA GGAAAATAC | 23 | ||
| EXON2 | F | TGAACAGAAGAACGTCATTG | 20 | 475 |
| R | CACCACACCTGGCTTTAT | 18 | ||
| EXON3 | F | TCATAGCTTACACGTA CTCTCACA | 24 | 381 |
| R | CACGCCTGGCTTCAACC | 17 | ||
| EXON4 | F | CATGGGTACAGCCTTTC | 17 | 384 |
| R | AGGGGTAGTGACTGATAAT | 19 | ||
| EXON5 | F | CTTGCTGCTAATCTTTCTGC | 20 | 383 |
| R | CCCAGGAGACATTGTTCAC | 19 | ||
| EXON6 | F | GAGCTTCATCAGCCATACTA | 20 | 546 |
| R | GTAGAGACGGGAGTTTTGC | 19 | ||
| EXON7 | F | TTCATGGAATCCTAGCCTAT | 20 | 504 |
| R | TACTTTCTCTGCTGGGTCA | 19 | ||
| EXON8 | F | CAAGTGCGTGTTAGAATCTG | 20 | 574 |
| R | AGCTGGTCTCGAACTCTTT | 19 | ||
| EXON9 | F | GGATTTACCACAGTTTACTT | 20 | 502 |
| R | CTGCTGCTAATAATGTTTC | 19 | ||
| EXON10 | F | TTGCTTACTTGGTATCAGAA | 20 | 352 |
| R | TTGATTTGTGGCATTAGG | 18 | ||
| EXON11 | F | GTAATCCTTGAAGCCTAAAA | 20 | 586 |
| R | GAAAGTTGGACCAT AAATAGAG | 22 | ||
| EXON12-1 | F | TTTCAGTAGGCTTAATTCACA | 21 | 1057 |
| R | CCTGCCACACCATTCAT | 17 | ||
| EXON12-2 | F | CCTTCCCAAGAGACCTACTA | 20 | 745 |
| R | TGGATCATTTACAAAAGTGG | 20 | ||
| EXON13 | F | GACCTTTTCTTGAGTTACATC | 21 | 1061 |
| R | TTTAGAGCCCTTTCAGAC | 18 |
BMPR2, bone morphogenetic protein receptor 2.
WES library preparation and sequencing
The quality and integrity of isolated genomic DNA were accessed on 0.8% agarose gel electrophoresis. DNA concentration was measured using Qubit 2.0 Fluorometer (Life Technologies, CA, USA). Exome sequences were enriched from 1.0 µg genomic DNA using an Agilent liquid captures system (Agilent SureSelect Human All Exon V6, CA, USA) according to the manufacturer’s protocol. Qualified genomic DNA was fragmented, end repaired, and phosphorylated, followed by A-tailing and ligation at the 3′ ends with paired-end adaptors (Illumina, CA, USA). DNA libraries were sequenced on Illumina HiSeq 4000 for paired-end 150 bp reads.
WES data analysis
The raw image files obtained from HiSeq 4000 were processed with Illumina Pipeline for base calling and raw data were stored as FastQ format. Further quality control was applied to guarantee high-quality clean data in downstream bioinformatics analyses (see Supplemental material). The cleaned-up sequence reads were aligned to the human reference genome (UCSC hg19) assembly with Burrows-Wheeler Aligner software.17 Reads that aligned to exon regions were collected for mutation identification with subsequent analysis. The splicing variants and variants located 2 bp to junction were also considered. Samtools mpileup, bcftools, and CoNIFER18 were used to access variant calling for single-nucleotide polymorphism, indels, and copy number variations. Only non-synonymous variants were retained. Functional annotation of variants was carried out using ANNOVAR.19 Variants obtained from previous steps were subsequently filtered out if the reported minor allele frequency was > 1% in the 1000 Genomes database (1000 Genomes Project Consortium).20 PolyPhen-2,21 SIFT,21 MutationTaster,22 and CADD23 were used to predict the effect of variants on the protein structure and functions. Variants were classified as pathogenic or likely pathogenic by at least half of the software.
Sanger sequencing re-validation
For the discordant results between Sanger sequencing and WES, Sanger sequencing was performed for further validation.
Statistical measures of performance
Statistical analyses were performed using SPSS version 20.0 statistical software (SPSS Inc.). Comparisons between WES and Sanger sequencing were analyzed using the McNemar–Bowker test. Concordance data between the two methods were obtained and κ statistic was used to measure the agreement of positive ratios between the two assays. The κ statistic evaluates the level of agreement between the two methods attributable to actual agreement rather than potential agreement by chance, with a κ-coefficient of > 0.75 indicating near-perfect agreement, 0.45–0.75 indicating moderate agreement, and < 0.45 indicating slight agreement, no agreement, or a random association.24 Test clinical sensitivity, specificity, and false-positive and false-negative rates were also calculated.25 Results with a P value < 0.05 were considered to be statistically significant.
Results
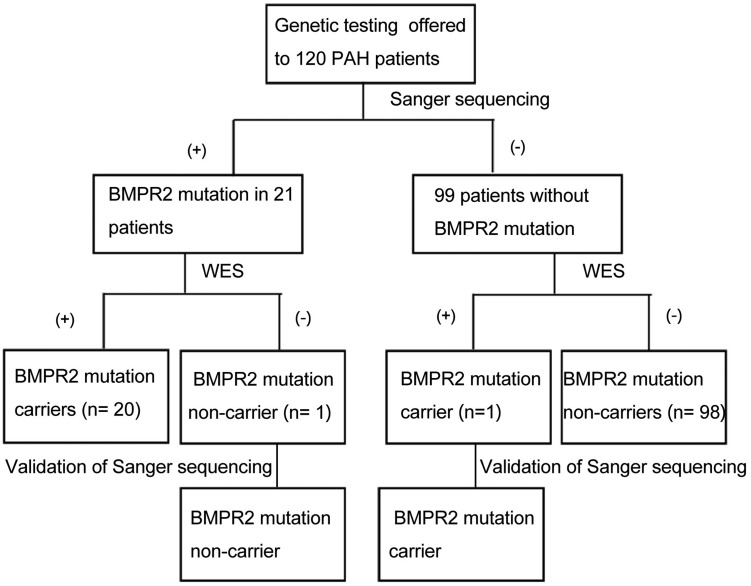
Between December 2013 and January 2015, genetic counselling and testing were performed in 120 PAH patients (mean age = 30.7 ± 6.6 years; 80% women), including ten heritable PAH and 110 IPAH. In this cohort, 96 (90%) patients were aged > 18 years. Twenty-one BMPR2 mutation carriers and 99 non-BMPR2 carriers were detected by Sanger sequencing (Fig.1). All patients were further sequenced by WES. After WES, an average of 8.8 Gb raw data and 154,888 variants per case, as paired-end 150 bp reads, were reported. On average, 99.69% of whole exome regions were sequenced, and the average depth of sequencing was 146.3X. As BMPR2 is the dominant genetic cause of PAH, we mainly focused on the variants of BMPR2.
One IPAH patient identified by Sanger sequencing as BMPR2 carrier (c.1040 T > A) was not found by WES. Another IPAH patient with variant (c.76 + 1 G > C) missed by Sanger sequencing was detected by WES. Overall, the rate of BMPR2 mutation was 17.5% in the overall cohort (14.5% in the IPAH group and 50.0% in the heritable PAH group) which was similar to our previous published data.26 Mutations in other PAH-predisposing genes (ENG, SMAD9, CAV-1, KCNK3 and ACVRL1, SMAD9, BMPR1B) were identified in 15% (n = 18) of patients who were considered to have IPAH or heritable PAH. These findings indicated that WES could detect the false-positive and false-negative results of Sanger sequencing. The genetic information of the false-positive/-negative results was summarized in Table 2.
Table 2.
Genetics information of false-positive and false-negative results.
| Patients ID | Exon number | Nucleotide change | Amid aid change | Mutation type |
|---|---|---|---|---|
| B792 | 8 | c.T1040 A | p.V713G | Missense mutation |
| 2028 | 1 | c.76 + 1 G > C | Splicing mutation |
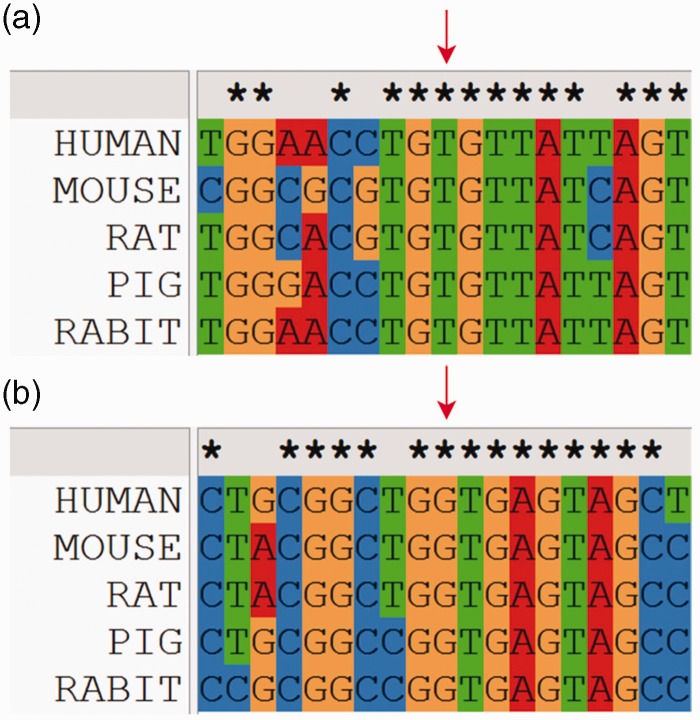
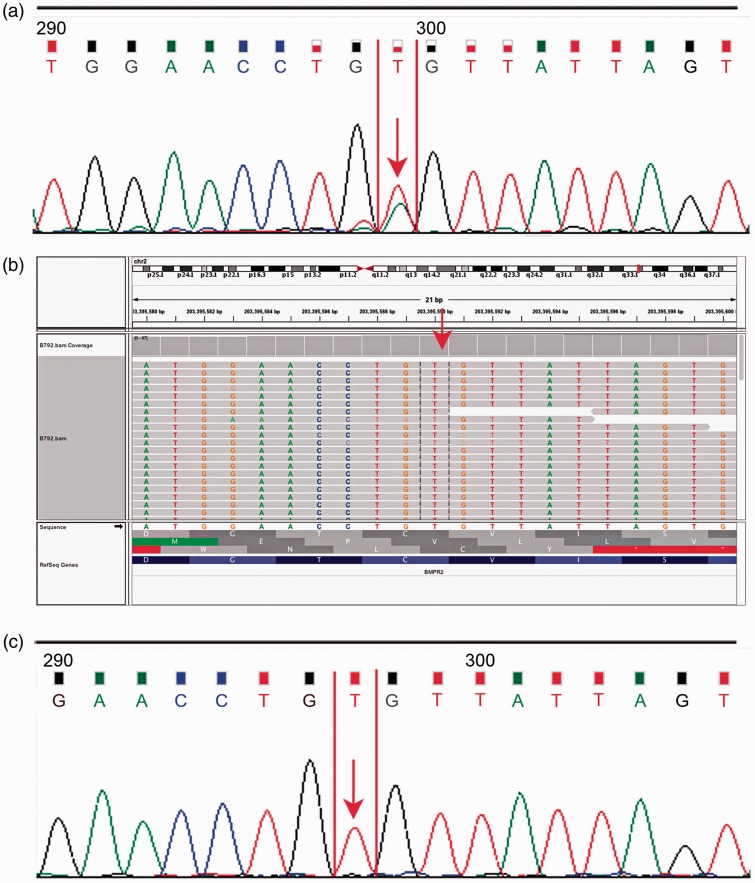
The missense variant c.1040 T > A, associated with an amino acid substitution Val347Glu (V1347G), was identified by Sanger sequencing but not found by WES. The variant was located in the eukaryotic protein kinase region. It was newly reported and highly conserved on the basis of homological sequence alignment (Fig. 2a). The electropherogram of previous Sanger sequencing demonstrated that the double peaks were typical (Fig. 3a). However, the mutation was absent in WES (Fig. 3b). After confirmation by Sanger sequencing, we found that the original double peaks were still absent (Fig. 3c).
Fig. 1.
Flow chart of BMPR2 mutations detection in PAH individuals. BMPR2, bone morphogenetic protein receptor 2; PAH, pulmonary artery hypertension; WES, whole-exome sequencing.
Fig. 2.
Homological sequence alignment of mutation site. (a) False-negative result; (b) false-positive result. Red arrow indicates the mutation site.
Fig. 3.
False-positive result of Sanger sequencing. (a) Electropherogram of previous Sanger sequencing; (b) profile obtained using WES. The mutation site is identified by the red arrow which indicates the mutation is absent in WES. (c) Electropherogram of Sanger sequencing for validation.
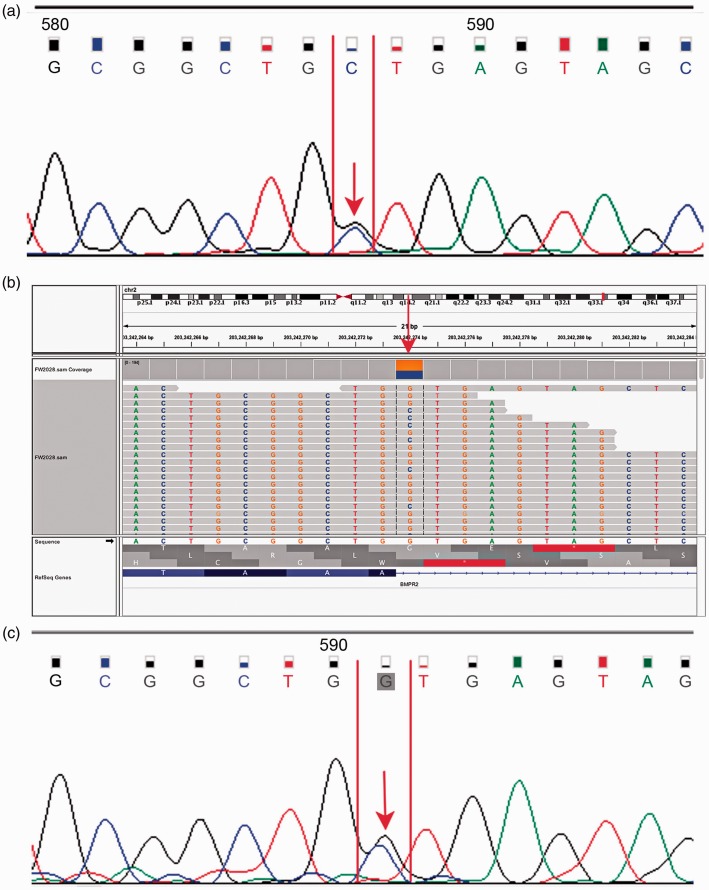
The splice mutation c.76 + 1 G > C missed by Sanger sequencing was located at the signal peptide domain. The variant was reported for the first time and highly conserved after homological sequence alignment (Fig. 2b). The double peaks in the electropherogram obtained from previous Sanger sequencing were ambiguities as a result of the peak distraction (Fig. 4a). Using WES, the deleterious splice mutation missed by Sanger sequencing was detected (Fig. 4b). We then re-sequenced the splice mutation site using Sanger sequencing for validation. There was still distraction from the anterior peak. However, the double peaks of the mutation site were more typical (Fig. 4c). The splice mutation c.76 + 1 G > C was classified as deleterious by MutationTaster and CADD (Table 2).
Fig. 4.
False-negative result of Sanger sequencing. (a) Electropherogram of previous Sanger sequencing; (b) profile obtained using WES. The splicing mutation was identified by the red arrow which indicates the mutation was detected by WES. (c) Electropherogram of Sanger sequencing for validation.
Statistical measures of performance
In the genetic testing for 120 PAH patients detected by Sanger sequencing and WES, the results were in agreement that 20 patients were BMPR2 carriers and 98 patients were BMPR2 non-carriers. One BMPR2 carrier in Sanger sequencing was indicated as negative, whereas another BMPR2 non-carrier was indicated as positive in WES. The agreement between Sanger sequencing and WES was near perfect at 98% (118/120), with a κ coefficient of 0.94. No significant difference was found between the two methods on the McNemar–Bowker test (P > 0.05).
After confirmation by Sanger sequencing, we found that WES detected the false-negative and false-positive results of Sanger sequencing. The sensitivity, specificity, false-positive rate, and false-negative rate of Sanger sequencing were 95.2% (20/21), 99.0% (98/99), 1% (1/99), and 4.8% (1/21), respectively. No false-positive and false-negative results of WES were identified in our analysis.
Discussion
In the present study, WES was used to validate BMPR2 mutations in 120 PAH individuals who were screened by Sanger sequencing. Our findings demonstrated that Sanger sequencing and WES agreement was 98.3% (118/120) with near-perfect agreement (κ coefficient = 0.848) with no significant difference between the two methods on the McNemar–Bowker test (P > 0.05). WES improved the accuracy of Sanger sequencing and detected 1% (1/99) false-positive and 4.8% (1/21) false-negative results of Sanger sequencing. No false-positive and false-negative results of WES were identified in our analysis. The present study indicated that WES is non-inferior to Sanger sequencing and may play a more important role in the genetic testing of PAH patients in the future.
Accurate identification of BMPR2 mutation is essential for clinical management and genetic consultation of PAH patients. Inaccurate genotype information, which masks the true correlation between genotype and phenotype, can have a harmful effect on PAH patients.4,27
WES, a “hypothesis-free” approach, is increasingly used in rare diseases that are clinically unspecific or involve a large number of genes.28 Rare diseases, such as PAH, seem rare individually, but they still affect numerous individuals and impose a significant clinical and economical challenge for society.29 Diagnostics of rare inherited diseases have entered a new era, in which WES has revealed several PAH causative genes such as CAV1 and KCNK39,10,30,31 and opens up a new realm of possibilities in future PAH research and clinical practice.
To our knowledge, this study represents the first clinical study that compares the accuracy of WES and Sanger sequencing in patients with PAH. As the dominant genetic cause of PAH, our analysis mainly focused on the validation of BMPR2 mutation. Importantly, with WES, we identified all previously detected BMPR2 mutations and identified a false-positive and a false-negative result of Sanger sequencing.
Our results revealed that WES could identify false-positive and false-negative results of Sanger sequencing. In Sanger sequencing, non-specific binding of the primers and the formation of DNA secondary structures may cause sequencing errors6,32 which may explain the false-positive result (c.1040 T > A) of Sanger sequencing. Human error is also increased especially when interpreting the raw sequencing results due to ambiguities in the capillary electrophoresis readouts.13 This factor could have caused the failure to identify the mutation site (c.76 + 1 G > C). Artifact misjudgment should be minimized, but it cannot be completely avoided. Some genotype information missed by Sanger sequencing might have a great influence on disease phenotype and drug resistance.4,27
As the “gold standard” in the routine PAH diagnostic, Sanger sequencing is straightforward to be performed and generally affordable. However, there are also other intrinsic limitations that widely impede the application of Sanger sequencing in the NGS era.4 First, the confidence read length of Sanger sequencing is relatively short, several exons of BMPR2 (exon 12, exon 13) are outside of the confidence read length of Sanger sequencing. Second, the accurate determination of each exon sequence should depend on both prime forward and reverse strands.6 BMPR2 gene has 13 exons with different exon lengths; even analyzing the mutation in one gene is time-consuming and labor-intensive. However, the use of WES in genetic testing simultaneously analyzes known mutations, as well as evaluates novel disease-modifying variants in patients with and without a family history of PAH.33 From the cost-effective standpoint, ability to generate protein-coding genome-wide sequencing data with less time in a short time frame, WES seems to be superior than traditional gold standard, Sanger sequencing, in genetic testing.12,34,35
It should be noted that genetic testing is a personal choice. Counsellors or geneticists should provide specific genetic counselling regarding the advantages and disadvantages of knowing their genetic status for symptomatic patients before they decide whether to undergo genetic testing.36 For genotype-positive asymptomatic individuals, routine longitudinal clinical follow-up is mandatory to enable early diagnosis and treatment. In addition, possible discrimination about genetic test results and the feelings of guilt for mutation carriers who have the possibility to pass the mutation to offspring should also be well considered by genetic counsellors or geneticists.36,37 The 12-year experiences from the French Referral Centre for Pulmonary Hypertension demonstrated that genetic counselling should be implemented in referral centers for PH.37
While WES is well indicated in disease-causing variants, it has some inevitable limitations. First, pathogenic mutations that occur outside the exome will be filtered out by WES. Second, other challenges of NSG, especially for whole-genome sequencing—including bioinformatics filtering techniques, software and hardware for data analysis, and the complexity of genome interpretation—should also be noted in WES.34 Third, although the platform costs for sequencing and hands-on time are decreased, the equipment and maintenance costs remain unaffordable for many laboratories.4 Fourth, WES cannot reliably detect triplet repeat changes, copy-number changes, and chromosomal rearrangements. Sanger sequencing and array comparative genomic hybridization are needed to complement the shortcomings of WES and expand the spectrum of WES-identified variants.38 However, given its versatility and cost-effectiveness, we believe that WES will become widely used for genetic testing in the next few years.
Limitations
Our analysis only focused on site mutations of BMPR2 and other PAH-causative genes and rearrangement of large fragments was not considered. Further clinical studies may be needed to elucidate the accuracy of WES in genetic testing of PAH beyond BMPR2.
Conclusions
WES improved the accuracy of Sanger sequencing and detected false-positive and false-negative results of Sanger sequencing in routine genetic testing of PAH. WES was non-inferior to Sanger sequencing and may play a more important role in genetic testing for PAH patients in the future. The use of WES in genetic testing for PAH patients has a certain guiding significance for clinical precision medicine.
Appendix
Quality control of whole-exome sequencing
The raw image files obtained from Hiseq 4000 were processed with Illumina pipeline for base calling and were stored as FastQ format (Raw data), which contain adapter contamination, low-quality nucleotide, and undetected nucleotide (N). These sequence artifacts can impose significant influence on downstream processing analysis. Hence quality control, which is listed below, is applied to guarantee the meaningful downstream analysis.
Quality control:
Filter reads with adapter contamination (> 10 nucleotide aligned to the adapter, allowing ≤ 10% mismatches).
Discard reads containing uncertain nucleotides > 10 percentage.
Discard the paired reads when single read has > 50 percentage low-quality (Phred quality < 5) nucleotides.
All the downstream bioinformatics analyses are based on the high quality clean data, which can be obtained after these steps. At the same time, QC statistics including total reads number, raw data, raw depth, sequencing error rate, percentage of reads with average quality > Q20, percentage of reads with average quality > Q30, and GC content distribution can be calculated.
Declaration of conflicting interests
The author(s) declare that there is no conflict of interest.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81320108005, 81630003, 81670052), the National Key Research and Development Program of China (2016YFC0901502), CAMS Innovation Fund for Medical Sciences (2016-I2M-1-002, 2016-I2M-4-003), and Beijing Natural Science Foundation (7172180) for the data analysis and interpretation.
References
- 1.Soubrier F, Chung WK, Machado R, et al. Genetics and genomics of pulmonary arterial hypertension. J Am Coll Cardiol 2013; 62: D13–21. [DOI] [PubMed] [Google Scholar]
- 2.Evans JD, Girerd B, Montani D, et al. BMPR2 mutations and survival in pulmonary arterial hypertension: an individual participant data meta-analysis. Lancet Respir Med 2016; 4: 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenzweig EB, Morse JH, Knowles JA, et al. Clinical implications of determining BMPR2 mutation status in a large cohort of children and adults with pulmonary arterial hypertension. J Heart Lung Transplant 2008; 27: 668–674. [DOI] [PubMed] [Google Scholar]
- 4.Casadella M, Paredes R. Deep sequencing for HIV-1 clinical management. Virus Res 2017; 239: 69–81. [DOI] [PubMed] [Google Scholar]
- 5.Doerks T, Copley RR, Schultz J, et al. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res 2002; 12: 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci 1977; 74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grada A, Weinbrecht K. Next-generation sequencing: methodology and application. J Invest Dermatol 2013; 133: e11. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen R, Paul JS, Albrechtsen A, et al. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 2011; 12: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, Roman-Campos D, Austin ED, et al. A novel channelopathy in pulmonary arterial hypertension. N Engl J Med 2013; 369: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Austin ED, Ma L, LeDuc C, et al. Whole exome sequencing to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ Cardiovasc Genet 2012; 5: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reuter MS, Tawamie H, Buchert R, et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatry 2017; 74: 293–299. [DOI] [PubMed] [Google Scholar]
- 12.Landouré G, Sullivan JM, Johnson JO, et al. Exome sequencing identifies a novel TRPV4 mutation in a CMT2C family. Neurology 2012; 79: 192–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vattulainen S, Aho J, Salmenpera P, et al. Accurate genetic diagnosis of Finnish pulmonary arterial hypertension patients using oligonucleotide-selective sequencing. Mol Genet Genomic Med 2015; 3: 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Biase D, Visani M, Baccarini P, et al. Next generation sequencing improves the accuracy of KRAS mutation analysis in endoscopic ultrasound fine needle aspiration pancreatic lesions. PLoS One 2014; 9: e87651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morandi L, de Biase D, Visani M, et al. Allele specific locked nucleic acid quantitative PCR (ASLNAqPCR): an accurate and cost-effective assay to diagnose and quantify KRAS and BRAF mutation. PLoS One 2012; 7: e36084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galie N, Humbert M, Vachiery JL, et al. 2015 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension: The Joint Task Force for the Diagnosis and Treatment of Pulmonary Hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS): Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC), International Society for Heart and Lung Transplantation (ISHLT). Eur Heart J 2016; 37: 67–119. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krumm N, Sudmant PH, Ko A, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res 2012; 22: 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genomes Project Consortium Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013; Chapter: Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz JM, Rodelsperger C, Schuelke M, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- 23.Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang H, Bai X, Meng F, et al. Comparison of immunohistochemistry and mRNA in situ hybridization in detecting thyroid transcription factor-1 expression in non-small cell lung carcinomas tissue. Oncol Lett 2015; 10: 3581–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawass NE. Comparing the sensitivities and specificities of two diagnostic procedures performed on the same group of patients. Br J Radiol 1997; 70: 360–366. [DOI] [PubMed] [Google Scholar]
- 26.Liu D, Liu QQ, Eyries M, et al. Molecular genetics and clinical features of Chinese idiopathic and heritable pulmonary arterial hypertension patients. Eur Respir J 2012; 39: 597–603. [DOI] [PubMed] [Google Scholar]
- 27.Kawashiri MA, Nomura A, Konno T, et al. Can next-generation sequencing replace Sanger sequencing for screening genetic variants? Circ J 2014; 78: 2845–2847. [DOI] [PubMed] [Google Scholar]
- 28.Tetreault M, Bareke E, Nadaf J, et al. Whole-exome sequencing as a diagnostic tool: current challenges and future opportunities. Expert Review Mol Diagn 2015; 15: 749–760. [DOI] [PubMed] [Google Scholar]
- 29.Hayes A, Costa T, Scriver CR, et al. The effect of Mendelian disease on human health. II: Response to treatment. Am J Med Genet 1985; 21: 243–255. [DOI] [PubMed] [Google Scholar]
- 30.Eyries M, Montani D, Girerd B, et al. EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat Genet 2013; 46: 65–69. [DOI] [PubMed] [Google Scholar]
- 31.Solomon BD, Pineda-Alvarez DE, Hadley DW, et al. Personalized genomic medicine: lessons from the exome. Mol Genet Metab 2011; 104: 189–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hert DG, Fredlake CP, Barron AE. Advantages and limitations of next-generation sequencing technologies: a comparison of electrophoresis and non-electrophoresis methods. Electrophoresis 2008; 29: 4618–4626. [DOI] [PubMed] [Google Scholar]
- 33.Kamps R, Brandao RD, Bosch BJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci 2017; 18: pii: E308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majewski J, Schwartzentruber J, Lalonde E, et al. What can exome sequencing do for you? J Med Genet 2011; 48: 580–589. [DOI] [PubMed] [Google Scholar]
- 35.Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 2012; 91: 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung WK, Austin ED, Best DH, et al. When to offer genetic testing for pulmonary arterial hypertension. Can J Cardiol 2015; 31: 544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girerd B, Montani D, Jaïs X, et al. Genetic counselling in a national referral centre for pulmonary hypertension. Eur Respir J 2016; 47: 541–552. [DOI] [PubMed] [Google Scholar]
- 38.Xue Y, Ankala A, Wilcox WR, et al. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med 2015; 17: 444–451. [DOI] [PubMed] [Google Scholar]