Abstract
The TORC1 pathway is necessary for ribosomal biogenesis and initiation of protein translation. Furthermore, the differentiation of Th1 and Th17 cells requires TORC1 activity. To investigate the role of the TORC1 pathway in the differentiation of Th1 and/or Th17 cells in more detail, we compared the differentiation capacity of naïve T cells from wild type and p70S6K1 knockout mice. Expression of many of the genes associated with Th17-cell differentiation, such as IL17a, IL17f, and IL-23R, were reduced in p70S6K1 knockout mice. In contrast, the development of Th1, Th2, and Treg cells was unaffected in the absence of p70S6K1. Furthermore, expression of the major transcription factor in Th17-cell differentiation, retinoic acid receptor-related orphan receptor gamma T, remained unchanged. However, the acetylation of histone 3 at the promoters of IL17a and IL17f was reduced in the absence of p70S6K1. In accordance with the in vitro data, the kinetics, but not the development, of EAE was affected with the loss of p70S6K1 expression. Collectively, our findings suggested that both in vitro and in vivo differentiation of Th17 cells were positively regulated by p70S6K1.
Keywords: IL-17, mTOR, p70S6K1, T-cell differentiation
Introduction
The differentiation of naïve T cells into various effector cells has been well described in numerous review articles [1, 2]. When this differentiation process is altered, pathological conditions may result. Hence the understanding of the factors involved in regulating the differentiation of T cells is important. The role of cytokines in skewing the differentiation process during T-cell activation has been well documented. Depending on the cytokines present in the milieu, the activated T cells can differentiate into various effector or regulatory T cells. More recently, there have been reports indicating the environmental cues and the metabolic status of the T cells also play a critical role in determining the fate of the T-cell differentiation process [3]. It has been proposed that T-cell differentiation is coordinated by cytokine signaling and cellular metabolism [4].
The serine/threonine kinase mammalian target of rapamycin (mTOR) signaling pathway plays a pivotal role in regulating cellular metabolism and proliferation [5]. mTOR exists in two independent kinase complexes that can be distinguished based on their components; TORC1 and TORC2. The TORC1 pathway is acutely sensitive to rapamycin, uniquely consisting of mTOR associated with raptor, and has been shown to be activated during cell proliferation and growth. As a consequence of TORC1 signaling, p70S6K1/2 and 4E-BP1 (eukaryotic translation initiation factor 4E-binding protein 1) are phosphorylated, which results in ribosome biogenesis and release from translation suppression, respectively. The activation of p70S6K1/2 results in the phosphorylation of the S6 ribosomal protein that is a core component of the ribosomal complex in protein translation. Since protein translation is important in the synthesis of cell components and effector molecules in T cells and other cell types, inhibitors of the TORC1 pathway, such as rapamycin, have been shown to block the activation of T cells [4].
Recent genetic ablation studies have further demonstrated that mTOR signaling is involved in T-cell differentiation (Supporting Information Fig. 1). In the absence of mTOR expression, the differentiation of T cells to the various effector T cells, such as Th1, Th2, and Th17 cells, is deficient [6]. However, an increase in the induction of regulatory T cells is observed. Subsequent studies have more specifically reported that the elimination of TORC1 and TORC2 in T cells results in the inhibition of Th1 and Th17-cell, and Th2-cell differentiation, respectively [7]. Lee et al. [8], however, have described the inhibition of the Th1 and Th2-cell development in mice lacking rictor expression, which is essential for TORC2 signaling. Hence the exact role of TORC1 and TORC2 in T-cell differentiation remains unclear.
One of the results of TORC1 signaling is the phosphorylation and activation of p70S6K1. It is not known if p70S6K1 is responsible for Th1 or Th17-cell differentiation. Here, we report that the elimination of p70S6K1 expression resulted in a decrease in Th17-cell development in T cells grown under Th17-cell skewing conditions in vitro. The development of Th1 and Th2 cells and the induction of regulatory T cells were unaffected in the absence of p70S6K1. In addition, p70S6K1 did not alter the signaling pathways and transcription factors that are responsible for Th17-cell development. However, the acetylation of histone 3 in the regulatory sequences near the IL17 gene was reduced in T cells deficient in p70S6K1 expression that suggested that p70S6K1 could block Th17-cell development by limiting chromatin accessibility. Taken together, p70S6K1 facilitates Th17-cell differentiation by promoting the expression of IL17 genes epigenetically.
Results
Th17-cell differentiation is reduced in the absence of p70S6K1
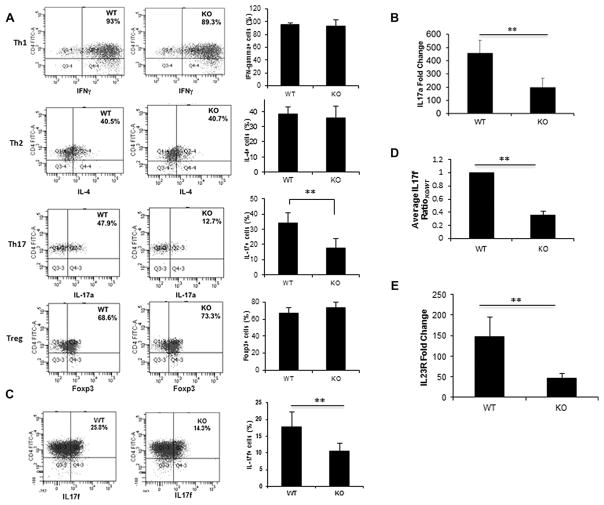
Previous studies [6] have shown that the differentiation of naïve T cells to certain effector cells was dependent on mTOR signaling. In particular, the differentiation of Th1 and Th17 effector cells were dependent on the TORC1 pathway. To investigate what role p70S6K1, a downstream component of the mTORC1 pathway, plays in T-cell differentiation, naïve T cells from p70S6K1 knockout mice were differentiated in vitro to the various effector T cells and were then compared with the in vitro differentiated T cells obtained from wild-type mice. In general, the activation of these cells did not significantly differ between the knockout mice and their wild-type counterpart (Supporting Information Fig. 2) indicating that p70S6K1 does not play a major role in T-cell activation and the generation of naïve T cells. However, when naïve T cells were activated in vitro under skewing conditions for various effector T cells, a reduction in Th17-cell differentiation was observed with T cells from p70S6K1 knockout mice, while no difference was observed between T cells from wild type and p70S6K1 knockout mice with regards to the differentiation of Th1, Th2, or Treg cells (Fig. 1A). In order to get the optimum differentiation for different subsets of T-helper cells, we used different lengths of time for different subsets of T-helper cells (6 or 7 days for Th1 and Th2 cells, and 3 days for Th17 and Treg cells). In Supporting Information Fig. 3, we analyzed the differentiation of Th1, Th2, and Th17 cells on Day 3, and we observed no difference in Th1 and Th2 cell differentiation between wild type and knockout cells, but the Th17-cell differentiation was reduced in knockout cells in comparison to wild type. In accordance with the intracellular cytokine staining results, a substantial reduction in the levels of IL17a transcript was observed in T cells from knockout mice compared to T cells from wild-type mice (Fig. 1B). Since mTOR is involved in cellular metabolism, we checked whether the difference in Th17 cell differentiation observed was due to the differential proliferation or survival of Th17 cells in knockout cells. As shown in Supplemental Information Fig. 4A, the viability (based on FVD dye exclusion) of in vitro differentiated T-helper cells was similar between wild type and knockout cells (81.9 versus 78.6%, respectively). With respect to proliferation, we did not observe any proliferation of in vitro differentiated Th17 cells as measured by Ki67, whereas Th0 cells proliferate vigorously in the same culture conditions (Supporting Information Fig. 4B). This could be due to the antiproliferative effect of TGFβ present in the Th17-cell-skewing culture conditions. In general, there was no difference in the proliferation of CD4+ T cells from wild type and knockout mice (Supporting Information Fig. 2). Moreover, the status of the phospho-p70S6K1 for all the in vitro differentiated T-helper subsets shown in Supporting Information Fig. 5 ruled out the possibility of preferential activation of p70S6K1 in Th17-cell subsets rendering these cells affected in knockout T cells. Collectively, these findings suggested that p70S6K1 may positively regulate the differentiation of Th17, but not Th1, Th2, or regulatory T cells.
Figure 1.
Role of p70S6K1 in in vitro differentiation of Th1, Th2, Th17, and Treg cells. (A) Naive splenic CD4+T cells (CD4+CD62L+) from WT and p70S6K1 KO mice were activated by plate-bound anti-CD3 and anti-CD28 (5 μg/mL each) under either Th1, Th2, Th17, or Treg skewing conditions. In vitro-differentiated T cells were stimulated with PMA and ionomycin for 4 h with last 2 h in the presence of monensin. Stimulated T cells were stained with the following antibodies: for Th1 cells, IFN-γ; for Th2 cells, IL-4; for Th17 cells, IL-17a; and for Treg cells, Foxp3. Data were obtained by FACS Canto II and analyzed by FACS DIVA. The numbers in the boxes indicate the percentages of antibody-stained cells relative to the total number of cells. Summary of three independent experiments for individual protein is shown (right). (B) RT-PCR analysis for IL-17a expression was done on complementary DNA from in vitro-differentiated Th17 cells. These experiments were normalized to a housekeeping gene (either β actin or GAPDH). Then the normalized results were compared to Th0 cells to determine the fold change. (C) Flow cytometry and (D) RT-PCR analyses for IL17f were done using in vitro-generated Th17 cells. The amount of IL-17f mRNA expression in WT and KO Th17 cells were normalized first to β actin, and then the normalized values were compared to wild type to determine the ratio. (E) RT-PCR analysis of IL-23 receptor (IL23R) levels was done using in vitro-differentiated Th17 cells. The amount of IL-23R mRNA expression in WT and KO Th17 cells were normalized first to β actin, and then the normalized values were compared to Th0 to determine the fold change. (B–D) Data are shown as mean + SEM from three independent experiments with total nine samples per group. **p < 0.05, Student’s t-test.
In association with IL17a production, the expression of IL-23 receptor plays a major role in Th17-cell differentiation and function [9]. Furthermore, IL17f, highly homologous to IL17a and another member of the IL17 protein family, has both similar and distinct roles in the immune system [10]. To investigate if p70S6K1 affects the expression of the other cytokines and receptors, we analyzed the expression of IL17f and IL-23R in Th17 cells from the wild type and knockout mice. As seen in Fig. 1C and D, the amount of both IL17f protein and transcript was decreased in T cells from p70S6K1 knockout mice that were differentiated under Th17-cell conditions. Similarly, a reduction of IL-23R transcripts was also observed in the induced Th17 cells from the knockout mice (Fig. 1E). These findings further support the role of p70S6K1 in Th17-cell differentiation under in vitro skewing conditions.
Absence of p70S6K1 does not affect the IL-6 signaling pathway
It has been well established that the role of the IL-6 signaling pathway is pivotal in the differentiation of Th17 cells. In particular, IL-6 treatment during T-cell activation results in the tyrosine phosphorylation of STAT3, which induces the expression of IL17 [11]. It has been reported that the inhibition of mTOR signaling by rapamycin resulted in a decrease in STAT3 phosphorylation [12]. To investigate if p70S6K1 contributes in activating the IL-6 signaling pathway, we compared the levels of STAT3 phosphorylation in T cells from wild type and p70S6K1 knockout mice after IL-6 treatment. As seen in Supporting Information Fig. 6A, an increase in STAT3 phosphorylation was seen in T cells treated with IL-6 from both wild type and knockout mice indicating that p70S6K1 was not required for IL-6-mediated STAT3 phosphorylation. We have also checked the phospho-STAT3 status after naïve CD4+ T cells were cultured for 24 h in Th17-cell skewing conditions followed by IL-6 starvation for 1 h, and subsequent treatment with IL-6 for 1 h. As shown in Supporting Information Fig. 6B, there was no difference in the status of Stat3 phosphorylation (Tyr705) between wild type and p70S6K1 knockout T cells. In addition, Delgoffe et al. [6] have shown that the TORC1 pathway negatively regulates the expression of the suppressor of cytokine signaling 3 (SOCS3) gene in T cells. Since p70S6K1 is one of the downstream targets of the TORC1 signaling pathway, it was also possible that p70S6K1 may negatively regulate SOCS3 expression to block IL-6 signaling. As seen in Supporting Information Fig. 6C, the amount of SOCS3 transcripts in differentiated Th17 cells from wild type and knockout mice was similar. Taken together, these findings indicated that the IL-6 signaling pathway was not dependent on p70S6K1 expression. Hence the reduction of Th17-cell differentiation with naïve T cells from p70S6K1 knockout mice was not mediated by blocking the IL-6 signaling pathway.
p70S6K1 does not affect the expression of the transcription factor RORγT
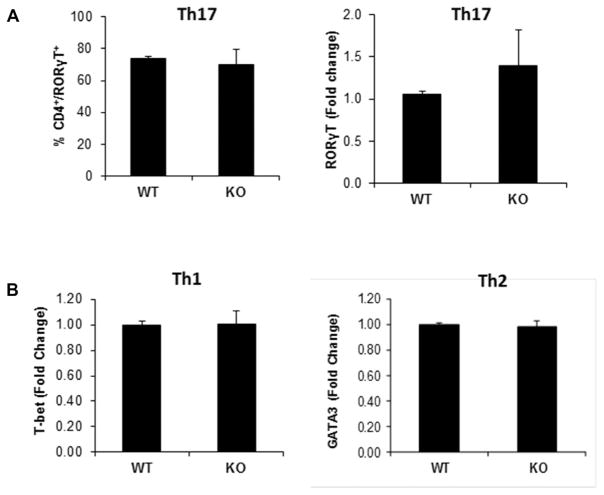
In addition to the IL-6 signaling pathway, the transcription factor retinoic acid receptor-related orphan receptor gamma T (RORγT) plays a major role in differentiating naïve T cells to the Th17 effector cells [13, 14]. It was possible that the absence of p70S6K1 could negatively affect the expression of RORγT. As seen in Fig. 2A (left panel, and Supporting Information Fig. 7), the levels of intracellular RORγT protein were not affected with the loss of p70S6K1 expression. Consistent with these results, the amount of RORγT mRNA transcript was not significantly different between T cells activated under Th17-cell skewing conditions from wild type and p70S6K1 knockout mice (Fig 2A, right panel). We also checked the status of other transcription factors that have been reported to be involved in Th17-cell differentiation [15, 16]. As shown in Supporting Information Fig. 8, we did not see any difference in the mRNA levels of Ahr, Batf, and Irf4 in in vitro differentiated Th17 cells from wild type and knockout T cells. We were unable to detect any RORα message in our experiment. Also the induction of the transcription factors T-bet and GATA3, which are responsible for Th1 and Th2 differentiation, respectively, was similar between wild type and knockout T cells (Fig. 2B). Hence, p70S6K1 does not play a significant role in the expression of the various transcription factors responsible for T-effector cell differentiation.
Figure 2.
Effect of p70S6K1 on the status of transcription factors involved in the differentiation of Th1, Th2, and Th17 cells. Naive splenic CD4+ T cells (CD4+CD62L+) from WT and p70S6K1 KO mice were activated by plate-bound anti-CD3 and anti-CD28 under either Th1, Th2, or Th17 skewing conditions. (A) Flow cytometry and RT-PCR analyses were performed for retinoic acid receptor-related orphan receptor gamma T (RORγT) using in vitro-differentiated Th17 cells. Following in vitro differentiation, Th17 cells were stimulated with PMA and ionomycin for 4 h, last 2 h with monensin. Stimulated cells were either stained with antibodies against CD4 and RORγT and analyzed by flow cytometry (left), or used for total RNA isolation (right). The cDNA was then analyzed by quantitative PCR to measure RORγT transcript levels (right). These experiments were first normalized to a housekeeping gene (either β actin or GAPDH). Then the normalized results were compared to wild-type Th17 cells to determine the fold change. (B) RT-PCR analyses of T-bet (left) and GATA3 (right) levels were done using in vitro-differentiated Th1 and Th2 cells, respectively. The amount of T-bet and GATA3 mRNA expression in WT and KO differentiated cells were normalized first to β actin, and then the normalized values were compared to wild type to determine the fold change. All data shown as mean + SEM from two independent experiments with total six samples per group.
Access to the IL-17 promoter is reduced in the absence of p70S6K1
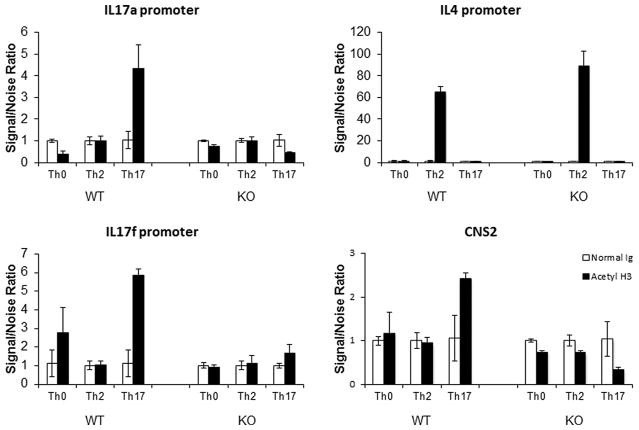
Several studies have shown that perturbations to the metabolic status of T cells during differentiation may epigenetically alter gene expression [17]. Furthermore, Wellen et al. [18] have shown that histone acetylation and gene expression were dependent on cellular metabolism. Since mTOR signaling affects cellular metabolism, it was plausible that the absence of p70S6K1 could have affected the gene expression epigenetically. To assess this possibility, we compared the status of histone 3 (H3) acetylation at the promoter of IL17 by chromatin immunoprecipitation (ChIP) assay using in vitro differentiated Th17 cells. As seen in Fig. 3, the level of acetylated H3 chromatin at the IL17a promoter was reduced in the p70S6K1 knockout T cells as compared to wild-type T cells under Th17-cell skewing conditions. These findings suggested that the chromatin near the IL17a promoter was inaccessible. It is possible that the differential H3 acetylation observed at the IL17 promoter of wild type and knockout T cells was due to the differences in the number of cells expressing IL17 between wild type and knockout cells. To establish the effect of p70S6K1-deficiency on the epigenetic imprinting of the IL17 gene, we used flow cytometry analyses where we determined the MFI of the IL17+ cells and compared these values between wild type and knockout in vitro differentiated Th17 cells. As shown in Supporting Information Fig. 9 (right panel), MFI of IL17+ cells obtained from knockout CD4+ T cells was reduced in comparison to MFI obtained from wild-type cells. These data along with the reduction in the percentage of Th17 cells obtained from knockout T cells (left panel) suggested that the p70S6K1-deficiency affected the epigenetic imprinting of the IL17 gene. Wang et al. [19] have shown that the noncoding sequence 2 (CNS2), that is juxtaposed between the IL17a and IL17f promoters, plays a key role in coordinating the expression of both IL17a and IL17f genes. As seen with the IL17a promoter, the acetylation of H3 at both the CNS2 and IL17f promoter was decreased in the knockout T cells as compared to wild-type T cells under Th17-cell skewing conditions. In contrast, the acetylation of the IL4 promoter was unaffected in both wild type and knockout T cells under Th2-cell skewing conditions (Fig. 3). Besides the H3 acetylation of IL17 promoter, the recruitment of RNA polymerase II to IL17a promoter was also decreased in knockout in comparison to wild-type T cells (Supporting Information Fig. 10). We have also tested the recruitments of Batf and Irf4 to IL17a and IL4 promoters. Based on our results (Supporting Information Fig. 11), we concluded that neither Batf nor Irf4 was recruited to IL17a promoters. The slight increase observed in the recruitment of Batf was not specific to IL17 promoter, since this increase was also observed not only in IL4 promoter but also in several other promoters tested (Supporting Information Fig. 11, and data not shown). Thus, these findings suggested that p70S6K1 may play a role in making the promoters of IL17a and IL17f, and the regulatory regions of CNS2, but not the IL4 promoter, accessible for transcription.
Figure 3.
Effect of p70S6K1 on the level of histone 3 acetylation of Th17-associated gene promoters. Chromatin immunoprecipitation (ChIP) analyses using in vitro-differentiated Th0, Th2, and Th17 cells obtained from either WT or KO mice were conducted for acetylated histone H3. Sheared chromatins from differentiated T cells were incubated with antibodies against either normal rabbit IgG or acetylated histone 3 overnight. Immune complexes were precipitated with Protein A/G agarose beads and washed twice with chromatin immunoprecipitation buffer. The beads were boiled and the captured DNA was precipitated with Chelex beads. The immunoprecipitated chromatin was used in quantitative PCR with the primers described in Table 2. The relative level of histone 3 acetylation at the indicated gene promoter expressed as a Signal/Noise Ratio was calculated as 2^(Ctnormal Ig − Ctanti-acetylated histone 3). Data shown as mean + SEM pooled from two independent experiments with total eight samples per group.
Delayed development of EAE in p70S6K1 knockout mice
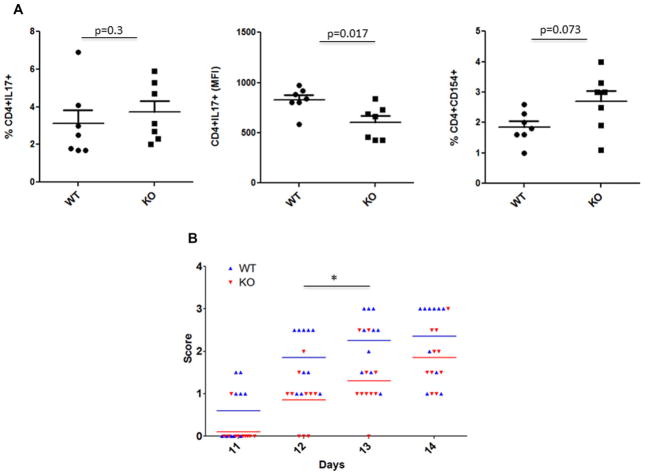
Th17 cells have been shown to play a major role in the development of experimental autoimmune encephalomyelitis in mice [20]. Based on our results, p70S6K1 plays a positive role in the differentiation of naïve T cells to Th17 cells in vitro. It was conceivable that the development of EAE in the p70S6K1 knockout mice would be decreased as compared to wild-type mice. Before we initiated the EAE experiment, we wanted to monitor the in vivo differentiation of myelin oligodendrocyte glycoprotein (MOG)-specific T cells in wild type and p70S6K1 knockout mice without inducing the pathology. For this purpose, both wild type and p70S6K1 knockout mice were immunized with MOG in CFA without administering pertussis toxin. Splenocytes from individual mice were harvested 10 days after immunization, and were cultured in the presence of MOG, IL-23, and anti-IFN-γ antibody for 3 days. These restimulated splenocytes were used to measure the MOG-reactive CD4+ T cells by determining the percentage of CD4+CD154+ T cells [21]. We also measured the percentage of CD4+ T cells that produced IL17, and the amount of IL17 production by CD4+ T cells was determined by measuring the MFI of CD4+IL17+ T cells. As shown in Fig. 4A and Supporting Information Fig. 12, although the percentage of MOG-reactive CD4+ T cells were the same in the wild type and knockout splenocytes (right panel), CD4+ T cells from the knockout splenocytes tended to produce less IL17 when compared to the wild type (middle panel). The percentage of CD4+ T cells that produced IL17 was comparable between wild type and knockout splenocytes (left panel). These in vivo differentiation results were different from the results obtained from the in vitro differentiation experiments in which the percentage of IL-17-producing cells was lower in knockout CD4+ T cells as compared with wild-type cells. This discrepancy could be due to the following reasons: first, differences in the mode of activation: polyclonal activation in the presence of Th17-cell-skewing conditions during in vitro differentiation versus antigen-specific activation during in vivo differentiation; and second, the role of other cytokines (e.g. IL-2, IL-23, IL-21, etc.) during the differentiation process. As far as the in vivo differentiation is concerned, our data suggests that p70S6K1 may be involved in IL-17 expression, but not in Th17-cell differentiation.
Figure 4.
Role of p70S6K1 in in vivo differentiation of MOG-specific Th17 cells and the onset of EAE. (A) Wild type and p70S6K1 knockout mice were immunized with MOG in complete Freund’s adjuvant (CFA) without administering pertussis toxin. Splenocytes from individual mice were harvested 10 days after immunization, and were cultured in the presence of MOG (20 μg/mL), IL-23 (20 ng/mL), and anti-IFN-γ antibody (10 μg/mL) for 3 days. These cultured splenocytes were stimulated with PMA/ionomycin for 4 h with last 2 h with monensin, and the percentage of CD4+ T cells produced IL-17 (left) was determined. The amount of IL-17 produced by CD4+ T cells (middle) was determined by measuring the MFI of CD4+IL17+ T cells. To measure the MOG-reactive CD4+ T cells (right), splenocytes were cultured overnight with monensin to determine the percentage of CD4+CD154+ T cells. p value was determined with Mann–Whitney test. (B) EAE was induced in WT and KO mice (10 mice/group) by immunizing with MOG35–55 peptide in CFA, followed by administration of pertussis toxin, and the animals were observed for disease progression (clinical score). Each symbol represents an individual animal and bars represent means. The differences in the neurological score along the disease progression were analyzed by two-way ANOVA followed by the Bonferroni post-hoc test. *p < 0.01.
Next, to monitor the EAE development, both wild type and p70S6K1 knockout mice were immunized with MOG35–55 peptide in CFA, followed by administration of pertussis toxin, and the animals were observed for the development of EAE. As seen in Fig. 4B, the progression of EAE, based on the EAE clinical score, was slower in the p70S6K1 knockout mice as compared to the wild-type mice, but eventually both groups of mice developed the full-blown disease. These findings suggested that the absence of p70S6K1 expression resulted in a delay of EAE development but did not prevent or suppress EAE. It is possible that other factors, such as IFN-γ expression, contribute to the pathology of EAE independently of p70S6K1. Next, to investigate whether the difference in the development of EAE observed in knockout mice was cell intrinsic, we performed an adoptive transfer experiment. Both wild type and p70S6K1 knockout mice were immunized with MOG in CFA without administering pertussis toxin. Splenocytes from individual mice were harvested 10 days after immunization, and were cultured in the presence of MOG, IL-23, and anti-IFN-γ antibody for 3 days. These restimulated splenocytes were then injected intraperitoneally to naïve wild-type mice, and the animals were observed for the development of EAE. Wild-type mice that received in vitro restimulated immunized splenocytes from knockout mice developed EAE slower than the mice that received in vitro restimulated immunized splenocytes from wild-type mice (data not shown). These data suggest that the cell intrinsic properties of knockout mice may be responsible for slower development of EAE. Collectively, these findings indicated that the in vivo differentiation of Th17 cells to MOG and the kinetics of progression of EAE were affected by the expression of p70S6K1.
Discussion
The consequence of TORC1 signaling is the activation of the p70S6K1 pathway and the inactivation of 4E-BP1, which results in ribosomal biogenesis and initiation of translation, respectively. Furthermore, it has been shown that the blocking the TORC1 signaling results in the inhibition of Th1 and Th17-cell development [7]. Here, we report the genetic inhibition of p70S6K1 expression resulted in the suppression of Th17-cell development. In contrast, the development of the other T-effector cells, such as Th1, Th2, and Treg cells was unaffected. The transcription factors and signaling pathways leading to Th17-cell differentiation were not dependent on p70S6K1. However, a decrease in chromatin accessibility to various promoters of the genes involved in Th17-cell differentiation was observed in the absence of p70S6K1. These findings indicated that p70S6K1 plays a pivotal role in the development of Th17 cells.
Kurebayashi et al. [22] have shown that the PI3K-AKT-mTOR-S6K1 pathway is involved in the differentiation of Th17 cells. Specifically, they have shown that the introduction of the constitutively active p70S6K1 could restore Th17-cell differentiation of T cells that were treated with the mTOR inhibitor rapamycin. The activation of p70S6K1 resulted in an increase in Egr2 expression, which has been reported to block the expression of Gfi1 [23]. Gfi1 blocks the activity of RORγT, a major regulator of Th17-cell development [24]. We have also investigated whether the absence of p70S6K1 could result in a decrease of Egr2 levels which would increase Gfi1 to block Th17-cells. However, we did not detect any difference in either Egr2 or Gfi1 transcripts between wild type and p70S6K1 knockout T cells under Th17 skewing conditions (Supporting Information Fig. 13). It is possible that the expression of the constitutively active p70S6K1 could have different effects than the ablation of p70S6K1 as in our findings.
The cellular metabolic status in T cells has been recently reported to dramatically change during activation, and agents that alter metabolism affect T-cell activation [25]. It has been reported by Magnuson et al. [26] that the TORC1 complex senses the cellular milieu and that TORC1-S6K1 regulates cellular function. In addition, mice in which the p70S6K1 gene has been disrupted, are resistant to insulin-induced diabetes [27] although normal levels of glucose are detected. It was later shown by Veilleux et al. [28] that the transport of glucose was inhibited when the expression of p70S6K1 was suppressed. Moreover, the transport of glucose for glycolysis during T-cell differentiation was required for the T-effectors Th1, Th2, and Th17 cells [29]. Furthermore, it has been reported that glucose metabolism epigenetically affects gene expression by modulating histone acetylation. Based on these findings, the inhibition of T-cell differentiation in our p70S6K1 knockout mice could be due to the suppression of glycolysis. However, we did not observe any inhibition in the differentiation of either Th1 or Th2 cells in our knockout mice. In accordance, the blockage of chromatin accessibility in the p70S6K1 knockout T cells was only seen in the promoters of the genes associated with Th17-cell differentiation. Since the differentiation of either Th1 or Th2 cells was not affected, this suggested that the suppression of Th17-cell differentiation may not be attributed to the lack of glycolysis.
p70S6K1 is one of many factors that are responsible for ribosomal formation [30]. Several studies have shown that perturbations to the process of ribosomal and polysome formation results in the inhibition of protein translation [31, 32]. Although only Th17-cell was affected, it is possible that the genetic removal of p70S6K1 reduced protein translation in T-cell differentiation. We compared the polysomal profile of the IL17a mRNA transcripts in activated T cells from wild type and p70S6K1 knockout mice which were differentiated under Th17-cell conditions, and did not observe any differences (Supporting Information Fig. 14). In accordance, the data in Fig. 1A indicated that p70S6K1 affects the transcription, but not the translation, of IL17a. Hence, the reduction in IL17 expression with the elimination of p70S6K1 expression was not due to either a change in IL17 translation, polysome formation or protein homeostasis.
It has been shown that Th1 and Th17 cells contribute to the development of EAE [33]. Furthermore, Delgoffe et al. [7] reported that the TORC1 pathway is required for the in vitro differentiation of Th1 and Th17 cells and the establishment of EAE. We demonstrated that the absence of p70S6K1 expression resulted in a delay in the progression, but not the development, of EAE. This could be due to the fact that T cells from p70S6K1 knockout mice produced reduced levels of IL17 while producing comparable levels of IFN-γ in comparison to their wild-type littermates. Thus reduced levels of IL-17 in the presence of IFN-γ slowed down the progression of EAE. Since the blockage of the TORC1 pathway inhibits both p70S6K1 and 4E-BP signaling, it is conceivable that both signals are required for the generation of encephalitis. This could explain our observation that the progression of EAE was slower when the expression of p70S6K1 was blocked. It remains to be investigated if 4E-BP plays a role in the development of EAE.
Hence, we have demonstrated that the in vitro differentiation of Th17 cells, but not the other T effector cells, was dependent on p70S6K1. In the absence of p70S6K1, acetylation of histone 3 within the promoters of genes that are associated with Th17-cell development was blocked. Moreover, the progression of EAE was slower without p70S6K1 expression. Taken together, these findings indicated that p70S6K1 positively regulated Th17-cell differentiation and development of EAE. It is not clear how p70S6K1 promotes the histone acetylation of Th17-associated genes.
Materials and methods
Animals
p70S6K1 knockout mice were generated by replacing exons 8–12 with a neo cassette flanked by loxP. These mice were then bred with a cre mouse to remove the neo cassette. These mice were generated by the Gene Targeting & Transgenic Facility in the University of Connecticut Health Center. A range of 8–10–week-old female mice were used for the differentiation experiments. All the mice used in the experiments were littermates generated by breeding heterozygous mice.
Antibodies and cytokines
Information regarding the antibodies and cytokines is listed in Table 1.
Table 1.
Antibodies and chemicals
| ANTIBODY | COMPANY | CATALOG # |
|---|---|---|
| Activation/differentiation antibodies | ||
| Anti-IFN-γ | BD Bioscience | 554408 |
| Anti-IL4 | BD Bioscience | 554432 |
| Anti-CD3 | BD Bioscience | 553057 |
| Anti-CD28 | BD Bioscience | 553294 |
| Cytokines | ||
| Recombinant TGFβ | R&D Systems | 240-B |
| Recombinant IL-4 | R&D Systems | 404-ML |
| Recombinant IL-12 | R&D Systems | 419-ML |
| Recombinant IL-6 | R&D Systems | 406-ML |
| Recombinant IL-2 | Roche | 23-6019 |
| Flow cytometry antibodies | ||
| Anti-CD4-FITC | eBioscience | 11-0041 |
| Anti-IL17F-PE | eBioscience | 12-7471 |
| Anti-Foxp3-PE | eBioscience | 12-5773 |
| Anti-IL4-APC | eBioscience | 17-7041 |
| Anti-IFNγAPC | BD Bioscience | 554413 |
| Anti-RORγ (T) | eBioscience | 17-6988 |
| Anti-IL17A-PE | eBioscience | 12-7177 |
| Chromatin Immunoprecipitation | ||
| anti-acetylated H3 Mab | Millipore | 17-615 |
| Western blot | ||
| anti-Phospho STAT3 (Y705) | Santa Cruz | sc-8059 |
| anti-Total STAT3 | Cell Signaling | 9139 |
| Chemicals | ||
| Phorbol-12-myristate-13-acetate | Calbiochem | 524400 |
| Ionomycin | Sigma | I9657 |
In vitro T-cell differentiation
Naïve CD4+T cells were differentiated in vitro based on the following protocol described by An et al. [34]. Briefly, naive CD4+T cells were enriched from murine splenocytes by negative selection for CD3/CD4 and then positive selection for CD62 ligand (Miltenyi Biotec Inc. San Diego, CA). The enriched T cells were then activated by plate-bound anti-CD3 and anti-CD28 (5 μg/mL each) under the following skewing conditions: for Th1, IL-12 (10 ng/mL), and anti-IL4 (10 μg/mL); for Th2, IL-4 (10 ng/mL) and anti-IFN-γ (10 μg/mL); for Th17, IL-6 (10 ng/mL), TGF-β (5 ng/mL), anti-IL4 (10 μg/mL), and anti-IFN-γ (10 μg/mL); for Treg cells, TGF-β (5 ng/mL), anti-IL-4 (10 μg/mL), and anti-IFN-γ (10 μg/mL). T cells were cultured under Th17-cell and Treg-cell skewing conditions for 3 days whereas Th1 and Th2 cells were cultured for an additional 3 or 4 days with IL-2 (10 ng/mL), respectively.
Intracellular staining by flow cytometry
Following in vitro differentiation, T cells were stimulated with PMA and ionomycin for 4 h, the last 2 h in the presence of monensin. Stimulated T cells were stained with the following antibodies CD4 (FITC), IL-17a (PE), IL-17f (PE), FOXP3 (PE), IL-4 (APC), RORγT (APC), and IFNγ (APC) using the BD Cytofix/Cytoperm fixation/permeablilization solution kit (BD Bioscience) as described by the manufacturer. Data were obtained by flow cytometry Canto II (BD Bioscience) and analyzed by FACS DIVA (BD Bioscience) or FlowJo software (Tree Star).
Gene expression analysis (RT-PCR)
Total RNA from differentiated cells was collected using the RNeasy Mini Kit (Qiagen) and used to generate complementary DNA with the VILO superscript kit (Invitrogen). The cDNA was then analyzed by quantitative PCR (BioRad) to measure transcript levels with the primers described in the supplemental material section.
Immunoblot
Cells were lysed with RIPA buffer and the proteins in the lysates were separated via SDS PAGE and transferred to a PVDF membrane. Proteins were visualized by enhanced chemifluorescence kit (GE Healthcare) and the Typhoon 9410 scanner (GE Health-care). Quantitation of the protein band density was done with the ImageQuant TL software (GE Healthcare).
ChIP analysis
Acetylated histone 3 (H3) was measured by chromatin immunoprecipitation as previously described [35] with the following exceptions. Sheared chromatins from differentiated T cells were incubated with antibodies against either normal rabbit IgG or acetylated histone 3 overnight. Immune complexes were precipitated with Protein A/G agarose beads and washed twice with chromatin immunoprecipitation buffer. The beads were boiled and the captured DNA was precipitated with Chelex beads. The immunoprecipitated chromatin was used in quantitative PCR with the primers described in Table 2. The relative level of histone 3 acetylation at the indicated gene promoter was expressed as a S/N that was calculated as 2^(Ctnormal Ig − Ctanti-acetylated histone 3).
Table 2.
Primers primers for ChIP assays
| Primers for ChIP assays | ||
| CNS2 [11] | ATGGGCCTCTCTTTCCACTGATG | GGAATTTGTGGTGGAAGGGAGTG |
| IL17p [11] | GCCTTTGTGATTGTTTCTTGCAG | CCTTGCCCAAAGAAACCTCTC |
| IL17Fp [11] | GGGAATCAAAGGGGACCCTAA | AAAGCAGAACCCACACGCAGAG |
| IL4p [36] | CTCATTTTCCCTTGGTTTCAGC | GATTTTTGTCGCATCCGTGG |
| Primers for RT-qPCR | ||
| RORγT [14] | CCGCTGAGAGGGCTTCAC | TGCAGGAGTAGGCCACATTACA |
| IL17a [14] | CTCCAGAAGGCCCTCAGACTAC | AGCTTTCCCTCCGCATTGACACAG |
| IL17f [14] | CTGTTGATGTTGGGACTTGCC | TCACAGTGTTATCCTCCAGG |
| IL23R [14] | GCCAAGAGAACCATTCCCGA | TCAGTGCTACAATCTTCAGAGGACA |
| GAPDH [14] | AGTATGACTCCACTCACGGCAA | TCTCGCTCCTGGAAGATGGT |
| Gfi1 | TTGGAGCTCTGACTGAAGGC | GGCAAAAGATTCCACCAGAA |
| Socs3 | AACTTGCTGTGGGTGACCAT | AAGGCCGGAGATTTCGCT |
| Egr2 | CAGAGATGGGAGCGAAGCTA | TTGACCAGATGAACGGAGTG |
| IL4 | CGAGCTCACTCTCTGTGGTG | TGAACGAGGTCACAGGAGAA |
| Ifnγ | GTCACCATCCTTTTGCCAGT | GCTTTGCAGCTCTTCCTCAT |
| Foxp3 | CCAGGGAGCCAGCTCTACTCT | CCAAAAGGTTGCTGTCTTTCCT |
| Tbx21 | ACCAGAACGCAGAGATCACTCA | CAAAGTTCTCCCGGAATCCTT |
| Gata3 | GAACCGGCCCCTTATCAAG | CAGGATGTCCCTGCTCTCCTT |
| Irf4 | CAAAGCACAGAGTCACCTGG | TGCAAGCTCTTTGACACACA |
| Ahr | CTCCTTCTTGCAAATCCTGC | GGCCAAGAGCTTCTTTGATG |
| Batf | GCGTTCTGTTTCTCCAGGTC | AGAGAGAAGAATCGCATCGC |
Source of the primer sequences are listed in the references in parentheses. The remaining primers were derived from “qPrimer Depot” [37].
Experimental autoimmune encephalomyelitis
EAE was induced in 10+ week-old female mice using EAE Induction kit from Hooke Laboratories (Lawrence, MA) according to the manufacturer’s protocol. Mice were scored on a scale of 0–4: 0—no clinical sign; 1—limp/flaccid tail; 2—limp tail and moderate hind limb weakness; 3—limp tail and complete hind limb paralysis; and 4—moribund state. To determine the in vivo differentiation of MOG-specific T cells, we followed the protocol described by Hooke Laboratories. In brief, both wild type and p70S6K1 knockout mice (7 mice/group) were immunized with MOG in CFA without administering pertussis toxin. Splenocytes from individual mice were harvested 10 days after immunization, and were cultured in the presence of MOG (20 μg/mL), IL-23 (20 ng/mL), and anti-IFN-γ antibody (10 μg/mL) for 3 days. In vitro cultured splenocytes were used for flow cytometry analyses to determine the production of IL17 by CD4+ T cells. To determine the intracellular staining for IL17, we stimulated in vitro cultured splenocytes with PMA/ionomycin for 4 h with last 2 h with monensin, and the amount of IL17 production by CD4+ T cells was determined by measuring the MFI of CD4+IL17+ T cells. To determine the levels of cell surface CD154 expression, same in vitro cultured splenocytes were incubated overnight with monensin according to the protocol described by Chattopadhyay et al. [21].
Supplementary Material
Acknowledgments
This research was supported (in part) by the Intramural Research Program of the NIH, National Institute on Aging.
Abbreviations
- ChIP
chromatin immunoprecipitation
- MOG
myelin oligodendrocyte glycoprotein
- mTOR
mammalian target of rapamycin
- RORγT
retinoic acid receptor-related orphan receptor gamma T
Footnotes
Conflict of interests: The authors declare no financial or commercial conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher’s web-site
References
- 1.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell JD, Delgoffe GM. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 2010;33:301–311. doi: 10.1016/j.immuni.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Kwon MJ, Ma J, Ding Y, Wang R, Sun Z. Protein kinase C-theta promotes Th17 differentiation via upregulation of Stat3. J Immunol. 2012;188:5887–5897. doi: 10.4049/jimmunol.1102941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J, Meng Y, Kwiatkowski DJ, Chen X, Peng H, Sun Q, Zha X, et al. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest. 2010;120:103–114. doi: 10.1172/JCI37964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruan Q, Chen YH. Nuclear factor-kappaB in immunity and inflammation: the Treg and Th17 connection. Adv Exp Med Biol. 2011;946:207–221. doi: 10.1007/978-1-4614-0106-3_12. [DOI] [PubMed] [Google Scholar]
- 15.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 17.Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Zhang Y, Yang XO, Nurieva RI, Chang SH, Ojeda SS, Kang HS, et al. Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity. 2012;36:23–31. doi: 10.1016/j.immuni.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med. 2005;11:1113–1117. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 22.Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, et al. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell Rep. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 24.Ichiyama K, Hashimoto M, Sekiya T, Nakagawa R, Wakabayashi Y, Sugiyama Y, Komai K, et al. Gfi1 negatively regulates T(h)17 differentiation by inhibiting RORgammat activity. Int Immunol. 2009;21:881–889. doi: 10.1093/intimm/dxp054. [DOI] [PubMed] [Google Scholar]
- 25.Barbi J, Pardoll D, Pan F. Metabolic control of the Treg/Th17 axis. Immunol Rev. 2013;252:52–77. doi: 10.1111/imr.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 27.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 28.Veilleux A, Houde VP, Bellmann K, Marette A. Chronic inhibition of the mTORC1/S6K1 pathway increases insulin-induced PI3K activity but inhibits Akt2 and glucose transport stimulation in 3T3-L1 adipocytes. Mol Endocrinol. 2010;24:766–778. doi: 10.1210/me.2009-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 31.Conn CS, Qian SB. Nutrient signaling in protein homeostasis: an increase in quantity at the expense of quality. Sci Signal. 2013;6:ra24. doi: 10.1126/scisignal.2003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheu S, Stetson DB, Reinhardt RL, Leber JH, Mohrs M, Locksley RM. Activation of the integrated stress response during T helper cell differentiation. Nat Immunol. 2006;7:644–651. doi: 10.1038/ni1338. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Weiner J, Liu Y, Smith AJ, Huss DJ, Winger R, Peng H, et al. T-bet is essential for encephalitogenicity of both Th1 and Th17 cells. J Exp Med. 2009;206:1549–1564. doi: 10.1084/jem.20082584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An J, Golech S, Klaewsongkram J, Zhang Y, Subedi K, Huston GE, Wood WH, 3rd, et al. Kruppel-like factor 4 (KLF4) directly regulates proliferation in thymocyte development and IL-17 expression during Th17 differentiation. FASEB J. 2011;25:3634–3645. doi: 10.1096/fj.11-186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasaki CY, Ghosh P, Longo DL. Recruitment of RelB to the Csf2 promoter enhances RelA-mediated transcription of granulocyte-macrophage colony-stimulating factor. J Biol Chem. 2011;286:1093–1102. doi: 10.1074/jbc.M110.119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 37.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–D809. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.