Abstract
Treatment switching in a randomised controlled trial occurs when participants change from their randomised treatment to the other trial treatment during the study. Failure to account for treatment switching in the analysis (i.e. by performing a standard intention-to-treat analysis) can lead to biased estimates of treatment efficacy. The rank preserving structural failure time model (RPSFTM) is a method used to adjust for treatment switching in trials with survival outcomes. The RPSFTM is due to Robins and Tsiatis (1991) and has been developed by White et al. (1997, 1999).
The method is randomisation based and uses only the randomised treatment group, observed event times, and treatment history in order to estimate a causal treatment effect. The treatment effect, ψ, is estimated by balancing counter-factual event times (that would be observed if no treatment were received) between treatment groups. G-estimation is used to find the value of ψ such that a test statistic Z(ψ) = 0. This is usually the test statistic used in the intention-to-treat analysis, for example, the log rank test statistic.
We present an R package that implements the method of rpsftm.
Introduction
In a two-arm randomised controlled trial, participants are randomly allocated to receive one of two treatments or interventions. Ideally, all participants would fully receive their allocated treatment and no other treatment. However, in a recent review, 98 of 100 trials published in four high quality general medical journals reported some form of departure from randomised treatment (Dodd et al., 2012). When treatment is ‘all-or-nothing’ (for example, a surgical procedure), possible departures include not receiving the allocated treatment, receiving the other trial treatment, or receiving a non-trial treatment. When treatment is given over time (for example, a drug for HIV treatment), departures also occur over time, and often include starting a new treatment in response to a disease-related event.
This paper specifically focuses on the case of treatment switching over time, where participants may switch to receive the other trial treatment during the trial (Latimer et al., 2014). A common example is in a trial of active treatment versus placebo where placebo arm participants may start the active treatment in response to disease progression.
A randomised controlled trial with departures from randomised treatment is commonly analysed by intention-to-treat, in which departures from randomised treatment are ignored and all randomised participants are compared in the groups to which they were randomised (Higgins et al., 2011). This provides an unbiased comparison of two treatment policies, which accept that treatment may be changed, but does not compare the efficacies of the treatments themselves (White et al., 1999).
In order to compare the efficacies of the treatments, analysts often use per-protocol analysis, which censors participants when they depart from randomised treatment. However, this loses the advantage that randomisation produces comparable groups and instead introduces selection bias. Another possibility is to include treatment as a time-dependent variable in a Cox regression model. The issue with this method is that the estimate of treatment effect may not have a causal interpretation when switchers are prognostically different to non-switchers (Robins and Tsiatis, 1991).
There has therefore been strong interest in causal inference methods including marginal structural models (MSM) (i.e. inverse probability of censoring weighting (IPCW)) (Hernán et al., 2001) and instrumental-variable-type methods (White, 2005; Watkins et al., 2013).
The IPCW method is an extension of the per-protocol censoring approach, whereby the bias associated with censoring participants that depart from randomised treatment is removed by weighting the remaining non-switchers (Latimer et al., 2016). A model is formed for the probability of switching using baseline and time-dependent covariates. Time-varying weights are then obtained for each participant based on the inverse probability of not switching until a given time (Watkins et al., 2013). The method relies on data on prognostic factors for mortality that also affect the probability of switching being collected at both baseline and over time. This is called the ‘no unmeasured confounders’ assumption. Since this assumption is untestable, the method relies on good planning at the study design stage in order to collect information on all possible confounders. The method also has two potential issues: a) if many prognostic factors are included in the model to calculate weights the model may fail to converge, and b) if only a few participants switch then large weights are assigned to the remaining participants, which may result in biased analyses. The method has already been implemented in R in the package ipw and so is not considered further here.
Instead, this paper focuses on a popular causal method - the rank preserving structural failure time model (RPSFTM) (Robins and Tsiatis, 1991). In contrast to the IPCW method which requires potential confounders to be collected over time, the RPSFTM is randomisation based and only requires information on the randomised treatment group, observed event times, and treatment history in order to estimate a causal treatment effect. The method has been used, for example, in submissions to the UK’s National Institute for Health and Clinical Excellence whose Decision Support Unit commissioned a guidance document (Latimer and Abrams, 2014). The RPSFTM has been implemented in Stata (White et al., 2002) but not in R.
This paper describes the theory of the RPSFTM and presents a new implementation of the method in R, using data based on a trial in HIV infection.
Theory
RPSFTM method and assumptions
The RPSFTM uses a causal model to produce counter-factual event times (the time that would be observed if no treatment were received) in order to estimate a causal treatment effect. Let be the observed event time for participant i, where and represent the time spent off and on treatment, respectively. The Ti are related to the counter-factual event times, Ui, via the following causal model:
| (1) |
where exp(-ψ0) is the acceleration factor associated with treatment and ψ0 is the true causal parameter.
A grid search (g-estimation) procedure is used to estimate the treatment effect that balances the counter-factual event times across randomised treatment groups. To estimate the causal treatment effect, ψ, we assume that the Ui are independent of randomised treatment group Ri, i.e. if the groups are similar with respect to all other characteristics except treatment, the average event times should be the same in each group if no participant were treated. In general, this assumption is plausible in a randomised controlled trial.
A g-estimation procedure is used to find the value of ψ such that Ui is independent of Ri. For a range of ψs, the hypothesis ψ0 = ψ is tested by computing Ui(ψ), subject to censoring, and calculating the test statistic Z(ψ). This is usually the same test statistic as for the intention-to-treat analysis, for example, the log rank test statistic to compare survival distributions between groups. In the rpsftm() function, the possible test options are the log rank, and the Wald test from a Cox or Weibull regression model. For the parametric Weibull test, the point estimate (ψ̂) is the value of ψ for which Z(ψ) = 0. For the non-parametric tests (log rank, Cox), ψ̂ is the value of ψ for which Z(ψ) crosses 0, since Z(ψ) is a step function. Confidence intervals are similarly found with the 100(1 − α)% confidence interval being the set {ψ : |Z(ψ)| < z1−α/2}, where z1−α/2 is the 1 − α/2 percentile of the standard normal distribution.
After finding ψ̂, an adjusted hazard ratio can be calculated. For example, by comparing counter-factual event times at ψ̂ in the control group to the observed event times in the experimental group we can estimate the treatment effect that would have been observed in the absence of switching (in the case where only the control group switch).
As well as assuming that the only difference between randomised groups is the treatment received, the RPSFTM also assumes a ’common treatment effect’. The common treatment effect assumption states that the treatment effect is the same for all participants (with respect to time spent on treatment) regardless of when treatment is received, which is implicit in equation (1).
Re-censoring
We assume that censoring occurs if participants survive to a specific calendar date (Robins and Tsiatis, 1991). Thus the potential censoring time Ci is known for all participants. The censoring indicators of the observed event times are initially carried over to the counter-factual event times. However, the uninformative censoring on the Ti scale may be informative on the Ui scale. Suppose we have two participants with the same Ui, one of whom receives the superior treatment. The participant receiving the superior treatment has their Ui extended so that they are censored whilst the other participant may observe the event.
In detail, Ci, the censoring times for the Ti, are transformed to Ci (1 − Pi + Pi exp(ψ)) when we work on the Ui scale, where Pi is a random variable, the proportion of time on treatment, This transformation of censoring times occurs by representing the censoring status in two equivalent ways: {Ci < Ti}, and re-scaling both sides {Ci (1 − Pi + Pi exp(ψ)) < Ti (1 − Pi + Pi exp(ψ)) = Ui}. We cannot assume the variable Pi is independent of Ui. For example, adherence to a protocolised treatment may depend on the underlying prognosis.
We can overcome this induced dependence by considering the sample space for Pi marginally over Ui and, optionally, conditioning on Ri. If we replace Pi with a function of its sample space then any dependency on Ui is thus removed. Any alternative transformed censoring times must be smaller than the original censoring times, else we may have to impute uncensored Ui values for censored Ti observations, which would be impossible. Hence taking the minimum value from the sample space for Pi meets both desiderata. If the sample space differs between arms, say switching is impossible in one arm, then this can be utilised to potentially observe more events with gains in efficiency.
Operationally, let Ci be the potential censoring time for participant i. A participant is then recensored at the minimum possible censoring time:
If then Ui is replaced by and the censoring indicator is replaced by 0. For treatment arms where switching does not occur, there can be no informative censoring and so re-censoring is not applied.
Sensitivity analysis
As previously mentioned, the RPSFTM has two assumptions:
The only difference between randomised groups is the treatment received.
The treatment effect is the same for all participants regardless of when treatment is received.
Whilst the first assumption is plausible in a randomised controlled trial, the latter may be unlikely to hold. For example, if control group participants can only switch at disease progression then the treatment benefit may be different in these participants compared to those randomised to the experimental treatment. The rpsftm() function allows for investigation of deviations from the common treatment effect assumption by featuring a treatment-effect modifier variable which means the treatment effect can be varied across participants. The assumption that defines the counter-factual treatment-free event times Ui is thus modified by multiplying ψ by some factor ki > 0:
The value taken by ki is derived from observed data and is left to the user to assign.
The package assumes that ki is determined at baseline, and re-censoring is undertaken in a similar way by re-censoring at the minimum possible censoring time:
Again, if then Ui is replaced by and the censoring indicator is replaced by 0. The case where ki is observed post randomisation is equally valid in terms of defining Ui, but more complicated re-censoring may be required, taking the minimum value of over the sample space of ki conditional on arm. This is not implemented in the package.
Using package rpsftm
The main function in the package rpsftm is of the same name, rpsftm(), which returns an object that has print, summary, and plot methods. The arguments to rpsftm() are given in table 1 below.
Table 1.
Arguments for the rpsftm() function
| rpsftm() arguments | |
|---|---|
| formula | a formula with a minimal structure of Surv(time,status) ~rand(arm,rx) where
|
| Further terms can be added to the right hand side to adjust for covariates. | |
| data | an optional data frame containing the variables |
| censor_time | variable or constant giving the time at which censoring would, or has occurred. This should be provided for all observations unlike standard Kaplan-Meier or Cox regression where it is only given for censored observations. If no value is given then re-censoring is not applied. |
| subset | an expression indicating which subset of the rows of data should be used in the fit. This can be a logical vector, a numeric vector indicating which observation numbers are to be included, or a character vector of row names to be included. All observations are included by default. |
| na.action | a missing-data filter function. This is applied to the model.frame after any subset argument has been used. Default is options()$na.action. |
| test | one of survdiff, coxph or survreg. Describes the test to be used in the estimating equation. Default is survdiff. |
| low_psi | the lower limit of the range to search for the causal parameter. Default is −1. |
| hi_psi | the upper limit of the range to search for the causal parameter. Default is 1. |
| alpha | the significance level used to calculate the confidence intervals. Default is 0.05. |
| treat_modifier | an optional variable that ψ is multiplied by on an participant observation level to give differing impact to treatment. Default is 1. |
| autoswitch | a logical to autodetect cases of no switching. Default is TRUE. If all observations in an arm have perfect compliance then re-censoring is not applied in that arm. If FALSE then re-censoring is applied regardless of perfect compliance. |
| n_eval_z | The number of points between hi_psi and low_psi at which to evaluate the Z-statistics in the estimating equation. Default is 100. |
Example
The rpsftm function will be illustrated using a simulated dataset immdef taken from (White et al., 2002), based on a randomized controlled trial (Concorde Coordinating Committee, 1994). The trial compares two policies (immediate or deferred treatment) of zidovudine treatment in symptom free participants infected with HIV. The immediate treatment arm received treatment at randomisation whilst the deferred arm received treatment either at onset of AIDS related complex or AIDS (CDC group IV disease) or development of persistently low CD4 count. The endpoint considered here was time from study entry to progression to AIDS, or CDC group IV disease, or death.
Data
The immdef data frame has 1000 rows of 9 variables as described in table 2
Table 2.
Description of simulated data set
| immdef variables | |
|---|---|
| id | participant ID number |
| def | indicator that the participant was assigned to the Deferred treatment arm |
| imm | indicator that the participant was assigned to the Immediate treatment arm |
| censyrs | censoring time, in years, corresponding to the close of study minus the time of entry for each participant |
| xo | an indicator that switching occurred |
| xoyrs | the time, in years, from entry to switching, or 0 for participants in the Immediate arm |
| prog | an indicator of disease progression (1), or censoring (0) |
| progyrs | time, in years, from entry to disease progression or censoring |
| entry | the time of entry into the study, measured in years from the date of randomisation |
The first six observations are given below
> library(rpsftm) > head(immdef) id def imm censyrs xo xoyrs prog progyrs entry 1 1 0 1 3 0 0.000000 0 3.000000 0 2 2 1 0 3 1 2.652797 0 3.000000 0 3 3 0 1 3 0 0.000000 1 1.737838 0 4 4 0 1 3 0 0.000000 1 2.166291 0 5 5 1 0 3 1 2.122100 1 2.884646 0 6 6 1 0 3 1 0.557392 0 3.000000 0
For example, participant 2 was randomised to the deferred arm, started treatment at 2.65 years and was censored at 3 years (the end of the study). Subject 3 was randomised to the immediate treatment arm and progressed (observed the event) at 1.74 years. Subject 5 was randomised to the deferred treatment arm, started treatment at 2.12 years and progressed at 2.88 years. The trial lasted 3 years with staggered entry over the first 1.5 years. The variable censyrs gives the time from entry to the end of the trial.
Intention-to-treat analysis
First, we estimate the effect of giving zidovudine ignoring any treatment changes during the trial. This is found by fitting an accelerated failure time model using the eha package:
> library(eha) > itt_fit <- aftreg(Surv(progyrs, prog) ~ imm, data = immdef) > itt_fit Call: aftreg(formula = Surv(progyrs, prog) ~ imm, data = immdef) Covariate W.mean Coef Time-Accn se(Coef) Wald p imm 0.510 -0.147 0.863 0.077 0.056 Baseline parameters: log(scale) 1.404 0.062 0.000 log(shape) 0.392 0.052 0.000 Baseline life expectancy: Events 312 Total time at risk 1932.5 Max. log. likelihood -855.14 LR test statistic 3.69 Degrees of freedom 1 Overall p-value 0.0548817
The intention-to-treat estimate is −0.147 which means that lifetime is used up exp(−0.147) = 0.863 times as fast when on zidovudine as when off zidovudine (or zidovudine extends lifetime by a factor of exp(0.147) = 1.16).
Fitting the RPSFTM
We now show how to use rpsftm with the immdef data. First, a variable rx for the proportion of time spent on treatment must be created:
rx <- with(immdef, 1-xoyrs/progyrs)
This sets rx to 1 in the immediate treatment arm (since no participants could switch to the deferred arm, and xoyrs=0 in such participants), 0 in the deferred arm participants that did not receive treatment (as xoyrs=progyrs in such participants) and 1-xoyrs/progyrs in the deferred arm participants that did receive treatment. Using the default options, the fitted model is
rpsftm_fit_lr <- rpsftm(formula=Surv(progyrs, prog) ~ rand(imm, rx),
data=immdef,
censor_time=censyrs)
The above formula fits an RPSFTM where progyrs is the observed event time, prog is the indicator of disease progression, imm is the randomised treatment group indicator, rx is the proportion of time spent on treatment and censyrs is the censoring time. The log rank test is used in finding the point estimate of ψ̂. Re-censoring is performed since the censor_time parameter is specified; if not specified then re-censoring would not be performed. After finding ψ̂, rpsftm refits the model at ψ̂ and produces a survdiff object of the counter-factual event times to be used in plotting Kaplan-Meier curves. The list of objects that the function returns is given in table 3.
Table 3.
Outputs from the rpsftm() function
| rpsftm() outputs | |
|---|---|
| psi | the estimated parameter |
| fit | a survdiff object to produce Kaplan-Meier curves of the estimated counter-factual event times in each treatment arm using plot() |
| CI | a vector of the confidence interval around psi |
| Sstar | the (possibly) re-censored Surv() data using the estimate value of psi to give counterfactual untreated failure times. |
| rand | the rand() object used to specify the allocated and observed amount of treatment. |
| ans | the values from uniroot.all used to solve the estimating equation, but embedded within a list as per uniroot, with an extra element root_all, a vector of all roots found in the case of multiple solutions. The first element of root_all is subsequently used. |
| eval_z | a data frame with the Z-statistics from the estimating equation evaluated at a sequence of values of psi. Used to plot and check if the range of values to search for solution and limits of confidence intervals need to be modified. Further elements corresponding to either a survdiff, coxph, or survreg object. This will always include: |
| call | the R call object |
| formula | a formula representing any adjustments, strata or clusters - used for the update() function |
| terms | a more detailed representation of the model formula |
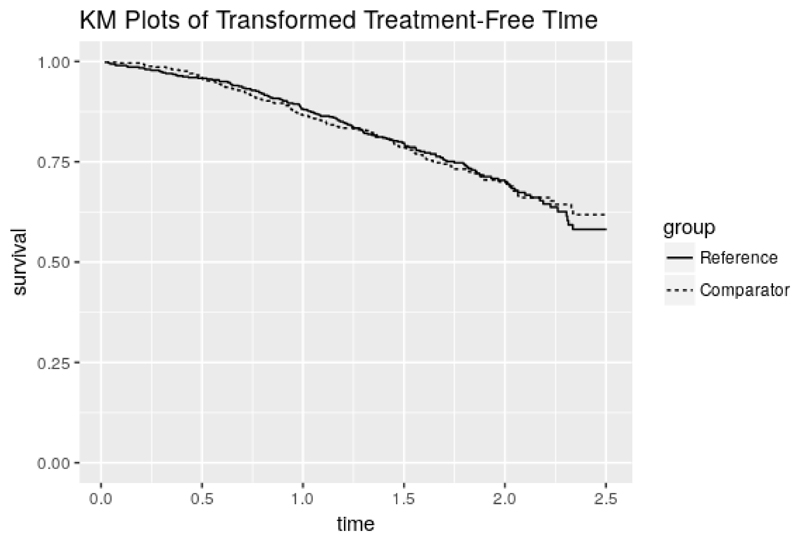
The point estimate and 95% confidence interval (CI) can be returned using rpsftm_fit_lr$psi and rpsftm_fit_lr$CI which gives ψ̂ = –0.181(–0.350, 0.002), slightly larger than in the intention-to-treat analysis. This means that lifetime is used up exp(–0.181) = 0.834 times as fast when on zidovudine as when off zidovudine, i.e. the time to progression to AIDS, or CDC group IV disease, or death is longer when on zidovudine (treatment is beneficial). However, the confidence interval contains zero, which suggests the treatment effect is non-significant. The function plot() produces Kaplan-Meier curves of the counter-factual event times in each group and can be used to check that the distributions are indeed the same at ψ̂ as shown in figure 1
Figure 1.
Output from plot(rpsftm_fit_lr)
We now provide examples of using the Cox regression model and the Weibull model in place of the log rank test. To use the Wald test from a Cox regression model, we specify test=coxph in the function parameters. Covariates can also be included in the estimation procedure by adding them to the right hand side of the formula. For example, baseline covariates that are included in the intention-to-treat analysis may also be incorporated into the estimation procedure of the RPSFTM. In the following example we add entry time as a covariate and use summary() to find the value of ψ̂ and its 95% CI.
> rpsftm_fit_cph <- rpsftm(formula=Surv(progyrs, prog) ~ rand(imm, rx) + entry,
+ data=immdef,
+ censor_time=censyrs,
+ test=coxph)
> summary(rpsftm_fit_cph)
arm rx.Min. rx.1st Qu. rx.Median rx.Mean rx.3rd Qu. rx.Max.
1 0 0.0000000 0.0000000 0.0000000 0.1574062 0.2547779 0.9770941
2 1 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000
n= 1000, number of events= 286
coef exp(coef) se(coef) z Pr(>|z|)
entry 0.1235 1.1315 0.1487 0.831 0.406
exp(coef) exp(-coef) lower .95 upper .95
entry 1.131 0.8838 0.8454 1.514
Concordance= 0.514 (se = 0.018)
Rsquare= 0.001 (max possible= 0.976)
psi: -0.1811697
exp(psi): 0.8342938
Confidence Interval, psi -0.3496874 0.003370267
Confidence Interval, exp(psi) 0.7049084 1.003376
For the Weibull model we specify test=survreg in the function parameters. :
> rpsftm_fit_wb <- rpsftm(formula=Surv(progyrs, prog) ~ rand(imm, rx) + entry,
+ data=immdef,
+ censor_time=censyrs,
+ test=survreg)
> summary(rpsftm_fit_wb)
arm rx.Min. rx.1st Qu. rx.Median rx.Mean rx.3rd Qu. rx.Max.
1 0 0.0000000 0.0000000 0.0000000 0.1574062 0.2547779 0.9770941
2 1 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000
Call:
rpsftm(formula = Surv(progyrs, prog) ~ rand(imm, rx) + entry,
data = immdef, censor_time = censyrs, test = survreg)
Value Std. Error z p
(Intercept) 1.3881 0.0857 16.197 5.34e-59
entry -0.0582 0.0906 -0.642 5.21e-01
Log(scale) -0.4176 0.0568 -7.349 2.00e-13
Scale= 0.659
Weibull distribution
Loglik(model)= -759.8 Loglik(intercept only)= -760
Number of Newton-Raphson Iterations: 6
n= 1000
psi: -0.1811851
exp(psi): 0.8342809
Confidence Interval, psi -0.3501459 0.005170935
Confidence Interval, exp(psi) 0.7045852 1.005184
As we can see from the output for the Cox and Weibull models, the estimates of ψ are similar to the estimate obtained from using the log rank test. Both fitted models could be used as inputs to the plot() function, which produce figures very similar to figure 1.
As a sensitivity analysis we can investigate what would happen to the estimate of ψ if the treatment effect in the deferred treatment group was half of that in the immediate treatment group by setting ki = 1 for participants in the latter group and ki = 0.5 for participants in the former group.
> weight <- with(immdef, ifelse(imm==1, 1, 0.5))
> rpsftm(Surv(progyrs, prog) ~ rand(imm, rx), data = immdef, censor_time = censyrs,
+ treat_modifier = weight
+ )
arm rx.Min. rx.1st Qu. rx.Median rx.Mean rx.3rd Qu. rx.Max.
1 0 0.0000000 0.0000000 0.0000000 0.1574062 0.2547779 0.9770941
2 1 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000
Call:
rpsftm(formula = Surv(progyrs, prog) ~ rand(imm, rx), data = immdef,
censor_time = censyrs, treat_modifier = weight)
N Observed Expected (O-E)^2/E (O-E)^2/V
.arm=0 500 157 157 5.75e-07 1.22e-06
.arm=1 500 143 143 6.31e-07 1.22e-06
Chisq= 0 on 1 degrees of freedom, p= 0.999
psi: -0.1704745
exp(psi): 0.8432646
In this case the estimate of treatment effect is reduced slightly to −0.170 and lifetime is used up 0.843 times as fast when on treatment as when off treatment.
Trouble Shooting
There is no guarantee that unique solutions exist to the estimating equations for the estimate and confidence interval limits, or that we have searched a wide enough interval to find them if they do exist and are unique.
There are three instances where rpsftm will produce warning messages due to the search interval. The function first evaluates Z(ψ) at low_psi and hi_psi and will produce a warning message if Z(ψ) is the same sign at these two points and no root exists within the interval.
Warning messages: 1: In rpsftm(formula = Surv(progyrs, prog) ~ rand(imm, rx), data = immdef, : The starting interval (-1, -0.9) to search for a solution for psi gives values of the same sign (7.53, 6.88). Try a wider interval. plot(obj$eval_z, type="s"), where obj is the output of rpsftm() 2: In rpsftm(formula = Surv(progyrs, prog) ~ rand(imm, rx), data = immdef, : Evaluation of the estimated values of psi failed. It is set to NA 3: In rpsftm(formula = Surv(progyrs, prog) ~ rand(imm, rx), data = immdef, : Evaluation of a limit of the Confidence Interval failed. It is set to NA 4: In rpsftm(formula = Surv(progyrs, prog) ~ rand(imm, rx), data = immdef, : Evaluation of a limit of the Confidence Interval failed. It is set to NA
This suggests widening the search interval via trial and error until a root for ψ can be found between low_psi and hi_psi. The second warning message occurs when uniroot.all, the function used to solve the estimating equation for ψ̂ and its 95% confidence interval limits, fails to find any one of these. It will set the value to NA and produce the following warning message
> rpsftm_fit <- rpsftm(formula=Surv(progyrs, prog) ~ rand(imm, rx), + data=immdef, + censor_time=censyrs, + low_psi=-1, + hi_psi=-0.1) Warning message: In rpsftm(formula = Surv(progyrs, prog) ~ rand(imm, rx), data = immdef, : Evaluation of a limit of the Confidence Interval failed. It is set to NA
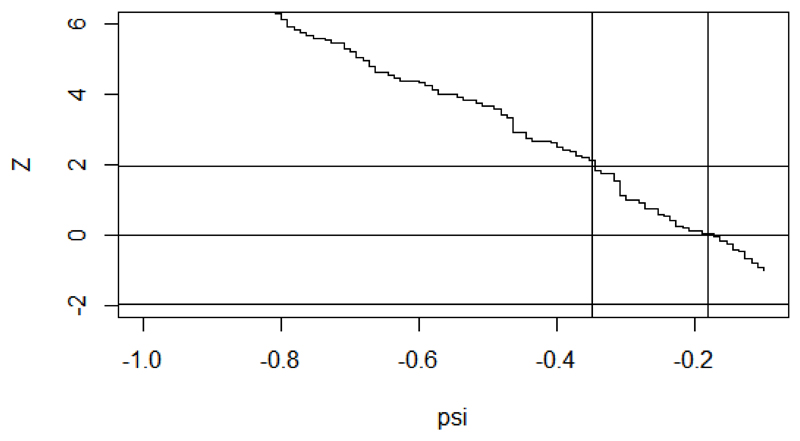
Investigation of a plot of Z(ψ) against ψ in figure 2 for a range of values of ψ could show why the function fails to find a root. The fitted object, rpsftm_fit, returns a data frame rpsftm_fit$eval_z with values of the Z-statistic evaluated at 100 points between the limits of the search interval. The data frame can be supplied as the argument to plot() to visualise the estimating equation.
Figure 2.
Plot of Z(ψ) against ψ to detect problems with the choice of search interval
plot(rpsftm_fit$eval_z, type="s", ylim=c(-2, 6)) abline(h=qnorm(c(0.025, 0.5, 0.975))) abline(v=rpsftm_fit$psi) abline(v=rpsftm_fit$CI)
In this case, we see that the search interval used was not wide enough to find the upper confidence limit. The third warning message occurs when multiple roots are found:
Warning message: In root(0) : Multiple Roots found
In order to show what happens when multiple roots are found, a subset of the immdef dataset was created and is stored within the test area of rpsftm. Whilst the function uniroot.all will return multiple roots, the function rpsftm will only display the first root found by uniroot.all. A plot of Z(ψ) against ψ can be used to find the other roots.
The limits of the confidence intervals may not exist at all if Z(·) reaches an asymptote before it hits the required quantile of the standard normal distribution, in which case the limits should be reported as ± ∞. This scenario would be illustrated by a similar plot to figure 2.
Another possibility is for the coxph function to fail to converge. This occurs when the maximum likelihood estimate of a coefficient is infinity, e.g. if one of the treatment groups has no events. The coxph documentation states that the Wald statistic should be ignored in this case and therefore the rpsftm output should be taken with caution.
Limitations
As well as the potential computational issues highlighted in the previous section, the RPSFTM itself has some limitations. The method relies on the assumption that the treatment effect is the same for all participants regardless of when treatment is received. We have allowed for investigation of deviations from this assumption within the rpsftm() function by adding a treatment-effect modifier variable. Another possible limitation of the model is its requirement for only the total amount of time spent on/off treatment. In instances where participants can switch back and forth between treatments the model may be inefficient.
Conclusion
The intention is to fill a void in the methodology available to R users by providing this package and thus facilitate the adoption of newer methods in application to real data in future. The sensitivity analyses, which generalise the definition of the counter-factual treatment-free event times, are an original development.
Further developments to the package may include expanding the functionality to allow multi-armed studies with three or more arms. Such an extension would have at its core a set of g-estimating equations to solve, one for each element of the vector of ψ parameters. Defining the syntax to capture the multi-dimensional metric of time on treatments is challenging, as will be the numerical computational details when the g-estimating equations are step functions.
Acknowledgement
Ian White was supported by the Medical Research Council Unit Programme MC_UU_12023/21. Simon Bond is a visiting worker at the MRC Biostatistics Unit, with primary affiliation to Cambridge Clinical Trials Unit.
Contributor Information
Annabel Allison, Medical Research Council Biostatistics Unit Forvie Site, Robinson Way, Cambridge, UK.
Ian R White, MRC Clinical Trials Unit at UCL 90 High Holborn, London, UK.
Simon Bond, Cambridge Clinical Trials Unit, Cambridge University Hospitals Foundation NHS Trust, Hills Rd, Cambridge UK.
Bibliography
- Concorde Coordinating Committee. Concorde: MRC/ANRS randomised double-blind controlled trial of immediate and deferred zidovudine in symptom-free HIV infection. Lancet. 1994;343:871–881. doi: 10.1016/S0140-6736(94)90006-X. [DOI] [PubMed] [Google Scholar]
- Dodd S, White IR, Williamson P. Nonadherence to treatment protocol in published randomised controlled trials: a review. Trials. 2012;13(1):84. doi: 10.1186/1745-6215-13-84. ISSN 1745-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernán MA, Brumback B, Robins JM. Marginal structural models to estimate the joint causal effect of nonrandomized treatments. Journal of the American Statistical Association. 2001;96(454):440–448. [Google Scholar]
- Higgins JP, Deeks JJ, Altman DG. Special Topics in Statistics. John Wiley & Sons, Ltd; Chichester, UK: 2011. ISBN 9780470712184. [DOI] [Google Scholar]
- Latimer NR, Abrams KR. Adjusting survival time estimates in the presence of treatment switching. Technical Report July, NICE DSU Technical Support Document. 2014;16 URL http://scharr.dept.shef.ac.uk/nicedsu/technical-support-documents/treatment-switching-tsd/ [PubMed] [Google Scholar]
- Latimer NR, Abrams K, Lambert P, Crowther M, Wailoo A, Morden J, Akehurst R, Campbell M. Adjusting for treatment switching in randomised controlled trials - A simulation study and a simplified two-stage method. Statistical Methods in Medical Research. 2014 doi: 10.1177/0962280214557578. ISSN 0962-2802. [DOI] [PubMed] [Google Scholar]
- Latimer NR, Henshall C, Siebert U, Bell H. Treatment switching: Statistical and decision-making challenges and approaches. International journal of technology assessment in health care. 2016;32(3):160–166. doi: 10.1017/S026646231600026X. [DOI] [PubMed] [Google Scholar]
- Robins J, Tsiatis A. Correcting for non-compliance in randomized trials using rank preserving structural failure time models. Communications in Statistics Theory and Methods. 1991;20:2609–2631. doi: 10.1080/03610929108830654. [DOI] [Google Scholar]
- Watkins C, Huang X, Latimer N, Tang Y, Wright EJ. Adjusting overall survival for treatment switches: commonly used methods and practical application. Pharmaceutical Statistics. 2013;12(6):348–357. doi: 10.1002/pst.1602. ISSN 1539-1612. [DOI] [PubMed] [Google Scholar]
- White I, Walker S, Babiker A, Darbyshire J. Impact of treatment changes on the interpretation of the Concorde trial. AIDS. 1997;11:999–1006. doi: 10.1097/00002030-199708000-00008. [DOI] [PubMed] [Google Scholar]
- White I, Babiker A, Walker S, Darbyshire J. Randomisation-based methods for correcting for treatment changes: examples from the Concorde trial. Statistics in Medicine. 1999;18:2617–2634. doi: 10.1002/(SICI)1097-0258(19991015)18:19<2617::AID-SIM187>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- White IR. Uses and limitations of randomization-based efficacy estimators. Statistical Methods in Medical Research. 2005 Aug;14(4):327–347. doi: 10.1191/0962280205sm406oa. ISSN 0962-2802. [DOI] [PubMed] [Google Scholar]
- White IR, Walker S, Babiker A. strbee: Randomization-based efficacy estimator. The Stata Journal. 2002;2:140–150. [Google Scholar]