SUMMARY
Lignocellulose degradation is central to the carbon cycle and renewable biotechnologies. The xyloglucan (XyG), β(1→3)/β(1→4) mixed-linkage glucan (MLG), and β(1→3) glucan components of lignocellulose represent significant carbohydrate energy sources for saprophytic microorganisms. The bacterium Cellvibrio japonicus has a robust capacity for plant polysaccharide degradation, due to a genome encoding a large contingent of Carbohydrate-Active Enzymes (CAZymes), many of whose specific functions remain unknown. Using a comprehensive genetic and biochemical approach we have delineated the physiological roles of the four C. japonicus Glycoside Hydrolase Family 3 (GH3) members on diverse β-glucans. Despite high protein sequence similarity and partially overlapping activity profiles on disaccharides, these β-glucosidases are not functionally equivalent. Bgl3A has a major role in MLG and sophorose utilization, and supports β(1→3) glucan utilization, while Bgl3B underpins cellulose utilization and supports MLG utilization. Bgl3C drives β(1→3) glucan utilization. Finally, Bgl3D is the crucial β-glucosidase for XyG utilization. This study not only sheds the light on the metabolic machinery of C. japonicus, but also expands the repertoire of characterized CAZymes for future deployment in biotechnological applications. In particular, the precise functional analysis provided here serves as a reference for informed bioinformatics on the genomes of other Cellvibrio and related species.
Keywords: β-glucosidase, Cellvibrio japonicus, lignocellulose, mixed-linkage glucan, xyloglucan
INTRODUCTION
Terrestrial biomass, specifically complex plant cell walls (lignocellulose), is a major reservoir in the global carbon cycle and a vast renewable resource for the production of food, chemicals, and materials. It is estimated that 1011 tons of plant biomass are broken down annually by a diverse array of bacteria and fungi (Lynd et al., 2002). Microbial lignocellulose degradation plays a key role in nutrient acquisition in animal digestive tracts and is therefore central to the health of humans, livestock, and wildlife (Krajmalnik-Brown et al., 2012; Fernandes et al., 2014; Martens et al., 2014; Grondin et al., 2017). Additionally, there is sustained interest in the development of lignocellulose bioconversion technologies for the synthesis of value-added products that can displace petrochemicals (Bokinsky et al., 2011; Bugg et al., 2011; Chundawat et al., 2011; Jordan et al., 2012).
The intrinsic recalcitrance of plant biomass, which stymies microbial degradation and biorefinery applications alike, arises from the chemically complex, structurally composite nature of the plant cell wall (Himmel et al., 2007). Terrestrial plant cell walls are constructed of paracrystalline cellulose fibers embedded in a matrix of amorphous polysaccharides (hemicellulose and pectins), cross-linked polyphenolics (lignins), and structural proteins. The composition of these components varies across plant species and tissues (Carpita and McCann, 2000).
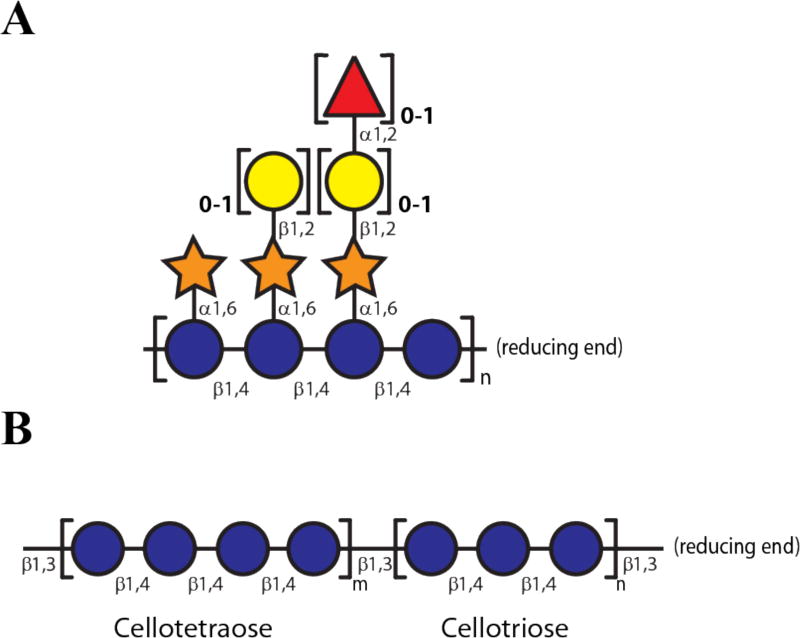
Historically, plant cell wall types have been delineated on the basis of hemicellulose composition (Carpita and McCann, 2000; Scheller and Ulvskov, 2010). In dicots and non-commelinid monocots, the xyloglucans (XyGs) are the predominant hemicellulosic polysaccharides, representing up to 25% of total dry weight of the wall (Carpita and McCann, 2000; Vogel, 2008; Scheller and Ulvskov, 2010). XyGs are characterized by a cellulose-like linear β(1→4)-D-glucose backbone, which is regularly decorated with α(1→6)-xylopyranosyl branches in two common motifs, XXGG and XXXG, where G represents unbranched D-Glcp-β(1→4) and X represents a [D-Xylp-α(1→6)]-D-Glcp-β(1→4) substitution (Tuomivaara et al., 2015). Depending on the species and tissue, the xylosyl side-chains can be further ramified by diverse monosaccharides, often galactopyranosyl, fucopyranosyl, and arabinofuranosyl residues, and specific residues may also be acetylated (Fig 1A) (Schultink et al., 2014).
Figure 1. Representative structure of xyloglucan (XyG) and mixed linkage β-glucan (MLG).
(A) Dicot (fucogalacto)xyloglucan indicating variable sidechain substitution. (B) General Poales MLG structure comprised of β(1→4)-linked cellotetraose and cellotriose units connected by β(1→3) linkages. Representations of monosaccharide residues are according to (Varki et al., 2009): glucose, blue circles; xylose, orange stars; galactose, yellow circles; L-fucose, red triangles.
In contrast, the XyG content of the cell walls of the Poales (e.g. grasses and cereals of the Poaceae) and Pteridophytes (represented by the horsetails) is low. Instead, xylans and mixed-linkage β-glucans (MLGs) predominate (Carpita and McCann, 2000; Vogel, 2008; Popper et al., 2011). These MLGs are thought to be comprised of predominantly cellotriosyl and cellotetraosyl moieties (β(1→4)-D-gluco-tri-and tetrasaccharides, respectively) that are connected by β(1→3) linkages to form a polysaccharide chain with an irregular sequence and three-dimensional structure. The relative ratios of the cellotriosyl and cellotetraosyl units vary depending upon the plant species, and the non-linear β(1→3) linkages typically represent ca. 30% of the total linkages within the polysaccharide (Fig 1B) (Fincher, 2009).
In addition to cellulose, XyG, MLG, and callose (β(1→3)-glucan (Ellinger and Voigt, 2014)) represent the major sources of glucose for primary metabolism by saprophytic microorganisms. Of these, the Gram-negative bacterium Cellvibrio japonicus (formerly Pseudomonas fluorescens subsp. cellulosa) has emerged as a powerful system for biomass enzyme discovery, due to its ability to degrade nearly all plant cell wall polysaccharides via the production of more than 150 Carbohydrate-Active enZymes (CAZymes) from 59 Glycoside Hydrolase (GH), Polysaccharide Lyase (PL), Carbohydrate Esterase (CE), and Auxiliary Activity (AA) families (Deboy et al., 2008; Lombard et al., 2014; Gardner, 2016). Moreover, robust reverse-genetic and transcriptomic tools for C. japonicus have been developed, which significantly enable systems biology and metabolic engineering for industrial applications (Gardner and Keating, 2010b, 2012; Gardner et al., 2014; Larsbrink et al., 2014a; Nelson and Gardner, 2015; Forsberg et al., 2016).
Using a comprehensive in vitro, in vivo, and in silico approach, we have recently revealed the central contributions of key GH5 and GH6 cellulases (Nelson and Gardner, 2015), an AA10 lytic polysaccharide mono-oxygenases (Gardner et al., 2014), and two GH3 β-glucosidases (Nelson et al., 2017) employed for efficient utilization of crystalline cellulose by C. japonicus. Specifically, in the latter case we were able to determine that two of four GH3 enzymes were important for cellodextrin utilization, yet the physiological roles for the remaining two GH3 members were unresolved. In this report, we further delineate the specific physiological functions of all four C. japonicus GH3 enzymes through a systematic combination of gene-deletion mutants, heterologous gene expression, recombinant enzyme production, and biochemical analysis. Our results reveal that these enzymes are not functionally redundant despite high sequence similarity (ranging from 45% to 60% between all four GH3s) (Fig S1), but instead have distinct roles in cleaving the diverse glucosidic linkages present in plant biomass in vivo.
RESULTS
Linkage specificity of C. japonicus GH3 members
Substrate choice for in vitro enzymology
Our previous studies on the functions of the C. japonicus GH3 enzymes tested activity exclusively on β(1→4)-linked gluco-oligosaccharides (cellodextrins). Although all GH3 members were competent β(1→4)-glucosidases in vitro and were able to confer growth on cellobiose when expressed heterologously in E. coli in vivo, individual activities on cello-oligosaccharides varied by orders-of-magnitude amongst the recombinant enzymes (Nelson et al., 2017). These results suggested that the true substrates for some of the GH3 enzymes might not be β(1→4)-linked gluco oligosaccharides. To explore the wider capacity of the C. japonicus GH3 enzymes to cleave other β-glucosidic linkages present in nature, we performed Michaelis-Menten kinetic analysis of β(1→2)-, β(1→3)-, and β(1→6)-linked disaccharides, as well as representative xylogluco-oligosaccharides (XyGOs) containing a β(1→4)-linked Glcp backbone (Table 1, Fig S2).
Table 1.
Kinetic parameters of C. japonicus GH3 enzymes against different gluco-disaccharides, cellotetraose and xyloglucan based oligosaccharides.a
| Enzyme | Substrate |
kcat (min−1) |
Km (mM) |
kcat/Km (min−1.mM−1) |
|---|---|---|---|---|
| GH3A | sophorose | 2.14 ± 0.05 × 103 | 3.36 ± 0.33 | 637 ± 64.6 |
|
|
||||
| laminaribiose | 9.23 ± 0.33 × 103 | 2.62 ± 0.33 | 3.52 ± 0.46 × 103 | |
|
|
||||
| cellobioseb | 117 ± 2.5 | 1.8 ± 0.09 | 65 ± 3.53 | |
|
|
||||
| cellotetraoseb | - | - | 5.31 ± 0.11 × 103 | |
|
|
||||
| GXXG | 37.6 ± 1.38 | 1.67 ± 0.13 | 22.5 ± 1.94 | |
|
|
||||
| GLLG | NDc | ND | ND | |
|
|
||||
| gentiobiose | 24.5 ± 1.04 | 11.3 ± 1.23 | 2.17 ± 0.25 | |
|
| ||||
| GH3B | sophorose | 19.9 ± 0.50 × 103 | 5.81 ± 0.46 | 3.43 ± 0.28 × 103 |
|
|
||||
| laminaribiose | 5.65 ± 0.19 × 103 | 2.39 ± 0.29 | 2.35 ± 0.30 × 103 | |
|
|
||||
| cellobioseb | 5.39 ± 0.18 × 103 | 1.5 ± 0.13 | 3.59 ± 0.33 × 103 | |
|
|
||||
| cellotetraoseb | - | - | 522 ± 12 | |
|
|
||||
| GXXG | 23.7 ± 3.53 | 5.15 ± 1.17 | 4.60 ± 1.25 | |
|
|
||||
| GLLG | ND | ND | ND | |
|
|
||||
| gentiobiose | 386 ± 29.8 | 9.31 ± 1.96 | 41.5 ± 9.30 | |
|
| ||||
| GH3C | sophorose | 3.46 ± 0.44 × 103 | 4.60 ± 0.97 | 752 ±185 |
|
|
||||
| laminaribiose | 3.14 ± 0.11 × 103 | 0.64 ± 0.07 | 4.91 ± 0.56 × 103 | |
|
|
||||
| cellobioseb | 168 ± 6 | 1.9 ± 0.14 | 88.4 ± 7.24 | |
|
|
||||
| cellotetraoseb | - | - | 856 ± 22 | |
|
|
||||
| GXXG | 27.0 ± 1.87 | 2.46 ± 0.33 | 11.3 ± 1.73 | |
|
|
||||
| GLLG | ND | ND | ND | |
|
|
||||
| gentiobiose | 161 ± 2.06 | 4.88 ± 0.20 | 33 ± 1.41 | |
|
| ||||
| GH3D | sophorose | 2.23 ± 0.13 × 103 | 28.7 ± 3.04 | 77.8 ± 9.37 |
|
|
||||
| laminaribiose | 2.85 ± 0.21 × 103 | 6.41 ± 1.13 | 445 ± 85.1 | |
|
|
||||
| cellobioseb | 10 ± 0.7 | 11.5 ± 1.5 | 0.87 ± 0.13 | |
|
|
||||
| cellotetraoseb | - | - | 91 ± 2.5 | |
|
|
||||
| GXXG | 1.33 ± 0.08 × 103 | 0.83 ± 0.13 | 1.60 ± 0.27 × 103 | |
|
|
||||
| GLLG | 749 ± 45.5 | 1.21 ± 0.14 | 619 ± 80.9 | |
|
|
||||
| gentiobiose | 39.7 ± 2.05 | 83.3 ± 5.85 | 0.48 ± 0.04 | |
Supporting initial-rate kinetic data is available in Fig S2.
Data from (Nelson et al., 2017). For cellotetrose, substrate saturation was not achieved and kcat/Km values were determined from the slope of linear velocity-[S] plots.
ND, not determined due to insufficient activity.
Specifically, sophorose (Glcpβ(1→2)Glcp) is a well-known inducer of cellulase gene expression in bacteria and filamentous fungi (Yamane et al., 1970; Castro et al., 2014). The cognate β(1→2) linkage is also found naturally in yeast sophorolipids (Sugisawa and Edo, 1966; Huang and Wages, 2016). Additionally, cyclic β(1→2)-glucans are involved in infection or symbiosis with plants and animals (Briones et al., 2001; Arellano-Reynoso et al., 2005; Rigano et al., 2007). Laminaribiose (Glcpβ(1→3)Glcp) is representative of exclusive β(1→3) backbone linkages found in bacterial exopolysaccharides (e.g. curdlan), yeast cell wall glucan, and algal cell wall laminarin (Bull and Chesters, 1966; Kollar et al., 1997; Zavaliev et al., 2011; Zhang and Edgar, 2014). Within plant biomass, β(1→3) linkages occur in callose (β(1→3)-glucan) and in grass and cereal MLGs (Fincher, 2009; Ellinger and Voigt, 2014). The β(1→6) linkages represented by gentiobiose (Glcpβ(1→6)Glcp) are comparatively rare, however β(1→6)-glucans are known to be an integral component of yeast cell wall polysaccharides (Kollar et al., 1997). Finally, the xyloglucan oligosaccharides GXXG and GLLG represent known intermediates in the complete saccharification of plant cell wall XyG by C. japonicus (Larsbrink et al., 2011; Larsbrink et al., 2014a).
As detailed below, all of the GH3 members exhibit β-glucosidase specificities that are neither necessarily restricted nor related to cellulose utilization. In recognition of the new biochemical and biological data presented herein, the individual enzymes will be henceforth denoted with the non-prescriptive identifiers Bgl3A, Bgl3B, Bgl3C, and Bgl3D (Bgl: β-glucosidase, cf. (Bhatia et al., 2005; Uchiyama et al., 2015), rather than the original (Deboy et al., 2008) Cel3A, Cel3B, Cel3C, and Cel3D, respectively (Cel: cellulase (Henrissat et al., 1998)).
Bgl3A exhibits β (1→3) and β (1→4) specificity in vitro
In our recent cellulose utilization study, Bgl3A exhibited the highest catalytic efficiency towards longer cello-oligosaccharides (degree-of-polymerization, DP: 3–5), as indicated by high kcat/km values among the four GH3 members (Nelson et al., 2017). Bgl3A exhibited a similarly high specificity for the β(1→3)-linked laminaribiose, with a kcat/km value ca. 1.5-fold lower than cellotetraose as a benchmark, indicating a potential biological role of the enzyme in β(1→3)-glucan and MLG saccharification. Bgl3A displayed ca. 8-fold less specificity towards the β(1→2) linkage of sophorose than cellotetraose, and was catalytically ineffective toward gentiobiose and the XyGOs, all of which exhibited lower activities than cellobiose (Table 1, Fig S2).
Bgl3B is agnostic toward β (1→2), β (1→3) and β (1→4) linkages in vitro
The highly constitutively expressed bgl3B has been shown to be fundamental in cellodextrin utilization by C. japonicus, and the gene product has a strong preference for cellobiose (Nelson et al., 2017). Commensurate with these observations, Michaelis-Menten analysis with alternate β-di-glucosides demonstrates superior specificity for cellobiose over longer cello-oligosaccharides. The Bgl3B kcat/km values also indicate high specificities for sophorose and laminaribiose, however, this enzyme was weakly active on gentiobiose. Bgl3B only cleaved the XyGO substrate GXXG weakly, and was not active against the bis-galactosylated GLLG XyGO (Table 1, Fig S2).
Bgl3C displays preferential β (1→3) specificity in vitro
Previous biochemical characterization and mutational analysis of Bgl3C indicated only modest activity toward β(1→4)-linked cello-oligosaccharides and suggested no physiological contribution to cellulose utilization (Nelson et al., 2017). Extending the biochemical analysis of Bgl3C to include alternate β-disaccharides revealed a predominant specificity for the β(1→3)-linked laminaribiose, with a kcat/km value 55- and 5.7-fold higher than for cellobiose and cellotetraose, respectively. Sophorose was hydrolyzed with a comparable kcat value to that of laminaribiose, although the km value for sophorose was significantly (7-fold) higher, resulting in a lower overall specificity constant. As also observed for Bgl3A and Bgl3B, Bgl3C was poorly active on gentiobiose and GXXG, and not active on GLLG (Table 1, Fig S2).
Bgl3D is a XyGO-specific β (1→4) glucosidase in vitro
Of the four GH3 members from C. japonicus, Bgl3D was catalytically weak against all tested cello-oligosaccharides. Specifically, the kcat/km value for cellobiose was ca. 1 mM−1 min−1, while the highest kcat/km value observed was 90 mM−1 min−1 for cellotetraose (Nelson et al., 2017). Although Bgl3D can hydrolyze sophorose and laminaribiose with weak to moderate specificities due to comparatively high km values, the data also indicate a uniquely high specificity for XyGOs (Table 1, Fig S2). Indeed, Bgl3D was the only GH3 enzyme from C. japonicus that was able to hydrolyze both GXXG and GLLG, which are key intermediates in the complete saccharification of XyG (Larsbrink et al., 2011; Larsbrink et al., 2014a). The kcat/km values of Bgl3D for GXXG and GLLG were ca. 17- and 7-fold higher, respectively, than for cellotetraose, which highlights the significance of side-chain branching to substrate recognition and catalysis. Enzyme product analysis revealed the specificity of Bgl3D for the non-reducing-terminal β(1→4)-Glcp residue of GXXG, as well as the ability of Bgl3D to work in concert with Xyl31A (Larsbrink et al., 2011; Larsbrink et al., 2014a) to effect the complete, stepwise hydrolysis of GXXG (Fig S3).
GH3 members are not universally nor equally sufficient for disaccharide utilization
We demonstrated previously that heterologous expression of individual C. japonicus GH3 genes were sufficient to confer the non-cellulolytic bacterium Escherichia coli with the ability to utilize cellobiose as a sole carbon source (Nelson et al., 2017). To further support in vitro enzyme specificity data and probe the potential sufficiency of the observed primary and side activities to support growth, we applied the same heterologous expression system using sophorose, laminaribiose, or gentiobiose as sole carbon sources (Fig 2).
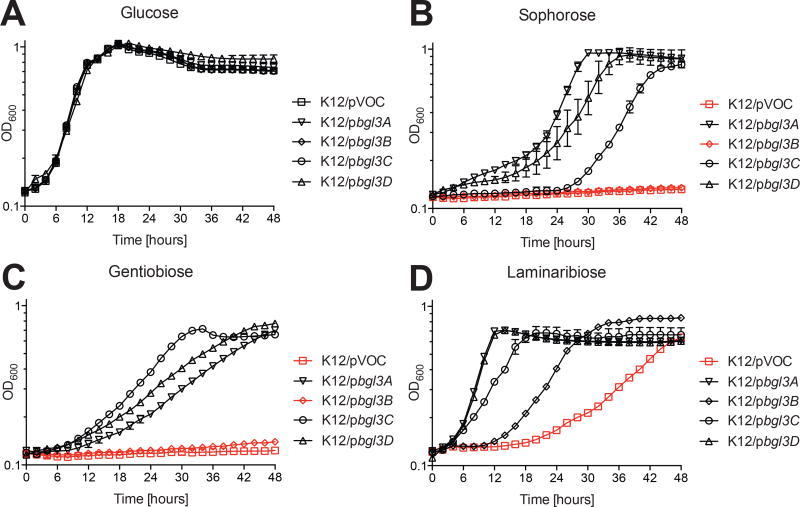
Figure 2. Growth of E. coli strains expressing individual C. japonicus GH3 genes using mono- and disaccharides.
(A) glucose, (B) sophorose, (C) gentiobiose, (D) laminaribiose. E. coli harboring the empty pBBRMCS-5 vector (pVOC) was included as a negative control. Experiments were performed in biological triplicate with the error bars representing the standard deviation. Growth rates and maximum optical density are summarized in Table S1.
Analysis of potential signal peptides using LipoP 1.0 (Rahman et al., 2008) to predict subcellular localization indicated that Bgl3A and BglC possess a Signal Peptidase II cleavage site, and are therefore likely to be anchored facing the periplasm or extracellular environment via N-terminal lipidation on Cys26 and Cys21, respectively (Fig S1) (Paetzel et al., 2002). Indeed, previous analysis indicated that recombinant expression of full-length Bgl3A (then known as CelD), but not an N-terminally truncated version, in E. coli results in membrane association (Rixon et al., 1992). LipoP 1.0 also indicated that Bgl3D possesses a predicted Signal Peptidase I cleavage site and is therefore likely to be secreted to the periplasm (Paetzel et al., 2002). In contrast, Bgl3B lacked a predicted signal peptide and is thus likely to be cytosolic.
Congruent with the kinetic analysis of recombinant enzymes, individual heterologous expression of the full protein coding sequences of all four GH3 genes enabled E. coli to more rapidly utilize laminaribose compared to the negative control strain (Fig 2D, Table S1). We attribute the eventual growth observed in the negative control strain after a prolonged lag period to the three GH1 enzymes and two GH3 enzymes encoded by E. coli K12 (Blattner et al., 1997), which may have sufficient side activity to support limited growth on laminaribiose.
Similarly, the heterologous expression of bgl3A, bgl3C, and bgl3D in E. coli conferred growth on both sophorose and gentiobiose (Fig 2B & 2C), although growth on gentiobiose was significantly slower in all cases (Table S1). For these GH3 genes, the growth rates of the engineered E. coli strains generally correlated with the kinetic data, specifically that laminaribose > sophorose > gentibiose (Fig 2). The growth of these strains is indicative of appropriate trafficking to the periplasm, as predicted by the aforementioned signal peptide analysis, where the substrates are accessible.
In contrast, the apparent failure of heterologous expression of the bgl3B gene to confer growth on gentibiose or sophorose despite the comparably high catalytic efficiency of the corresponding enzyme on both substrates was striking, but not unexpected due to its predicted cytosolic location (vide supra). As such, Bgl3B would be unable to confer growth to E. coli in the absence of suitable inner membrane PTS transporters for sophorose and gentibiose. Indeed, E. coli is currently only known to contain a β(1→4)-specific diglucoside transporter, and the import of other β-diglucosides has not been characterized (Tchieu et al., 2001).
Functional roles of GH3 members in alternate β-glucan utilization in C. japonicus
Possessing a broader understanding of the catalytic potential of its GH3 members, we then sought to ascertain the individual contributions of these β-glucosidases to the physiology of C. japonicus. Despite the apparent substrate promiscuity observed in assays in vitro, we anticipated that the controlled expression and localization of these enzymes in their native environment strongly affects their individual contributions to the utilization of specific β-glucans. Building upon our previous reverse-genetic analysis of cellulose utilization, we employed a suite of individual (Δbgl3A, Δbgl3B, Δbgl3C, Δbgl3D) and combinatorial gene deletion mutants, including a GH3 quadruple mutant (Δ4βG) (Nelson et al., 2017).
β(1→6)-glucosides
As described above, three of four engineered E. coli strains were able to leverage the expression of GH3 genes to grow using gentiobiose as a sole carbon source, despite weak kinetics on this disaccharide. However, in vivo analysis of genetic mutants is ultimately essential to reveal the actual physiologically-relevant contributions of individual GH3 members to β(1→6)-glucoside utilization by C. japonicus. We observed that all of the single gene deletion mutants grew like wild type when gentiobiose was the sole carbon source. Strikingly, all of the multiple mutants, including the 4βG strain, also had no growth defect on gentiobiose (Fig S4), clearly indicating that at least one non-GH3 CAZyme is primarily responsible for cleaving the β(1→6) linkages of this disaccharide in C. japonicus.
β(1→2)-glucosides
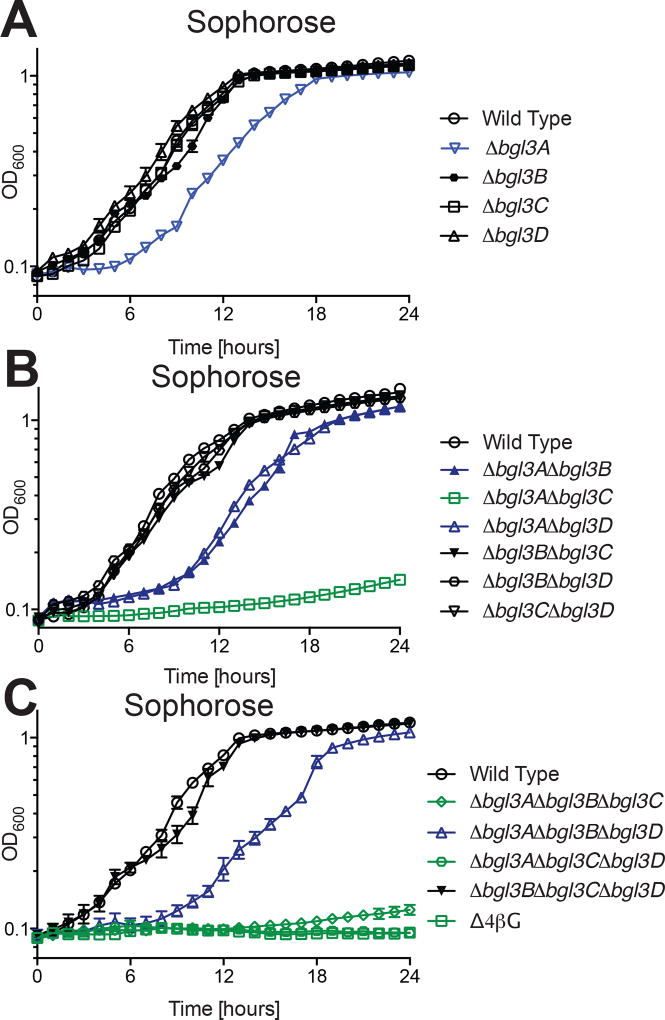
Mutational analysis suggested that the bgl3A and bgl3C gene products contribute synergistically to sophorose utilization (Fig 3). Specifically, when grown on this β(1→2)-linked diglucoside the Δbgl3A single deletion strain had a reproducible growth defect. The growth rate and maximum growth obtained was mirrored in the Δbgl3A Δbgl3B and Δbgl3A Δbgl3D double mutant strains, indicating that the Bgl3B or Bgl3D do not work in concert with Bgl3A to hydrolyze sophorose. In contrast, the Δbgl3A Δbgl3C double mutant strain was unable to grow, despite the absence of a growth phenotype for the Δbgl3C single mutant. The importance of Bgl3C and Bgl3A for sophorose utilization may be explained by comparable kinetics against this disaccharide and identical predicted trafficking to the periplasm (Table 1).
Figure 3. Growth of C. japonicus wild-type and GH3 gene deletion mutants on the β(1→2) linked dissacharide sophorose.
(A) single, (B) double, (C) triple and quadruple mutants versus wild type. Experiments were performed in biological triplicate with the error bars representing the standard deviation. Growth rates and maximum optical density are summarized in Table S1. All mutants grew as wild type on glucose, as shown previously (Nelson et al., 2017).
Notably, strains containing the Δbgl3B deletion grew like wild type using sophorose, indicating that the cytoplasmic GH3 does not play a role in sophorose utilization despite the high activity of Bgl3B against this substrate. In light of the demonstrable action of Bgl3A and Bgl3C, it is likely that all exogenous sophorose is completely hydrolyzed in the periplasm under the experimental conditions and transported into the cytoplasm as glucose. As sophorose is an inducer for cellulase gene expression in C. japoncius (Yamane et al., 1970), rapid degradation of this disaccharide may be an example of a metabolic mechanism to regulate cellulase expression.
β(1→3)-glucosides
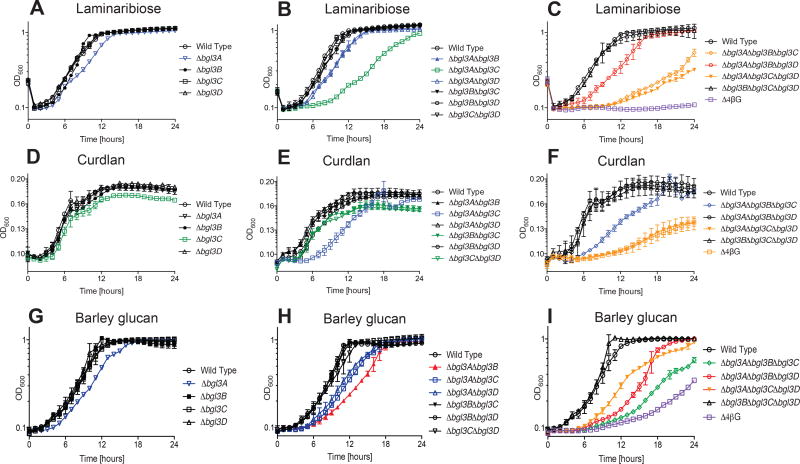
When grown on the β(1→3)-linked diglucoside laminaribiose, the Δbgl3A single deletion had a slight but reproducible growth defect, which was essentially recapitulated in the Δbgl3A Δbgl3B and Δbgl3A Δbgl3D double mutants (Fig 4A). Similar to the trend observed with sophorose, a Δbgl3A Δbgl3C double mutant had a longer lag period and a decreased growth rate compared to wild type (Fig 4B, Table S1). Commensurate with the primary role of Bgl3A in the utilization β(1→3)-linked saccharides, the Δbgl3B Δbgl3C Δbgl3D triple mutant grew as wild type (Fig 4C), which was also true with the polysaccharides curdlan and MLG (vide infra). The Δbgl3A Δbgl3C Δbgl3D and Δbgl3A Δbgl3B Δbgl3C triple mutants displayed increased lag periods, decreased growth rate, and did not achieve wild type final cell density (Table S1). Finally, the Δ4βG quadruple mutant was unable to grow using laminaribiose as a sole carbon source, which correlated with the sufficiency of all four GH3s toward this substrate (Fig 2D).
Figure 4. Growth of C. japonicus wild-type and GH3 gene deletion mutants on the β(1→3) linkage-containing substrates.
(A–C) laminaribiose, (D–F) curdlan, (G–I) MLG. Experiments were performed in biological triplicate with the error bars representing the standard deviation. Growth rates and maximum optical density are summarized in Table S1. All mutants grew as wild type on glucose, as shown previously (Nelson et al., 2017).
During growth on curdlan as a representative all-β(1→3)-glucan, the Δbgl3C single mutant strain displayed a slight, but reproducible growth defect (Fig 4D). This growth defect was further exacerbated in the Δbgl3A Δbgl3C double mutant (Fig 4E). The Δbgl3A Δbgl3B Δbgl3C triple mutant had a decreased growth rate (Fig 4F), and behaved similarly to the Δbgl3A Δbgl3C double mutant (Table S1). Finally, both the Δbgl3A Δbgl3C Δbgl3D triple mutant and Δ4βG quadruple mutant had extended lag phases, decreased growth rates, and were not able to grow to wild type levels of final cell density (Fig 4F). Notably, there is an absence of a growth rate defect for the Δbgl3A single mutant on curdlan when one is observed on laminaribiose. However, a growth rate defect on curdlan only emerges with the Δbgl3A Δbgl3C double mutant. The Δbgl3C single mutant defect is one of maximum growth, not growth rate, which may be a consequence of the generally poor growth of all strains on curdlan (Table S1). The decreased growth rates and overall lower cell densities achieved with curdlan compared to the other substrates is likely due to carboxymethylation of the commercial substrate to improve solubility (Zhang and Edgar, 2014). Similar reductions in C. japonicus growth have been observed with the artificial β(1→4)-glucan carboxymethylcellulose (Gardner and Keating, 2010a).
As a representative of the matrix glycan abundant in the cell walls of grasses, cereals, and horsetails, we tested the growth of our suite of GH3 mutants on barley MLG. Similar to laminaribiose, the only single mutant that displayed a growth rate defect on MLG was Δbgl3A (Fig 4G). Although the Δbgl3B single mutant grew as wild-type on MLG, the Δbgl3A Δbgl3B double mutant exhibited a more exaggerated growth defect than the Δbgl3A single mutant, suggesting that these two GH3 enzymes work synergistically for MLG utilization (Fig 4H, Table S1). Similar synergy was observed during growth on β(1→4)-linked cellodextrins (Nelson et al., 2017). Most of the GH3 triple mutants had growth defects of moderate severity, but interestingly the Δbgl3B Δbgl3C Δbgl3D triple mutant grew like wild type, which further suggested that the bgl3A gene product is the main driver of MLG oligosaccharide hydrolysis (Fig 4I, Table S1). The Δ4βG quadruple mutant was still able to grow on MLG, albeit with a distinctly long lag phase (Fig 4I), suggesting the presence of additional β-glucosidases. Analogously, an identical growth profile was observed for this mutant on cellobiose (Nelson et al., 2017).
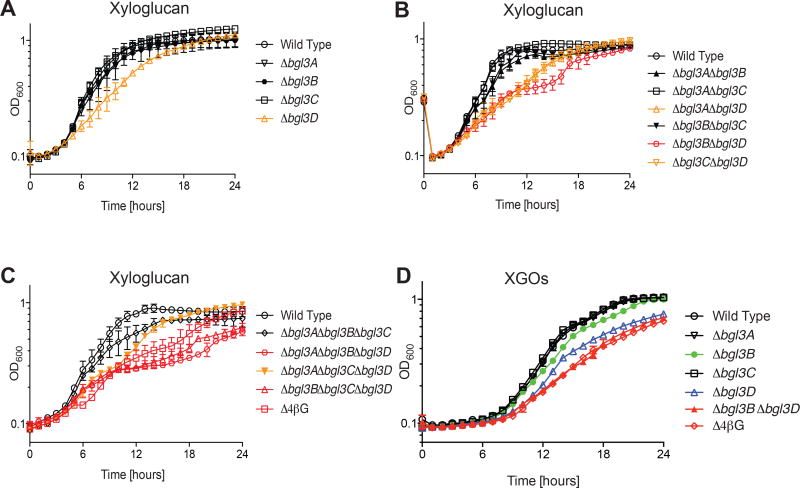
Xyloglucan β(1→4)-glucosides
As described in the Introduction, XyG is an abundant cell wall matrix glycan built on an all-β(1→4)-linked glucan backbone, which is found in essentially all terrestrial plants (Vogel, 2008; Scheller and Ulvskov, 2010). To explore the potential contribution of C. japonicus GH3 β-glucosidases to the utilization of this ubiquitous polysaccharide, we examined the growth of our suite of mutants grown with XyG and XyGOs. Strikingly, the Δbgl3D single mutant exhibited a reduced growth rate on XyG (Fig 5A), which was slightly exacerbated in the Δbgl3B Δbgl3D double mutant (Fig 5B). None of the GH3 triple mutants nor the quadruple mutant displayed growth defects more severe than the Δbgl3B Δbgl3D double mutant (Fig 5C). The importance of the bgl3D and bgl3B gene products for XyG utilization was directly recapitulated during growth on XyGOs, with the additional observation that the subtle role of Bgl3B was more directly exposed in the growth profiles of the Δbgl3B single and Δbgl3B Δbgl3D double mutants (Fig 5D, Table S1).
Figure 5. Growth of C. japonicus wild-type and GH3 gene deletion mutants on xyloglucan and xylogluco-oliogsaccharides.
(A) single, (B) double, (C) triple and quadruple mutants versus wild type on XyG. (D) Growth of select strains on XyGOs. Experiments were performed in biological triplicate with the error bars representing the standard deviation. Growth rates and maximum optical density are summarized in Table S1. All mutants grew as wild type on glucose, as shown previously (Nelson et al., 2017).
We hypothesized that the ability of the Δ4βG mutant to grow on XyG was due in part to xylose utilization arising from the functional α-xylosidase Xyl31A encoded by the xyloglucan utilization locus (XyGUL) of C. japonicus (Larsbrink et al., 2014a). To test this hypothesis, we deleted the xylose isomerase gene xylA, from the Δ4βG mutant strain, as it was previously shown that a C. japonicus ΔxylA mutant was unable to utilize xylose as a carbon source (Nelson et al., 2016). The quintuple mutant had a reduced growth rate compared to the Δ4βG mutant and achieved three-fold lower maximum density (Fig S5, Table S1). The residual growth of the Δ4βG ΔxylA quintuple mutant is most likely due to the action of the XyGUL GH35 β-galactosidase on the galactosyl sidechains, which substitute approximately 50% of the xylose residues on tamarind seed XyG (Larsbrink et al., 2014a), and possibly by the action of cryptic β-glucosidases, as indicated by our results for this mutant when using cellobiose or MLG.
DISCUSSION
The high-throughput (meta)genomics of saprophytic microorganisms of ecological and biotechnological interest has generated a vast abundance of sequence data on the diverse suites of CAZymes and other proteins that drive biomass degradation (Medie et al., 2012; El Kaoutari et al., 2013; Kunath et al., 2017; Mukherjee et al., 2017). A common observation is that most saprophytes encode multiple homologs from individual CAZyme families within their genomes, and even the most exacting biochemical analysis performed in vitro can fail to reveal enzyme performance in complex, biologically relevant situations (Zhang et al., 2002; Cartmell et al., 2011; Naas et al., 2014), particularly as all physiological and regulatory context is removed (Forsberg et al., 2016; Nelson et al., 2017). Therefore, a combinatorial approach that synthesizes in vitro and in vivo methods can be a powerful tool to achieve a deeper understanding of CAZyme function. As such, we have leveraged C. japonicus as a model saprophytic organism to delineate the individual contributions of four prima facie similar GH3 β-glucosidases to environmental polysaccharide utilization using an integrated systems biology approach.
Bgl3A has a major role in MLG and sophorose utilization and supports curdlan degradation
Our previous study indicated a supporting role of Bgl3A in cellulose (all-β(1→4)-glucan) utilization and a substrate preference for cello-oligosaccharides of DP >2 (Nelson et al., 2017). Our current biochemical data demonstrated that Bgl3A can also hydrolyze the β(1→3) linkage of laminaribiose with a comparably high specificity and that heterologous expression in E. coli was sufficient to confer growth on this disaccharide. Commensurately, C. japonicus the Δbgl3A single deletion mutant was the only one to exhibit slowed growth on laminaribiose and MLG as sole carbon sources, while no growth defect was observed for the triple mutant Δbgl3B Δbgl3C Δbgl3D (Fig 4). These data suggest the primacy of Bgl3A in the degradation of MLG oligosaccharides arising from Poales cell walls. The observed growth defects from double mutants (Fig 4B, 4E, & 4H) revealed that bgl3A and bgl3C gene products were the main drivers of curdlan utilization (all-β(1→3)-glucoside). The requirement for two GH3 enzymes for effective substrate utilization was also observed with Bgl3A and Bgl3B, which were primarily responsible for the consumption of MLG that comprises both β(1→3) and β(1→4)-linkages. This latter observation is concordant with our previous study indicating that Bgl3B is the single greatest contributor to β(1→4)-linked cellobiose utilization (Nelson et al., 2017), and indicates particular synergy in MLG oligosaccharide utilization.
Growth analysis of single mutants indicates that the bgl3A gene product is also the primary enzyme responsible for sophorose utilization by C. japonicus. The loss of Bgl3A can only be compensated by Bgl3C, despite Bgl3B also possessing excellent activity toward this substrate (Bgl3D is hobbled by a km value of ca. 30 mM). The abundance of β(1→2)-glucans in the natural environment (vide supra) and their corresponding importance to the growth of saprophytes is presently unclear. It is worth noting, however, this disaccharide has been shown to induce cellulase production in C. japonicus (Yamane et al., 1970), and Bgl3A may function together with Bgl3C (vide infra) in signal attenuation.
Bgl3B underpins cellodextrin degradation and supports MLG utilization
Biochemical and reverse genetic analyses have previously shown the essential contribution of Bgl3B in cellulose utilization with an exquisite specificity towards cellobiose (Nelson et al., 2017). Present biochemical characterization and heterologous expression in E. coli also revealed comparable activity of the enzyme towards laminaribiose (Table 1, Fig 2), which suggested a potential physiological role in the utilization of β-glucans containing (1→3) linkages. As discussed above, Bgl3B displayed a distinct supporting role to Bgl3A in MLG utilization, as evidenced by the enhanced growth defect of the double mutant Δbgl3A Δbgl3B (Fig 4H). This observation can be explained by the abundance of β(1→4) linkages in the MLG chain, together with the high activity of Bgl3B on cello-oligosaccharides (Nelson et al., 2017). Conversely, mutational analysis evidenced only a minor contribution of Bgl3B for the utilization of the strictly β(1→3)-linked substrates laminaribiose and curdlan. Instead, such substrates are degraded by the more β(1→3)-specialized enzymes Bgl3A (vide supra) and Bgl3C (vide infra). Bgl3B has no contribution to sophorose utilization, as revealed by the lack of growth of the triple mutant Δbgl3A Δbgl3C Δbgl3D, in addition to the inability to confer growth on this substrate when heterologously expressed in E. coli (Fig 3). The cytosolic Bgl3B also does not appear to contribute to XyG utilization, presumably due to the fact that XyGO degradation occurs in the periplasm (Larsbrink et al., 2014a).
Bgl3C drives β(1→3)-glucan utilization
The Bgl3C enzyme contributes minimally to cellulose utilization in C. japonicus and the bgl3C gene was not up-regulated during growth on cellobiose as the sole carbon source (Nelson et al., 2017). Although the enzyme is active against cello-oligosaccharides (Nelson et al., 2017), further biochemical analysis revealed particular specificity toward β(1→3)-linkages, as represented by laminaribiose, which had a notably low Km value (Table 1). Deletion of the bgl3C gene alone failed to produce a growth defect on laminaribiose, but did result in slightly impaired growth on curdlan. As discussed above, mutants also lacking bgl3A exhibited large growth defects, and it appears that Bgl3A and Bgl3C work synergistically on all-β(1→3)-glucan (Fig 4). All C. japonicus GH3 enzymes, however, have at least a partial role in β(1→3)-glucan utilization, as the Δ4βG quadruple mutant was unable to grow on laminaribiose and curdlan. Moreover, the promiscuous activity against β(1→3) and β(1→4) linkages of Bgl3C also makes it a significant contributor to MLG utilization. Finally, Bgl3C also plays a role in β(1→2)-glucoside degradation by C. japonicus, as it is the only GH3 member able to compensate for the loss of Bgl3A in sophorose utilization, and was also sufficient to support growth when expressed in E coli.
Bgl3D is the crucial β-glucosidase for XyG utilization
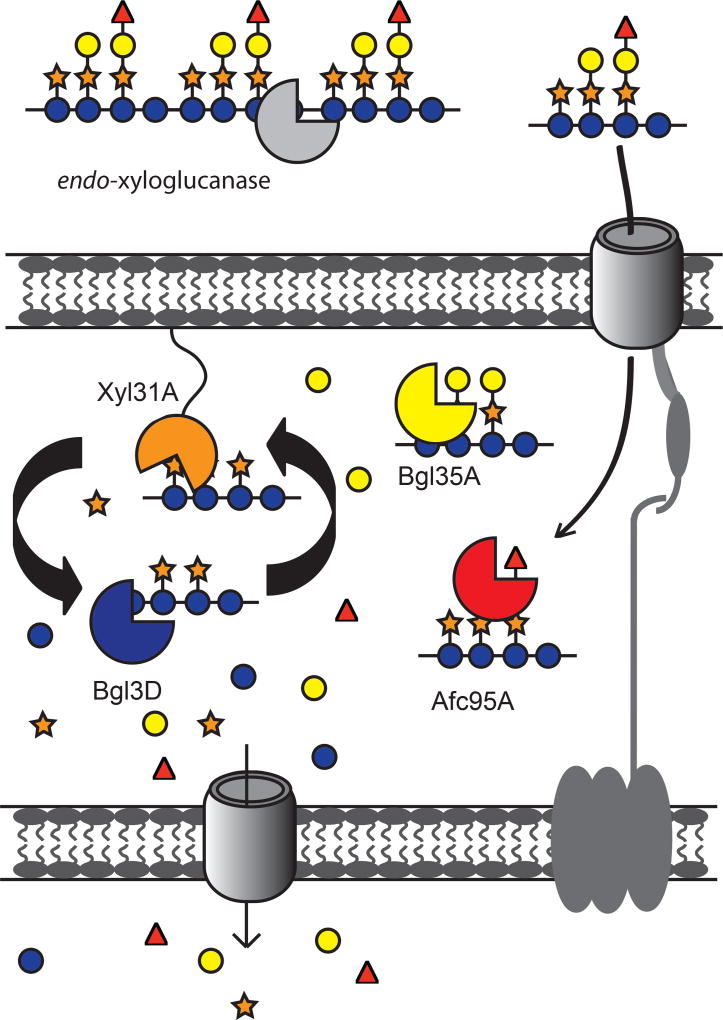
The present model for XyG utilization by C. japonicus involves a xyloglucan utilization locus (XyGUL) (Fig 6) that encodes three periplasmic, side-chain-cleaving GHs (a GH31 α-xylosidase, a GH35 β-galactosidase, and a GH95 α-L-fucosidase) and a predicted outer-membrane TonB dependent transporter (TBDT) for periplasmic uptake and saccharification of XyGOs produced by extracellular endo-xyloglucanases (Larsbrink et al., 2011; Larsbrink et al., 2014a; Attia et al., 2016; Attia and Brumer, 2016). Despite extensive efforts illuminating the concerted action of these players, the identity of the β-glucosidase(s) necessary for the complete deconstruction of the β(1→4)-linked XyGO backbone was heretofore unknown. Enzyme kinetic data (Table 1) distinctly identified Bgl3D as the only GH3 member with high catalytic efficiency toward the XyGOs GXXG and GLLG, which are the products of the XyGUL-encoded periplasmic α-xylosidase, Xyl31A. Notably, Xyl31A is highly specific for the non-reducing terminal α(1→6)-Xylp residue of XyGOs and is unable to remove internal xylosyl sidechains from the backbone (Larsbrink et al., 2011; Silipo et al., 2012; Larsbrink et al., 2014a). Thus, β(1→4)-glucosidase activity is essential for continued degradation of XyGOs in the C. japonicus periplasm.
Figure 6. Updated model of xyloglucan utilization by C. japonicus.
XyG hydrolysis is initiated outside of the cell by endo-xyloglucanases (e.g. CjGH74 (Attia et al., 2016)), followed by XyGO transport into the periplasm by a TonB-dependent transporter (TBDT). The C. japonicus α-L-fucosidase Afc95A removes terminal fucosyl residues, enabling full access of the β-galactosidase Bgl35A to both pendant galactosyl residues (Larsbrink et al., 2014a) Activity of the α-xylosidase Xyl31A is restricted to terminal non-reducing-end xylosyl residues (Larsbrink et al., 2014a), such that cycling between the α-xylosidase and the primary XyGO-specific β-glucosidase Bgl3D is required for complete saccharification to monosaccharides for primary metabolism (see Fig S3).
In vivo, previous RNAseq data and mutational analysis indicated that bgl3D was not involved in cellobiose utilization (Nelson et al., 2017). Here, analysis of the single and multiple GH3 mutants directly implicated Bgl3D as the primary β-glucosidase responsible for growth on XyG and XyGOs, which could be assisted to a very limited extent by the predominant β(1→4)-glucosidase Bgl3B (Fig 5). Additional recombinant enzyme product analysis in vitro has also provided direct evidence that Bgl3D can fulfill this role (Table 1). Collectively, detailed biochemical and physiological data were both essential to resolve the key outstanding question regarding the identity of the β(1→4)-glucosidase required for XyG utilization.
The sequestration of oligosaccharides by transport into the cell is a strategy used by many environmental bacteria (Grondin et al., 2017; Reintjes et al., 2017). The co-localization of Bgl3D with the sidechain-cleaving exo-glycosidases (Xyl31A, Bgl35A, and Afc95A) of the XyGUL in the periplasm of C. japonicus gives evidence of a unified strategy for competitive oligosaccharide acquisition and utilization by this saprophytic bacterium. This elegant system is analogous to that used by the human gut symbiont Bacterioides ovatus, in which two periplasmic GH3s operate in concert with an α-xylosidase, a β-galactosidase, and two α-L-arabinofuranosidases to saccharify dietary (arabinogalacto)xyloglucans (Larsbrink et al., 2014b; Hemsworth et al., 2016).
CONCLUSION
Using a combination of biochemical and physiological approaches, we have determined that the four GH3 members of Cellvibrio japonicus play unique roles in targeting different glucosidic linkages in diverse polysaccharides. Our work further illuminates the mechanism by which this model saprophyte, which has served as a treasure trove for CAZyme discovery for decades (Hazlewood and Gilbert, 1998; Gardner, 2016), utilizes the ubiquitous plant cell wall matrix glycans MLG, XyG, and callose. As such, the use of truly systems biology approaches is proving essential to disentangle apparent redundancy in microbial genomes encoding multiple members of single CAZyme families. The current study not only sheds the light on the metabolic capacity of C. japonicus, but also expands the accessible CAZyme repertoire for future deployment in biotechnological applications. For example, in light of recent demonstrations (Jabbour et al., 2013) that α-xylosidase addition to enzyme cocktails can improve ultimate glucose release, possibly through addressing tightly-bound XyG in cellulose (Pauly et al., 1999), we anticipate that additional combinations with a XyG-specific β-glucosidase (e.g. Bgl3D) may further improve saccharification. Not least, our functional analysis provides a gold-standard reference for informed bioinformatics on the genomes of other Cellvibrio and related species (Xie et al., 2015).
EXPERIMENTAL PROCEDURES
Recombinant protein production and purification
The expression vectors pET21a::GH3A, pET21a::GH3B, pET28a::GH3C, and pET28a::GH3D, produced as previously described without the predicted native signal peptide-encoding sequences (Nelson et al., 2017), were generously provided by Prof. Harry Gilbert (Newcastle University, UK). These constructs included fusion of a C-terminal 6× His-Tag (GH3A and GH3B) or an N-terminal 6× His-Tag (GH3C and GH3D) to aid purification. Constructs were transformed to the chemically competent BL21 E. coli strain. Colonies were grown on LB solid media containing 50 µg ml−1 ampicillin (GH3A and GH3B) or 50 µg ml−1 kanamycin (GH3C and GH3D). One colony was selected from each plate, inoculated in 15 mL LB liquid medium containing the same antibiotic, and grown overnight at 37 °C (200 RPM). The entire overnight cultures were used to inoculate 1 liter LB media containing the proper antibiotic before they were grown at 37 °C (200 RPM) until OD600= 0.8. In order to induce the overexpression, IPTG to a final concentration of 1 mM was added before the growing flasks were transferred to a 16 °C cooling shaker for an overnight incubation (200 RPM). Cultures were then centrifuged and pellets were re-suspended in 20 mL of E. coli lysis buffer containing 20 mM HEPES, pH 7.0, 500 mM NaCl, 40 mM imidazole, 5% glycerol, 1 mM DTT and 1 mM PMSF. Sonication was used to disrupt the cells and the clear supernatants were separated by centrifugation at 4 °C (4220 g for 60 min). In order to purify recombinant proteins from the clear soluble lysates, a BioRad® FPLC system with a Ni+2– affinity column was used. The system utilized a gradient elution up to 100% elution buffer containing 20 mM HEPES, pH 7.0, 100 mM NaCl, 500 mM imidazole, and 5% glycerol. Pure fractions verified by SDS-PAGE were pooled, concentrated, and buffer exchanged against 50 mM phosphate buffer (pH 7.5). Protein concentrations were finally determined using Epoch Micro-Volume Spectrophotometer System (BioTek®, USA) at 280 nm. Typical production yields of 2.5, 5, 22, and 15 mg per liter of culture medium were obtained for GH3A, GH3B, GH3C, and GH3D, respectively.
Carbohydrate sources used in kinetic experiments
Tamarind seed xyloglucan, sophorose, and laminaribiose were purchased from Megazyme® (Bray, Ireland). Gentiobiose was purchased from Carbosynth (Berkshire, UK).
Preparing the XyG-based oligosaccharide substrates
Both GXXG and GLLG were obtained using the XyG-degrading enzymes from C. japonicus. For making GXXG one gram of tamarind seed XyG was dissolved in 100 mL of double distilled water. To speed up dissolution, continuous stirring at 60 °C was employed for 2–3 hours. XyG solution was then cooled down to room temperature and a phosphate buffer (pH 7.5) to final concentration of 1 mM was added. One mg of the endo-xyloglucanase C. japonicus GH5E was then added and the digestion reaction was incubated at 40 °C overnight while stirring. The limit-digest reaction gave the XyGOs XXXG, XLXG, XXLG, and XLLG. After the completion of the reaction was verified using HPAEC-PAD and MALDI-TOF analyses, the reaction was stopped by boiling for 15 min. The XyGOs mixture was then cooled down to room temperature and 0.2 mg of the C. japonicus β-galactosidase Bgl35A (Larsbrink et al., 2014a) was added. Reaction was then incubated overnight while stirring at room temperature. After the reaction was complete, only the sole product XXXG was identified. In order to obtain GXXG, 0.5 mg of the C. japonicus α-xylosidase Xyl31A (Larsbrink et al., 2011) was added to the XXXG solution before the reaction mixture was incubated overnight while stirring at room temperature. After confirming the identity of the GXXG product, the solution was boiled to inactivate the enzymes, and then GXXG was freeze-dried after flash freezing in liquid nitrogen. For purifying GXXG, ~230 mg of the freeze-dried powder were dissolved in ~ 2.5 mL of ultra-pure water before they were loaded on 90-cm BioGel® P-2 Gel (Bio-Rad, California) column (XK 26/100, GE Healthcare). Fractions collected from the isocratic elution at 0.5 ml/min were screened for the presence of GXXG using HPAEC-PAD and MALDI-TOF analyses. Pure fractions were pooled, flash frozen and freeze-dried.
To produce GLLG, the XyGO mixture XXXG, XLXG, XXLG, and XLLG was obtained following the same protocol described above. The crude oligosaccharide mixture was peracetylated and separated according the previously described protocol (Ibatullin et al., 2008). XLLG was obtained after performing the deacetylation as described previously (Greffe et al., 2005). In order to obtain GLLG, 30–40 mg of XLLG were incubated overnight at room temperature with 0.5 mg of the C. japonicus α-xylosidase Xyl31A (Larsbrink et al., 2011) in a 4 mL reaction volume containing 25 mM citrate buffer (pH 6). Product identity and reaction completion were confirmed by HPAEC-PAD and MALDI-TOF analyses before the mixture was loaded on 90-cm BioGel® P-2 Gel (Bio-Rad, California) column (XK 26/100, GE Healthcare) for purification (vide supra).
Enzyme kinetics
All enzyme assays against the different disaccharides and XyG-based oligosaccharides were performed using a glucose detection kit (d-glucose-HK, Megazyme®). Enzyme assays were executed following manufacturer’s protocol with a minor modification: each recombinant β-glucosidase was added to a freshly prepared mixture of the tested substrate with the kit components (Buffer pH 7, NADP+ + ATP, hexokinase + glucose-6-phosphate dehydrogenase). Subsequently, the rate of glucose release, corresponding to the rate of enzymatic activity, can be monitored in a continuous assay format using Cary50 UV–visible spectrophotometer (Varian) at 340 nm. The enzyme concentration (0.004 to 0.81 µM) to be used was determined based on the resulting activity on the tested substrate and so that less than 10% conversion of the substrate is achieved within the measurement time. For calculation, a molar extinction coefficient of 6220 M−1cm−1 was used for NADPH at 340 nm.
Carbohydrate analytics
High Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) was performed on a Dionex ICS-5000 DC HPLC system operated by the Chromeleon software version 7 (Dionex) using a Dionex Carbopac PA200 column. Method of analysis followed the previously published protocol (Attia et al., 2016). Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) was performed on a Bruker Daltonics Autoflex System (Billerica, USA) and using the matrix, 2,5-dihydroxy benzoic acid as previously described (Attia et al., 2016).
Bgl3D product analysis
Six µg of the recombinant Bgl3D was incubated overnight at 37 °C with GXXG (0.25 mM final concentration) in a 400 uL reaction volume containing 50 mM phosphate buffer (pH 7.5). The enzyme was then inactivated by boiling for 15 min to stop the reaction. After the mixture was cooled down, 10 µg of the C. japonicus α-xylosidase Xyl31A was added and the reaction was incubated at 37 °C for 6 hours. By repeating the aforementioned steps, products were subjected to sequential degradation using GH3D and Xyl31A with a heat-inactivation step in between. Reactions were always monitored using HPAEC-PAD and the identity of the product from each step was verified using MALDI-TOF analysis as described above.
Growth Conditions
Growth experiments with E. coli and C. japonicus strains used MOPS (3-(N-morpholino)propanesulfonic acid) defined media (Neidhardt et al., 1974) with 0.25% (w:v) glucose, 0.25% (w:v) xylose, 0.5% (w:v) sophorose, 0.5% (w:v) laminaribiose, 0.5% (w:v) gentiobiose, 0.5% (w:v) CM-curdlan, 0.5% (w:v) barley glucan, 0.5% (w:v) xyloglucan, or 0.5% (w:v) xyloglucan oligosaccharides as carbon sources. Disaccharides, curdlan, and MLG were purchased from MegaZyme (Ireland, UK). Antibiotics were used at the following concentrations, kanamycin (50 µg ml−1) and gentamycin (15 µg ml−1). All growth experiment parameters including inoculation, temperature, aeration level, measurement of growth rate and maximum optical density were identical to what has been previously described (Nelson and Gardner, 2015; Nelson et al., 2017). All experiments were performed in biological triplicate and standard deviation was calculated using the GraphPad Prism 6 software (CA, USA).
Genetic techniques
The construction of the Δ4βG ΔxylA quintuple mutant and subsequent verification by PCR was identical to methods previously described (Gardner and Keating, 2012; Nelson and Gardner, 2015; Nelson et al., 2017). A complete list of strains, plasmids, and primers can be found in Table S2.
Supplementary Material
ORIGINALITY - SIGNIFICANCE STATEMENT.
The soil saprophyte Cellvibrio japonicus has emerged as a genetically tractable model for elucidating the molecular details of terrestrial biomass degradation. Here, we reveal the individual, specific contributions of the four Glycoside Hydrolase Family 3 β-glucosidases encoded by C. japonicus in the saccharification of the ubiquitous plant glycans callose, mixed-linkage glucan, and xyloglucan. This work highlights that systems-based approaches harnessing reverse genetics and recombinant enzymology are essential to disentangling apparent redundancy and delineating unique physiological roles of prima facie similar Carbohydrate-Active Enzymes (CAZymes).
Acknowledgments
Work at UMBC was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0014183. Work in Vancouver was supported by the Natural Sciences and Engineering Research Council of Canada (via Strategic Partnership Grant for the Industrial Biocatalysis Network, http://www.ibnet.ca/), the Canada Foundation for Innovation, and the British Columbia Knowledge Development Fund. M.A. and H.B. thank Prof. Harry Gilbert (Newcastle University) for providing plasmids for recombinant enzyme production in E. coli. M.A. is the recipient of post-graduate funding from the Michael Smith Laboratories. H.B. and J.G.G. thank the UMBC Eminent Scholar program for supporting the ongoing collaboration between the UMBC and UBC groups.
Footnotes
AUTHOR CONTRIBUTIONS
C.E.N. generated C. japonicus mutants and performed growth analysis experiments, generated heterologous expression plasmids for experiments in E. coli, and contributed to writing the manuscript. M.A.A. prepared the XyG-based oligosaccharide substrates, produced and biochemically characterized recombinant enzymes in vitro, and contributed to writing the manuscript. C.M. produced plasmids for recombinant protein production, and A.R. and C.M. contributed enzyme kinetic data for BglA and BglB on disaccharides. H.B. supervised the recombinant enzymology and contributed to writing the manuscript. J.G.G. designed the study, provided overall project direction, and contributed to writing the manuscript. All authors read and approved the final submitted version of the manuscript.
DISCLAIMER
This report was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any studies using human participants or animals.
In addition, the authors declare that they have no conflict of interest.
References
- Arellano-Reynoso B, Lapaque N, Salcedo S, Briones G, Ciocchini AE, Ugalde R, et al. Cyclic beta-1,2-glucan is a brucella virulence factor required for intracellular survival. Nature Immunology. 2005;6:618–625. doi: 10.1038/ni1202. [DOI] [PubMed] [Google Scholar]
- Attia M, Stepper J, Davies GJ, Brumer H. Functional and structural characterization of a potent GH74 endo-xyloglucanase from the soil saprophyte Cellvibrio japonicus unravels the first step of xyloglucan degradation. FEBS J. 2016;283:1701–1719. doi: 10.1111/febs.13696. [DOI] [PubMed] [Google Scholar]
- Attia MA, Brumer H. Recent structural insights into the enzymology of the ubiquitous plant cell wall glycan xyloglucan. Curr Opin Struct Biol. 2016;40:43–53. doi: 10.1016/j.sbi.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Bhatia Y, Mishra S, Bisaria VS. Purification and characterization of recombinant Escherichia coli-expressed Pichia etchellsii beta-glucosidase II with high hydrolytic activity on sophorose. Applied Microbiology and Biotechnology. 2005;66:527–535. doi: 10.1007/s00253-004-1754-8. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, 3rd, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered. Escherichia coli. PNAS. 2011;108:19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones G, De Iannino NI, Roset M, Vigliocco A, Paul PS, Ugalde RA. Brucella abortus cyclic beta-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infection and Immunity. 2001;69:4528–4535. doi: 10.1128/IAI.69.7.4528-4535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg TD, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bioproduct formation. Current Opinion in Biotechnology. 2011;22:394–400. doi: 10.1016/j.copbio.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Bull AT, Chesters CG. The biochemistry of laminarin and the nature of laminarinase. Adv Enzymol Relat Areas Mol Biol. 1966;28:325–364. doi: 10.1002/9780470122730.ch5. [DOI] [PubMed] [Google Scholar]
- Carpita N, McCann M. The Cell Wall. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and Molecular Biology of Plants. Somerset, NJ: John Wiley & Sons; 2000. pp. 55–108. [Google Scholar]
- Cartmell A, McKee LS, Pena MJ, Larsbrink J, Brumer H, Kaneko S, et al. The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J Biol Chem. 2011;286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LDS, Pedersoli WR, Antonio ACC, Steindorff AS, Silva-Rocha R, Martinez-Rossi NM, et al. Comparative metabolism of cellulose, sophorose and glucose in Trichoderma reesei using high-throughput genomic and proteomic analyses. Biotechnology for Biofuels. 2014;7 doi: 10.1186/1754-6834-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chundawat SPS, Beckham GT, Himmel ME, Dale BE. Deconstruction of Lignocellulosic Biomass to Fuels and Chemicals. Annual Review of Chemical and Biomolecular Engineering, Vol 2. 2011;2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- Deboy RT, Mongodin EF, Fouts DE, Tailford LE, Khouri H, Emerson JB, et al. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. Journal of Bacteriology. 2008;190:5455–5463. doi: 10.1128/JB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nature Reviews Microbiology. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Ellinger D, Voigt CA. Callose biosynthesis in arabidopsis with a focus on pathogen response: what we have learned within the last decade. Annals of Botany. 2014;114:1349–1358. doi: 10.1093/aob/mcu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Su W, Rahat-Rozenbloom S, Wolever TMS, Comelli EM. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutrition and Diabetes. 2014;4:e121. doi: 10.1038/nutd.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB. Exploring the evolution of (1,3;1,4)-beta-D-glucans in plant cell walls: comparative genomics can help! Current Opinion in Plant Biology. 2009;12:140–147. doi: 10.1016/j.pbi.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Forsberg Z, Nelson CE, Dalhus B, Mekasha S, Loose JS, Crouch LI, et al. Structural and Functional Analysis of a Lytic Polysaccharide Monooxygenase Important for Efficient Utilization of Chitin in Cellvibrio japonicus. J Biol Chem. 2016;291:7300–7312. doi: 10.1074/jbc.M115.700161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG. Polysaccharide degradation systems of the saprophytic bacterium Cellvibrio japonicus. World Journal of Microbiology & Biotechnology. 2016;32:121. doi: 10.1007/s11274-016-2068-6. [DOI] [PubMed] [Google Scholar]
- Gardner JG, Keating DH. Requirement of the type II secretion system for utilization of cellulosic substrates by Cellvibrio japonicus. Appl Environ Microbiol. 2010a;76:5079–5087. doi: 10.1128/AEM.00454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG, Keating DH. Requirement of the Type II Secretion System for Utilization of Cellulosic Substrates by Cellvibrio japonicus. Applied and Environmental Microbiology. 2010b;76:5079–5087. doi: 10.1128/AEM.00454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG, Keating DH. Genetic and functional genomic approaches for the study of plant cell wall degradation in Cellvibrio japonicus. Methods in Enzymology. 2012;510:331–347. doi: 10.1016/B978-0-12-415931-0.00018-5. [DOI] [PubMed] [Google Scholar]
- Gardner JG, Crouch L, Labourel A, Forsberg Z, Bukhman YV, Vaaje-Kolstad G, et al. Systems biology defines the biological significance of redox-active proteins during cellulose degradation in an aerobic bacterium. Mol Microbiol. 2014 doi: 10.1111/mmi.12821. [DOI] [PubMed] [Google Scholar]
- Greffe L, Bessueille L, Bulone V, Brumer H. Synthesis, preliminary characterization, and application of novel surfactants from highly branched xyloglucan oligosaccharides. Glycobiology. 2005;15:437–445. doi: 10.1093/glycob/cwi013. [DOI] [PubMed] [Google Scholar]
- Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. Polysaccharide Utilization Loci: Fuelling microbial communities. J Bacteriol. 2017 doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlewood GP, Gilbert HJ. Structure and function analysis of Pseudomonas plant cell wall hydrolases. Biochemical Society Transactions. 1998;26:185–190. doi: 10.1042/bst0260185. [DOI] [PubMed] [Google Scholar]
- Hemsworth GR, Thompson AJ, Stepper J, Sobala LF, Coyle T, Larsbrink J, et al. Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut. Open Biol. 2016;6 doi: 10.1098/rsob.160142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Teeri TT, Warren RAJ. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. Febs Letters. 1998;425:352–354. doi: 10.1016/s0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- Huang TT, Wages JM. New-to-nature sophorose analog: a potent inducer for gene expression in Trichoderma reesei. Enzyme Microb Technol. 2016;85:44–50. doi: 10.1016/j.enzmictec.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Ibatullin FM, Baumann MJ, Greffe L, Brumer H. Kinetic analyses of retaining endo-(xylo)glucanases from plant and microbial sources using new chromogenic xylogluco-oligosaccharide aryl glycosides. Biochemistry. 2008;47:7762–7769. doi: 10.1021/bi8009168. [DOI] [PubMed] [Google Scholar]
- Jabbour D, Borrusch MS, Banerjee G, Walton JD. Enhancement of fermentable sugar yields by alpha-xylosidase supplementation of commercial cellulases. Biotechnology for Biofuels. 2013;6:58. doi: 10.1186/1754-6834-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Bowman MJ, Braker JD, Dien BD, Hector RE, Lee CC. Plant cell walls to ethanol. Biochem, J. 2012;424:241–252. doi: 10.1042/BJ20111922. [DOI] [PubMed] [Google Scholar]
- Kollar R, Reinhold BB, Petrakova E, Yeh HJC, Ashwell G, Drgonova J, et al. Architecture of the yeast cell wall: beta(1–6)-glucan interconnects mannoprotein, beta(1–3)-glucan, and chitin. Journal of Biological Chemistry. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- Krajmalnik-Brown R, Ilhan Z, Kang D, DiBaise JK. Effects of Gut Microbiomes on Nutrient Absorption and Energy Regulation. Nutr Clin Pract. 2012;27:201–214. doi: 10.1177/0884533611436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath BJ, Bremges A, Weimann A, McHardy AC, Pope PB. Metagenomics and CAZyme Discovery. Methods Mol Biol. 2017;1588:255–277. doi: 10.1007/978-1-4939-6899-2_20. [DOI] [PubMed] [Google Scholar]
- Larsbrink J, Thompson AJ, Lundqvist M, Gardner JG, Davies GJ, Brumer H. A complex gene locus enables xyloglucan utilization in the model saprophyte Cellvibrio japonicus. Mol Microbiol. 2014a;94:418–433. doi: 10.1111/mmi.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsbrink J, Izumi A, Ibatullin FM, Nakhai A, Gilbert HJ, Davies GJ, Brumer H. Structural and enzymatic characterization of a glycoside hydrolase family 31 alpha-xylosidase from Cellvibrio japonicus involved in xyloglucan saccharification. Biochemical Journal. 2011;436:567–580. doi: 10.1042/BJ20110299. [DOI] [PubMed] [Google Scholar]
- Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014b;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Kelly AG, Tauzin AS, Brumer H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. 2014;426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medie FM, Davies GJ, Drancourt M, Henrissat B. Genome analyses highlight the different biological roles of cellulases. Nature Reviews Microbiology. 2012;10:227-U. doi: 10.1038/nrmicro2729. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Seshadri R, Varghese NJ, Eloe-Fadrosh EA, Meier-Kolthoff JP, Goker M, et al. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nature Biotechnology. 2017;35:676-+. doi: 10.1038/nbt.3886. [DOI] [PubMed] [Google Scholar]
- Naas AE, Mackenzie AK, Mravec J, Schuckel J, Willats WG, Eijsink VG, Pope PB. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? MBio. 2014;5:e01401–01414. doi: 10.1128/mBio.01401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Gardner JG. In-frame deletions allow functional characterization of complex cellulose degradation phenotypes in Cellvibrio japonicus. Applied and Environmental Microbiology. 2015;81:5968–5975. doi: 10.1128/AEM.00847-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Beri NR, Gardner JG. Custom fabrication of biomass containment devices using 3-D printing enables bacterial growth analyses with complex insoluble substrates. J Microbiol Methods. 2016;130:136–143. doi: 10.1016/j.mimet.2016.09.013. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Rogowski A, Morland C, Wilhide JA, Gilbert HJ, Gardner JG. Systems analysis in Cellvibrio japonicus resolves predicted redundancy of beta-glucosidases and determines essential physiological functions. Molecular Microbiology. 2017;104:294–305. doi: 10.1111/mmi.13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. Signal peptidases. Chemical Reviews. 2002;102:4549–4579. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- Pauly M, Albersheim P, Darvill A, York WS. Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant Journal. 1999;20:629–639. doi: 10.1046/j.1365-313x.1999.00630.x. [DOI] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Herve C, Domozych DS, Willats WGT, Tuohy MG, et al. Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants. Annual Review of Plant Biology, Vol 62. 2011;62:567–588. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- Rahman O, Cummings SP, Harrington DJ, Sutcliffe IC. Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World Journal of Microbiology & Biotechnology. 2008;24:2377–2382. [Google Scholar]
- Reintjes G, Arnosti C, Fuchs BM, Amann R. An alternative polysaccharide uptake mechanism of marine bacteria. Isme Journal. 2017;11:1640–1650. doi: 10.1038/ismej.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigano LA, Payette C, Brouillard G, Marano MR, Abramowicz L, Torres PS, et al. Bacterial cyclic beta-(1,2)-glucan acts in systemic suppression of plant immune responses. Plant Cell. 2007;19:2077–2089. doi: 10.1105/tpc.106.047944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon JE, Ferreira LM, Durrant AJ, Laurie JI, Hazlewood GP, Gilbert HJ. Characterization of the gene celD and its encoded product 1,4-beta-D-glucan glucohydrolase D from Pseudomonas fluorescens subsp. cellulosa. Biochem J. 1992;285(Pt 3):947–955. doi: 10.1042/bj2850947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annual Review of Plant Biology. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- Schultink A, Liu L, Zhu L, Pauly M. Structural diversity and function of xyloglucan sidechain substituents. Plants. 2014;3:526–542. doi: 10.3390/plants3040526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A, Larsbrink J, Marchetti R, Lanzetta R, Brumer H, Molinaro A. NMR Spectroscopic analysis reveals extensive binding interactions of complex xyloglucan oligosaccharides with the Cellvibrio japonicus glycoside hydrolase family 31 α-xylosidase. Chemistry-A European Journal. 2012;18:13395–13404. doi: 10.1002/chem.201200488. [DOI] [PubMed] [Google Scholar]
- Sugisawa H, Edo H. Thermal Degradation of Sugars.I. Thermal Polymerization of Glucose. Journal of Food Science. 1966;31:561-&. [Google Scholar]
- Tchieu JH, Norris V, Edwards JS, Saier MH. The complete phosphotransferase system in Escherichia coli. Journal of Molecular Microbiology and Biotechnology. 2001;3:329–346. [PubMed] [Google Scholar]
- Tuomivaara ST, Yaoi K, O'Neill MA, York WS. Generation and structural validation of a library of diverse xyloglucan-derived oligosaccharides, including an update on xyloglucan nomenclature. Carbohydrate Research. 2015;402:56–66. doi: 10.1016/j.carres.2014.06.031. [DOI] [PubMed] [Google Scholar]
- Uchiyama T, Yaoi K, Miyazaki K. Glucose-tolerant beta-glucosidase retrieved from a Kusaya gravy metagenome. Frontiers in Microbiology. 2015;6 doi: 10.3389/fmicb.2015.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Freeze HH. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Plainview, NY: 2009. e.a. [PubMed] [Google Scholar]
- Vogel J. Unique aspects of the grass cell wall. Current Opinion in Plant Biology. 2008;11:301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Xie ZZ, Lin WT, Luo JF. Genome sequence of Cellvibrio pealriver PR1, a xylanolytic and agarolytic bacterium isolated from freshwater. Journal of Biotechnology. 2015;214:57–58. doi: 10.1016/j.jbiotec.2015.07.021. [DOI] [PubMed] [Google Scholar]
- Yamane K, Suzuki H, Hirotani M, Ozawa H, Nisizawa K. Effect of nature and supply of carbon sources on cellulase formation in Pseudomonas fluorescens var. cellulosa. J Biochem. 1970;67:9–18. doi: 10.1093/oxfordjournals.jbchem.a129238. [DOI] [PubMed] [Google Scholar]
- Zavaliev R, Ueki S, Epel BL, Citovsky V. Biology of callose (beta-1,3-glucan) turnover at plasmodesmata. Protoplasma. 2011;248:117–130. doi: 10.1007/s00709-010-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Edgar KJ. Properties, chemistry, and applications of the bioactive polysaccharide curdlan. Biomacromolecules. 2014;15:1079–1096. doi: 10.1021/bm500038g. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogowski A, Zhao L, MG H, Avci U, Knox JP, Gilber HJ. Understanding How Complex Molecular Architecture of Mannan-degrading Hydrolysis Contributes to Plant Cell Wall Degradation. Journal of Biological Chemistry. 2002;289:2002–2012. doi: 10.1074/jbc.M113.527770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.