Abstract
Immune-compromised mouse models allow for testing the preclinical efficacy of human cell transplantations and gene therapy strategies before moving forward to clinical trials. However, CRISPR/Cas9 gene editing of the Wsh/Wsh mouse strain to create an immune-compromised model lacking function of Rag2 and Il2rγ led to unexpected morbidity and mortality. This warranted an investigation to ascertain the cause and predisposing factors associated with the outbreak. Postmortem examination was performed on 15 moribund mice. The main lesions observed in these mice consisted of ascending urogenital tract infections, suppurative otitis media, pneumonia, myocarditis, and meningoencephalomyelitis. As Escherichia coli strains harboring polyketide synthase (pks) genomic island were recently isolated from laboratory mice, the tissue sections from the urogenital tract, heart, and middle ear were subjected to E. coli specific PNA-FISH assay that revealed discrete colonies of E. coli associated with the lesions. Microbiological examination and 16S rRNA sequencing confirmed E. coli-induced infection and septicemia in the affected mice. Further characterization by clb gene analysis and colibactin toxicity assays of the pks+ E. coli revealed colibactin-associated cytotoxicity. Rederivation of the transgenic mice using embryo transfer produced mice with an intestinal flora devoid of pks+ E. coli. Importantly, these barrier-maintained rederived mice have produced multiple litters without adverse health effects. This report is the first to describe acute morbidity and mortality associated with pks+ E. coli urosepsis and meningitis in immunocompromised mice, and highlights the importance of monitoring and exclusion of colibactin-producing pks+ E. coli.
Introduction
Conversion of common murine models to an immune-compromised state has become highly desirable for translational research. The creation of immune-compromised mouse strains allows for the preclinical efficacy of human cell transplantations and gene therapy strategies to be tested in small rodent systems before moving forward to human clinical trials. Recently, we have utilized the Wsh/Wsh mouse strain to examine human neural crest cell contribution during embryonic and post-natal mouse development [1]. In order to increase human cell incorporation into the Wsh/Wsh mice, as well as provide a future system for human disease modeling, we used CRISPR (clustered regularly interspaced short palindromic repeat) and Cas (CRISPR-associated) proteins to knockout interleukin 2 receptor subunit gamma (Il2rγ) and recombination activating gene-2 (Rag2) in the Wsh/Wsh mice. The conversion of the Wsh/Wsh mice to an immune-compromised state led to unexpected morbidity and mortality. This warranted an investigation to ascertain the cause and predisposing factors associated with the outbreak.
Escherichia coli (E. coli) strains have been frequently isolated from rodents, but are not routinely or completely characterized; these bacteria are commonly considered commensals and are not currently excluded from specific pathogen-free mouse colonies. However, pathogenic E. coli can encode various virulence factors, which are classified into different subtypes, such as enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAggEC), diffusely adhering E. coli (DAEC) and adherent-invasive E. coli (AIEC) [2]. Another group called extraintestinal pathogenic E. coli (ExPEC) belongs to the B2 E. coli phylogroup and are associated with human cases of meningitis, septicemia, and urinary tract infections (ExPEC) [3,4].
The polyketide synthase (pks) is a 54-kb genomic island that encodes colibactin (Clb) gene cluster [5,6]. The genotoxic metabolite colibactin acts as a cyclomodulin that induces DNA damage and cell cycle arrest in mammalian cells. [5–9]. The pks genomic island is highly conserved in Enterobacteriaceae and has been isolated from commensal E. coli strains (B2 and Nissle 1917 E. coli), Citrobacter koseri, Klebsiella pneumoniae, and Enterobacter aerogenes [5,7]. A study identified that IL10-/- mice with experimentally-induced chronic lower bowel inflammation were colonized with a specific E. coli (O7:H7:K1) strain of phylogenetic group B2 that encodes high number of virulence associated genes [10]. The tumor promoting effects of pks+ NC101 E. coli strain (O2:H6/41) were identified in germfree IL10-/- mice carcinogenicity studies Further, carcinogenicity studies in germfree IL10-/- mice monoassociated with the pks+ NC101 E. coli strain (O2:H6/41) identified its tumor promoting effects [11,12]. The pks+ NC101 E. coli strain associated promotional events were specifically attributed to the pks genomic island and excluded the role of inflammation in the formation of invasive carcinoma [11]. IL10-/- mice monoassociated with a murine E. coli strain from wildtype mice (later identified as NC101) induced typhlitis due to increased synthesis of interferon γ and IL4 by CD4+ T cells [13]. pks+ E. coli strains have also been utilized experimentally in rodent models to study urosepsis and septicemia [14,15].
We recently identified pks+ E. coli strains from the gastrointestinal tract of commercially available mice, as well as mice maintained in a large biomedical research institute, and demonstrated the cytotoxic effects of colibactin produced by the isolated pks+ E. coli in a cell culture system [16]. These pks+ E. coli strains, which colonize asymptomatic humans, are also associated with inflammation, septicemia, meningitis, and urinary tract infections [3,4], as well as being commonly isolated in colon cancer patients [17]. The purpose of this report is to describe acute morbidity and mortality in immunocompromised mice, from which colibactin producing E. coli was isolated from blood, genitourinary tract, and brain. As genome-engineering technologies have allowed for an increase in the efficiency and reduction in the time it takes to generate knockout mouse models, many murine models used in human research are becoming immune-compromised in order to test human cell transplantations and viral gene therapies for translational research. The abrupt increase in mortality and morbidity associated with pks+ E. coli in immune-compromised mice entails routine and complete characterization of these bacteria to ensure exclusion of colibactin-producing pks+ E. coli strains from specific pathogen-free mouse colonies.
Materials and methods
Ethics statement
All animal experiments were approved by the Massachusetts Institute of Technology Committee on Animal Care (Protocol #0916-058-19: Gene Disease, Cancer, and Mammalian Development) and all animal procedures were performed following the National Institute of Health guidelines. Our facility conforms to Federal guidelines and has a PHS Approved Animal Welfare Assurance (#A3125-01). Mice were monitored at least twice daily, and any adverse conditions (e.g. difficulty with ambulation, hunched posture, body condition score (BCS) < 2, ruffled fur) were immediately brought to the attention of the veterinary staff. The mice for this study displayed normal behavior and health until suddenly showing signs of morbidity, and would succumb within 24 hours. Mice that became moribund/severely ill during this study were euthanized with CO2 exposure followed by cervical dislocation, and tissues were collected for analysis.
Production of Cas9 mRNA and sgRNA
Bicistronic expression vector px330 expressing Cas9 and sgRNA [18] was digested with BbsI and treated with Antarctic Phosphatase, and the linearized vector was gel purified. A pair of oligos (Table 1) for each targeting site was annealed, phosphorylated, and ligated to the linearized vector. T7 promoter was added to the Cas9 coding region by PCR amplification using primers Cas9 F and R (Table 1). The T7-Cas9 PCR product was gel purified and used as the template for in vitro transcription (IVT) using the mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies). The T7 promoter was added to sgRNA templates by PCR amplification using the primers listed in Table 1 of a previously published paper by the Jaenisch Lab [19]. The T7-sgRNA PCR product was gel purified and used as the template for IVT using the MEGAshortscript T7 kit (Life Technologies). Both the Cas9 mRNA and the sgRNAs were purified using MEGAclear kit (Life Technologies) and eluted in RNase-free water.
Table 1. Oligonucleotides used in this study.
| Gene | Oligonucleotide | Cloning Step |
|---|---|---|
| Rag2 | CACCGTATTGTGGGTGGTTATCAGC | sgRNA |
| Rag2 | CACCGCCCTCAGCAGGAGCAGCTGA | sgRNA |
| Il2rγ | AAACGCTGATAACCACCCACAATAC | sgRNA |
| Il2rγ | AAACTCAGCTGCTCCTGCTGAGGGC | sgRNA |
| Rag2 | TTAATACGACTCACTATAGTATTGTGGGTGGTTATCAGC | in vitro Transcription |
| Il2rγ | TTAATACGACTCACTATAGCCCTCAGCAGGAGCAGCTGA | in vitro Transcription |
| Rag2 | GGAAAAGCATGGGTGTTCTC | RFLP Assay |
| Rag2 | TCCTGGTATGCCAAGGAAAA | RFLP Assay |
| Il2rγ | TCTCCCTGGGGACTTAGCTT | RFLP Assay |
| Il2rγ | AGGGGCAGAGTAGGAGCACT | RFLP Assay |
One-cell embryo injection
Mice were obtained from the Jackson Laboratory and maintained in the Whitehead Institute animal facility. Wsh/Wsh female mice on a C57BL/6 background and ICR mice were used as embryo donors and foster mothers, respectively. Superovulated female Wsh/Wsh mice (7–8 weeks old) were mated to Wsh/Wsh stud males, and fertilized embryos were collected from oviducts. Cas9 mRNAs (from 20 ng/μL to 200 ng/μL) and sgRNAs (from 20 ng/μL to 50 ng/μL), as previously described [19,20], were injected into the cytoplasm of fertilized eggs with well-recognized pronuclei in M2 medium (Sigma). Per previous methods, the injected zygotes were cultured in KSOM with amino acids at 37°C under 5% CO2 in air until blastocyst stage by 3.5 days. Thereafter, 15–25 blastocysts were transferred into the uterus of pseudo-pregnant ICR females at 2.5 days post-coitum [19]. Following blastocyst transfer, all pseudo-pregnant females and resulting Wsh/Wsh immunocompromised mice were housed in autoclaved, sterile environments.
Surveyor assay and RFLP analysis for genome modification
The Surveyor assay was performed as described [21]. Genomic DNA from targeted and control mice or blastocysts was extracted and PCR was performed using gene-specific primers (Table 1) under the following conditions: 95°C for 5 min; 35 × (95°C for 30 s, 60°C for 30 s, 68°C for 40 s); 68°C for 2 min; hold at 4°C. PCR products were then denatured, annealed, and treated with Surveyor nuclease (Transgenomics). The DNA concentration of each band was measured on an ethidium-bromide-stained 10% acrylamide Criterion TBE gel (BioRad) and quantified using ImageJ software. For RFLP analysis, 10 μL of Il2rγ and 10 μL of Rag2 PCR product were digested with PvuII. Digested DNA was separated on an ethidium-bromide-stained agarose gel (2%). For sequencing, PCR products were cloned using the Original TA Cloning Kit (Invitrogen), and mutations were identified by Sanger sequencing.
Flow cytometry
Blood was collected via the submandibular vein using a sterile Goldenrod animal lancet (Medipoint Inc.) and collected into a BD Microtainer MAP containing 1.0mg of K2 EDTA (Becton, Dickinson and Company). Additionally, mice were euthanized with CO2 according to institutional guidelines and their spleens were harvested and processed. Single-cell suspensions were generated from the blood and spleen samples and cells were stained using empirically determined concentrations of antibodies for 25 min on ice. Flow cytometry was used to separate T-cells (TCR-Beta, APC-Cy7), B cells (IgM, Alexa Fluor 488), Natural Killer (NK) cells (NK1.1, DsRed) and lymphocytes (FSC/SSC) from either blood or spleen samples. Flow cytometry data analysis was performed using FlowJo software.
Tumor assay
Human breast carcinoma cell line MCF7 was cultured (a gift from the laboratory of Dr. Robert Weinberg, Whitehead Institute for Biomedical Research) [22] and 1x105 MCF7 cells were resuspended in 250 μL of Dulbecco’s Modified Eagle’s Medium (DMEM; Sigma) containing 10% fetal calf serum (FCS, Sigma). The MCF7 cell suspension was co-injected subcutaneously with 250 μL of matrigel into the flank of NOD SCID gamma mice, Wsh/Wsh mice, and Wsh/Wsh mice with CRISPR deletions of Rag2 and Il2rγ. Before injection, mice were anesthetized with isoflurane, and allowed to recover to full ambulation immediately after injection. Mice were monitored 2x per day for any signs of adverse health and/or tumor growth. Tumors developed within 6 weeks of injection and animals were euthanized before tumor size exceeded 3 cm in diameter, or inhibited the ability of the mouse to ambulate, following institutional guidelines.
Necropsy
A total of 15 Wsh/Wsh adult mice with knockouts of Rag2 and Il2rγ, comprising 12 female and 3 males from the breeding colony housed in autoclaved cages with autoclaved food, water, and hardwood bedding were submitted for necropsy because of unanticipated increased incidence of morbidity and mortality.
Culture
At necropsy, brain, blood, kidney, and uterus were harvested for culture using sterile techniques. Tissue samples were plated on chromID™ CPS® agar plates (Biomérieux) [23]. E. coli colonies growing on CPS or MacConkey agar plates were initially identified by their ability to ferment lactose. Cultures were characterized as E. coli by API® 20 E (Biomérieux). Feces from an additional 17 immunocompromised mice without clinical signs were cultured for the presence of pks+ E. coli. Nine rederived mice (6 female and 3 male, Rag2 and Il2rγ Wsh/Wsh) were also cultured for pks+ E. coli.
DNA extraction and PCR amplification
Thirteen E. coli isolates were collected with sterile plastic loops placed in sterile PBS in microfuge tubes and used for DNA extraction. The Roche High Pure PCR Template Preparation Kit was used for bacterial DNA extraction. DNA concentration was measured using NanoDrop 2000c (Thermo).
Serotyping
Four E. coli isolates from affected mice were submitted to the E. coli Reference Center at Pennsylvania State University for complete typing which included: O and H typing and analyses and PCR for heat-labile toxin (LT), heat-stabile toxin (estA and estB), Shiga-type toxin 1 and 2 (Stx1 and Stx2), intimin gamma (eae), and cytotoxic necrotizing factor 1 and 2 (cnf1 and cnf2).
Clb genes and phylogenetic group identified by PCR
PCRs were performed to detect pks (clbA and clbQ) and cdtB genes with primers JPN42 and JPN46 for clbA, JPN55 and JPN56 for clbQ, and cdt1 and cdt2 for cdtB gene, respectively [16]. Phylogenetic groups of the isolates were determined with a set of primers published previously [16].
Cell culture conditions and gentamicin protection assay for colibactin cytotoxicity
E. coli strains used for cytotoxicity assay included K12 (negative control), NC101 (colibactin positive control), and novel mouse isolates 1512290008 [24], 1512290026 [24], 1601050009, and 1601060011. The cell culture assay for colibactin cytotoxicity was performed as described previously with modifications [5]. HeLa S3 cells (ATCC CCL2.2) were grown and maintained in Eagle's Minimum Essential Medium (EMEM, ATCC) containing fetal calf serum (FCS, Sigma) and 1% Antibiotic-Antimycotic solution (Gibco) at 37°C with 5% CO2. Fifty-thousand cells were seeded onto 12-well cell culture plates and incubated at 37°C with 5% CO2 for 24 h. Overnight cultures of E. coli strains were grown for 2 h at 37°C and then adjusted to a multiplicity of infection (MOI, number of bacteria per cell at the onset of infection) of 25 and 100. Following inoculation, plates were centrifuged at 200 g for 10 minutes to facilitate bacteria interaction and then incubated at 37°C with 5% CO2 for 4 h. Cells were then washed with EMEM and replaced with EMEM containing 10% fetal calf serum (FCS, Sigma) and 200 μg/mL gentamicin (Gibco). Following 72 h incubation, plates were stained with Diff-quick stain (ThermoScientific). Cells were then inspected under a microscope for confluence and morphological changes. Images were captured with Axiovert-10 microscope (Zeiss) using Image Pro-Plus software version 7.0 at 20x magnification. Cell viability was estimated in the Diff-quick stained plates by quantifying signal intensity at 700 nm using an Odyssey CLx plate reader (LI-COR) and Image Studio version 5.2 software (LI-COR).
Histopathology and peptic nucleic acid (PNA) fluorescent in situ hybridization
Tissue sections of kidney, heart, lung, brain, spinal cord, ear, and reproductive tract from clinically affected mice were fixed in 10% formalin, embedded in paraffin, sectioned at ~4 μm, and stained with hematoxylin and eosin (H&E). The tissue sections from these clinically affected mice were subjected to PNA fluorescent in situ hybridization by GNR Traffic LightTM PNA FISH® probe to detect E. coli [16]. The tissue sections were incubated with the E. coli specific PNA FISH probe (AdvanDx, Inc., Woburn, MA) for hybridization at 55°C for 105 min and examined for fluorescence with a Zeiss Axioskop 2 plus microscope. Digital images were acquired with a QImaging-QIClick camera (QImaging, Surrey, BC, Canada).
Rederivation of Rag2 and Il2rγ Wsh/Wsh mice
Mice were rederived by embryo transfer using standard techniques and recipient female Swiss mice free of pks+ E. coli and other murine pathogens. These mice have been maintained in a separate room under barrier conditions, which consists of maintaining the mice in autoclaved caging with autoclaved bedding, water, and pelleted diet.
Statistical analyses
Differences in experimental groups were determined by the Student’s t test as appropriate. p values <0.05 were considered significant.
Results
CRISPR/Cas9 deletion of Rag2 and Il2rγ in the Wsh/Wsh mouse background
Deletion of the host immune response is crucial for survival of human cells after transplantation into murine models, as well as allowing for gene therapy and other clinical approaches to be applied to the animal model without risk of an adverse systemic immune response to the therapy being tested for efficacy. The onset of efficient and rapid genome editing technologies has increased the ability for different mouse strains to become immune-compromised for translational research studies. As an example, CRISPR and Cas proteins have been demonstrated as an efficient gene targeting technology [18, 19]. In our study, capped polyadenylated Cas9 mRNA was produced by in vitro transcription and co-injected into pro-nuclear stage one-cell Wsh/Wsh mouse embryos with sgRNAs to knockout Rag2 (Fig 1A) and Il2rγ (Fig 1D). Rag2 encodes a protein that is involved in the initiation of V(D)J recombination during B and T cell development while Il2rγ encodes a protein involved in the growth and maturation of T-cells, B-cells, and NK cells. Therefore, CRISPR/Cas9-induced in-dels or insertions in Il2rγ and Rag2 that cause a loss of function of the protein should create an immunocompromised Wsh/Wsh mouse strain for further research studies.
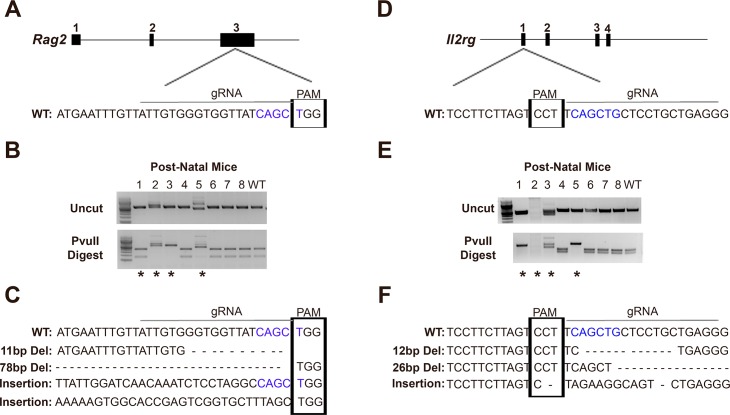
Fig 1. Knockout of Rag2 and Il2rγ in the Wsh/Wsh mouse background.
(A) Schematic of CRISPR/Cas9 targeting of Rag2 exon 3. (B) Rag2 DNA from post-natal mice both uncut (top gel) and cut with PvuII (bottom gel). (C) Rag2 wild-type (WT) DNA sequence followed by mutations after CRISPR/Cas9 delivery. (D) Schematic of CRISPR/Cas9 targeting of Il2rγ exon 1. (E) Il2rγ DNA from post-natal mice both uncut (top gel) and cut with PvuII (bottom gel). (F) Il2rγ WT DNA sequence followed by mutations found after CRISPR/Cas9 delivery. Straight line above WT genomic sequences, site of guide RNA; Boxed region of genomic sequences, PAM site; Blue font DNA, PvuII cut site; Asterisks, mice chosen as breeders for colony.
RFLP assays on DNA from post-natal mice displayed 3 out of 8 (37.5%) mice had altered Rag2 alleles (Fig 1B), while 4 out of 8 (50%) mice had altered alleles for Il2rγ (Fig 1E). Two males (#1: WT for Rag2 and homozygous mutant for Il2rγ; and #2: homozygous mutant for Rag2 and unknown, likely a large deletion mutant, for Il2rγ) and two females (#3: homozygous mutant for both Rag2 and Il2rγ; and #5: homozygous mutant for both Rag2 and Il2rγ) were chosen for continued breeding to create further generations of Rag2 and Il2rγ knockout mice (Fig 1B and 1E, asterisks). Sanger sequencing of the F1 generation displayed four mutant alleles in the population of Rag2 (Fig 1C) and 3 mutant alleles in the population for Il2rγ (Fig 1F) that were maintained through generations in the colony.
Loss of the B, T, and NK cells
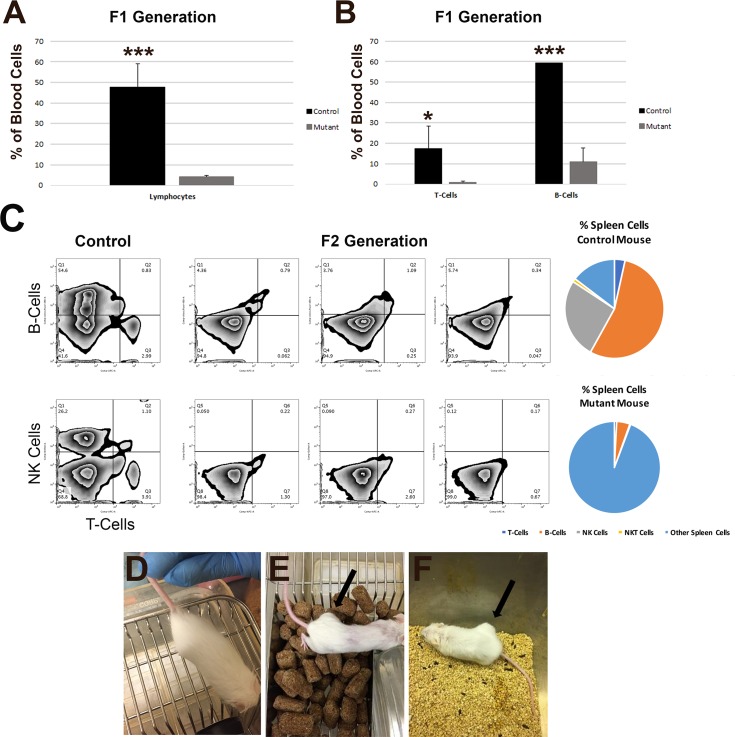
Blood samples were collected from Wsh/Wsh mice and first generation (F1) Wsh/Wsh mice with double allelic mutations and heterozygous mutations in Rag2 and Il2rγ. As loss-of-function of Rag2 and Il2rγ should deplete lymphocytes, and in particular the lymphocyte-derived B-, T-, and NK cells, blood cells were sorted by flow cytometry to quantify the numbers of lymphocytes in the F1 generation compared to control mice (Fig 2A). There was a significant reduction in the number of lymphocytes (47.85% for control mice, 4.30% for mutant mice; p = 0.0002) in the blood from the F1 generation (N ≥ 4) compared to the control mouse cohort (N ≥ 2). Closer examination of the blood to ascertain specific lymphocyte-derived cell types revealed a significant reduction in both T-cells (17.26% for control mice, 0.96% for mutant mice; p = 0.0297) and B-cells (59.29% for control mice, 10.91% for mutant mice; p < 0.0001) in the samples from the F1 generation in comparison to control mice (Fig 2B). NK cells were not present via staining with NK1.1 antibody in any of the F1 generation mice (N = 6; data not shown). Thus, within the first generation of mice, loss of Rag2 and Il2rγ even in heterozygotes led to a reduction in the presence of B-, T-, and NK cells.
Fig 2. Immunocompromised state of the Rag2 and Il2rγ knockout Wsh/Wsh mice.
(A) Flow cytometry from blood for lymphocytes in Wsh/Wsh control mice and the first (F1) generation of Rag2 and Il2rγ mutant mice. (B) Flow cytometry from blood for the presence of T-cells and B-cells in Wsh/Wsh control mice and the F1 generation of Rag2 and Il2rγ mutant mice. *, p < 0.05; *** p < 0.001; N ≥ 2 control mice; N ≥ 4 mutant mice. (C) Flow cytometry of spleen cells in two control (far left) mice compared to three F2 generation Rag2 and Il2rγ mutant mice displaying no detectable populations of B-cells (top row, y-axis), T-cells (top and bottom rows, x-axis), or NK cells (bottom row, y-axis). Percentages of cell types from the flow cytometry analysis are shown for both the control mouse (top right) and mutant moue (bottom right). (D-F) MCF7 human mammary carcinoma cells were injected subcutaneously into the flank of four Wsh/Wsh mice (D), an immune-compromised NOD SCID gamma mouse (E), and five Wsh/Wsh Rag2 and Il2rγ mutant mice (F). Human MCF7 cells survived and proliferated only in the immunocompromised NOD SCID gamma mouse and Wsh/Wsh Rag2 and Il2rγ mutant mice after 6 weeks post-injection. No human cells displayed growth and proliferation in Wsh/Wsh mice by 7 weeks post-injection. Arrows, tumor-like growth sites.
Wsh/Wsh F2 mice carrying double allelic mutations in Rag2 and Il2rγ were euthanized and spleen samples were taken for further quantification of immune cells. No lymph nodes were detectable for dissection in any of the F2 generation mice (N = 3; data not shown). Flow cytometry was performed on all three F2 mutant mice compared to two controls, and no visible populations of T-cells, B-cells, NK cells, or NKT cells were detected (Fig 2C). Percentages of each of the B-cells, T-cells, NK cells, NKT-cells, and other, non-immune spleen cells from the flow cytometry analysis are displayed in pie charts for the control mouse (top right) and immune-compromised mutant mouse (bottom right; only a small background of B-cells are visible; Fig 2C). Thus, F2 mice carrying mutations in both alleles of Rag2 and Il2rγ have a complete loss of B-, T-, and NK cells as expected with loss-of-function of Rag2 and Il2rγ.
A functional immunocompromised state in the Rag2 and Il2rγ knockout Wsh/Wsh mice
To ensure that the loss of B-, T-, and NK cells led to a functional immunocompromised state, MCF7 cells were injected subcutaneously into the flank of 5 (3 males and 2 females) Wsh/Wsh mice with mutations on both alleles of Rag2 and Il2rγ. Additionally, MCF7 cells were injected subcutaneously into the flank of 1 NOD SCID gamma mouse, a known immunocompromised mouse model, as well as 4 (2 males and 2 females) Wsh/Wsh mice with functional immune systems. Six weeks post-injection, tumors developed in all mice (100%, N = 5) with different Rag2 and Il2rγ knockout mutant alleles and a tumor was also noted in the NOD SCID gamma control mouse with a defective immune system which allows proliferation of human mammary carcinoma cells(Fig 2E and 2F, arrows). No tumors formed in any of the Wsh/Wsh mice at 7 weeks post-injections, as expected in mice with a functional immune system (Fig 2D). The rejection of MCF7 cells in the parental strain (Wsh/Wsh mice) without CRISPR deletion of Rag2 and Il2rγ and proliferation of MCF7 cells in Rag2 and Il2rγ knockout Wsh/Wsh mice suggests that these mice are in an immune-compromised state.
E. coli isolation from the Rag2 and Il2rγ knockout Wsh/Wsh mice
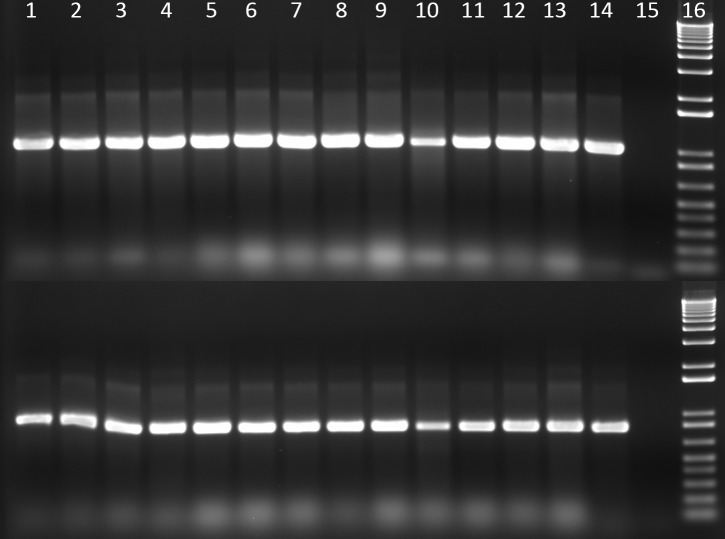
A random survey of the feces from 17 immunocompromised mice being maintained in the colony indicted that 100% of the mice were colonized with pks+ E. coli (S1 Fig). All 17 pks+ E. coli isolates belonged to phylogenetic group B2 (S2 Fig).
A sudden increase in morbidity and mortality developed within the Rag2 and Il2rγ knockout Wsh/Wsh mice, where mouse colony numbers dramatically declined and it was uncertain whether or not the colony could be maintained. This rapid increase in morbidity and mortality warranted an investigation, as these mice were housed in sterile, autoclaved environments within the barrier animal facility and were not removed at any time. E. coli, Enterococcus faecalis, Enterococcus faecium, and Staphylococcus xylosus were isolated from multiple tissues and blood of the mice submitted for evaluation. The presence of E. coli was confirmed in 93% of the cases either by culture or FISH analysis. Enterococcus spp. (40%) and S. xylosus (7%) were concurrently isolated along with E. coli from these affected mice (Table 2).
Table 2. Characterization of E. coli isolates from Rag2 and Il2rγ knockout Wsh/Wsh mice.
| Isolate ID | Serotype | Culture Sample | Phylogenetic Group | clbA | clbQ | cdtB | CYT |
|---|---|---|---|---|---|---|---|
| 1512290008 | O2:H6 | Blood | B2 | Positive | Positive | Negative | Positive |
| 1512290026 | O2:H6 | Blood | B2 | Positive | Positive | Negative | Positive |
| 1601050009 | O2:H6 | Blood | B2 | Positive | Positive | Negative | Positive |
| 1601060011 | O2:H6 | Blood | B2 | Positive | Positive | Negative | Positive |
| 1601190003 | NT | Uterus | B2 | Positive | Positive | Negative | NT |
| 1601190003 | NT | Kidney | B2 | Positive | Positive | Negative | NT |
| 1601260012 | NT | Uterus | B2 | Positive | Positive | Negative | NT |
| 1601260012 | NT | Kidney | B2 | Positive | Positive | Negative | NT |
| 1602030009 | NT | Uterus | B2 | Positive | Positive | Negative | NT |
| 1606150011 | NT | Kidney | B2 | Positive | Positive | Negative | NT |
| 1608160086 | NT | Kidney | B2 | Positive | Positive | Negative | NT |
| 1609230019 | NT | Uterus | B2 | Positive | Positive | Negative | NT |
| 1609290016 | NT | Uterus | B2 | Positive | Positive | Negative | NT |
CTY, Colibactin cytotoxicity assay; NT, not tested
Serotyping and analyses of other virulence factors in E. coli
Four isolates of E. coli from four mice were determined to be serotype O2:H6 (Table 2). The 4 isolates were positive for pks (clbA and clbQ) and colibactin cytotoxicity assay. None of these 4 isolates were positive for heat-labile toxin (elt), heat-stable enterotoxin a and b (estA and estB), Shiga-like toxin types 1 and 2 (stx1 and stx2), intimin-γ (eae), and cytotoxic necrotizing factors 1 and 2 (cnf1 and cnf2).
Rederivation of Rag2 and Il2rγ knockout Wsh/Wsh mice
Rederivation of the transgenic mice using embryo transfer produced mice with an intestinal flora devoid of pks+ E. coli. Six adult female and 3 adult male Rag2 and Il2rγ Wsh/Wsh mice were cultured for the presence of pks+ E. coli strains using standard culture methods. Importantly, these barrier-maintained rederived mice have produced multiple litters without adverse health effects.
Clb genes and phylogenetic group identified by PCR
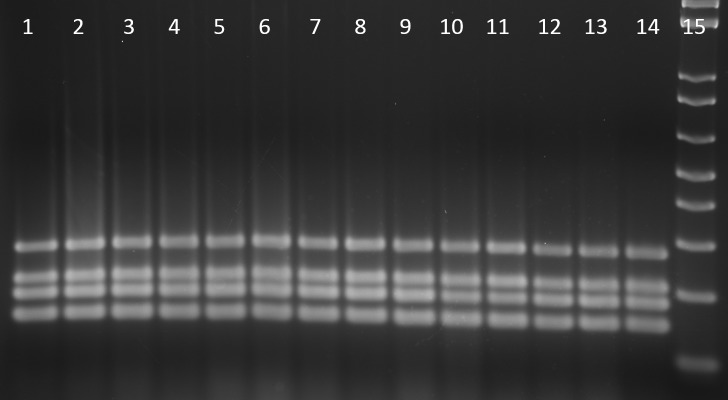
Thirteen E. coli strains from ten mice were positive for clbA and clbQ genes and negative for cdtB (Table 2 and Fig 3). All isolates belong in phylogenetic group B2 (Table 2 and Fig 4).
Fig 3. Amplification of clbA and clbQ in DNA from mouse E. coli isolates.
Top row: clbA gene, bottom row: clbQ gene. Lane 1 to lane 13, 13 E. coli isolates from mice samples; line 14, NC101 (positive control); line 15, no DNA control; line 16, 1 Kb plus molecular marker.
Fig 4. Phylogenetic group determination of E. coli.
Lane 1 to lane 13, 13 E. coli isolates from mice samples; line 14, NC101 (positive control); line 15, 1 Kb plus molecular marker. All samples were determined to be phylogenetic group B2.
Clb-encoding mouse E. coli isolates exert colibactin cytotoxicity
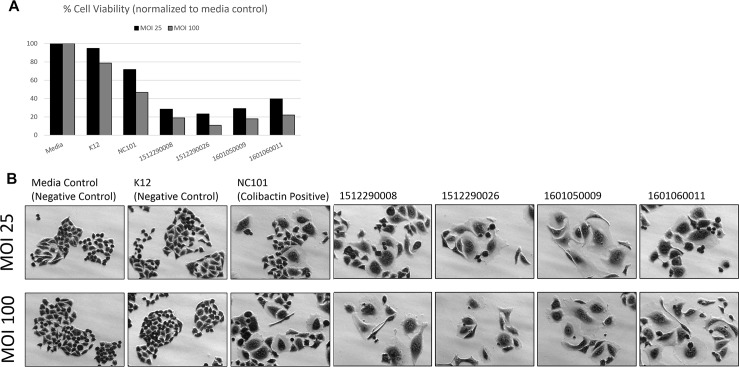
To confirm cytotoxic colibactin activity in the clb-encoding novel mouse E. coli isolates, HeLa cells were transiently infected with live bacteria at MOI 25 and 100. Novel mouse isolates 1512290008, 1512290026, 1601050009, and 1601060011 induced dose-dependent cytotoxicity. About 30–40% of cells survived after MOI 25, while less than 20% survived after MOI 100 (Fig 5A). Surviving cells appeared megalocytic and were phenotypically indistinguishable to that caused by Clb-encoding NC101 infection (Fig 5B). Media and K12 negative controls maintained similar cell confluence and morphology (Fig 5B).
Fig 5. HeLa cell colibactin cytotoxicity assay.
HeLa cells were inoculated with E. coli at a multiplicity of infection (MOI) of 25 and 100 for 4 h followed by a 72 h incubation in gentamicin-containing media. Megalocytosis (enlargement of the cell body and nucleus) was observed in cells infected with the colibactin (Clb)-encoding novel mouse E. coli isolates 1512290008, 1512290026, 1601050009, and 1601060011. NC101 is a Clb-encoding mouse E. coli isolate (positive control). K12 is a non-pathogenic E. coli strain (negative control). A) Estimation of cell viability. B) Representative images were taken at 20X magnification.
Histopathological evaluation
A total of fifteen Rag2 and Il2rγ knockout Wsh/Wsh mice comprising 12 females and 3 males were submitted for post mortem examination. Both males and females became moribund during our study, usually before 1 month of age; however, breeding females became ill at a rapid rate, either during the final week of pregnancy or within 1 week of nursing pups, and therefore more severely ill female mice were available for analysis. Table 3 summarizes the major organ systems affected, nature of the lesion and bacteria isolated and identified in different organ systems by microbial culture and FISH assay. The three affected male mice had pale tan shrunken kidneys with irregularly pitted capsular surface. Histological changes in the kidneys from the affected male mice were consistent with severe necrotizing granulocytic pyelonephritis with abundant bacteria and tubular degeneration and necrosis. Further characterization was performed on tissues/organs from the 12 female mice, to avoid any confounding factors related to gender.
Table 3. Pathology of spontaneous infection by E. coli in Rag2 and Il2rγ knockout Wsh/Wsh mice.
| Accession # | Sex | Microbial Culture | Histologic lesions | FISH- E. coli | |||
|---|---|---|---|---|---|---|---|
| E. coli | E. faecalis | E. faecium | S. xylosus | ||||
| 1512290008 | Male | bl,n,lu,s,k | l | n | Pyelonephritis and interstitial pneumonia | ND | |
| 1512290026 | Male | bl,n,lu,k | bl | Pyelonephritis | Kidney | ||
| 1601050009 | Female | bl | Metritis with macerated fetus, pyelonephritis, pneumonia, otitis media (bilateral), and epicarditis |
Uterus and placenta | |||
| 1601190003 | Female | lu, li, k,u | Placentitis, metritis, and pneumonia | Uterus and placenta | |||
| 1601260012 | Female | lu,li,k,u | lu,k,u | Metritis, vaginitis myocarditis, meningoencephalitis, otitis media(unilateral), and pneumonia |
Kidneys, uterus, vagina, and heart | ||
| 1602030009 | Female | lu,u | lu | Metritis, vaginitis, myocarditis, meningoencephalomyelitis, necrotizing hepatitis, unilateral otitis media and rhinitis | Uterus and placenta | ||
| 1606150011 | Male | k, br,bl | k, br |
br | Nephropathy (intralesional bacteria) | Kidneys | |
| 1608160086 | Female | k | Metritis and necrotizing suppurative pyelonephritis | Kidneys | |||
| 1609230019 | Female | u | Metritis, nephritis, interstitial pneumonia, and meningitis | Uterus | |||
| 1609290016 | Female | k |
k,u | k,u | Pyelonephritis and interstitial pneumonia | ND | |
| ND16-00081 | Female | No bacterial isolation was performed, as the animals were found dead |
Metritis with abundant intralesional bacteria and macerated fetus and pneumonia | Uterus and placenta | |||
| ND16-00082 | Female | Cystitis and vaginitis with intralesional bacilli |
Bladder and urogenital tract | ||||
| ND16-00083 | Female | Metritis and unilateral otitis media with intrahistiocytic bacteria | Uterus | ||||
| ND16-00094 | Female | Endometritis with macerated fetus and unilateral otitis media | ND | ||||
| ND16-00164 | Female | Pyelonephritis cystitis, endometritis, bilateral otitis media, rhinitis with intralesional bacteria, interstitial pneumonia, and meningitis |
Uterus | ||||
blood-bl, brain-br, kidneys-k, lungs-lu, liver-li, nares-n, spleen-s and uterus-u and ND: Not detected.
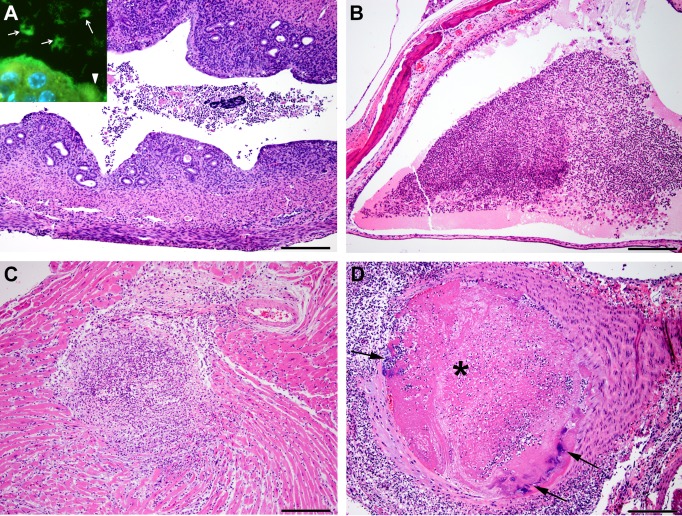
On gross examination, 4 of the 12 female mice were diagnosed with vaginal septum and pyometra. The vaginal orifice of the affected female mice was longitudinally bisected by a thin dorsoventral band of tissue. The urogenital tract and uterus contained pale tan to yellow, turbid, viscous material. Histopathological examination of the uterus and urogenital tract of 11 female mice revealed intraluminal cellular debris admixed with degenerate granulocytes, mononuclear cells, and myriad of bacteria (Fig 6A). The endometrium and underlying myometrium were also infiltrated by inflammatory aggregates, in a multifocal pattern.
Fig 6.
Histopathological lesions induced by pks+ E. coli (A) Mild subacute endometritis. The endometrium is disrupted by low numbers of inflammatory cells and ectatic glands that rarely contain pale eosinophilic material. The uterine lumen contains cellular debris and degenerate inflammatory cells. H&E Scale: 200μm. Inset: PNA-FISH assay with E. coli specific probe reveals colonies of fluorescent green E. coli bacteria lining the endometrium (white arrow head) and within the uterine lumen (white arrows). (B) Moderate focally extensive subacute otitis media. The middle ear lumen contains moderate amounts of pale esoinophilic fibrillar material, cellular debris, degenerate granulocytes, and mononuclear cells. H&E Scale: 200μm. (C) Moderate focally extensive subacute necrotizing myocarditis. The myocardium was focally disrupted by granulocytic and mononuclear cell infiltration, necrosis, and degeneration of the cardiomyocytes. H&E Scale: 200μm. (D) Aorta: Severe subacute necrotizing valvulitis and arteritis with fibrin thrombi (asterisk) and colonies of bacteria (black arrows). The aortic valve is focally disrupted by necrosis, few bacterial colonies, granulocytes, and mononuclear cell infiltrates.The aortic lumen is occluded by thrombus composed of fibrin, inflammatory cells, and bacteria. H&E Scale: 200μm.
The ear canal was affected by mild to moderate unilateral or bilateral otitis media in 6 out of 12 female mice. The otitis media was characterized by loss of ciliated epithelial lining and the lumen contained cellular debris, pale eosinophilic material, and low numbers of granulocytes and histiocytes admixed with extracellular and intrahistiocytic bacilli (Fig 6B).
The heart was affected by necrotizing granulocytic myocarditis (Fig 6C) and epicarditis with mural septic thrombi in 3 out of 12 female mice. The aortic valve was focally disrupted by necrotizing valvulitis characterized by loss of fibrous connective tissue and cartilage, with granulocytic and mononuclear cell infiltrates admixed with bacterial colonies. The aorta was completely occluded by septic thrombi and transmurally, the aortic wall was infiltrated by granulocytes and mononuclear cells (Fig 6D).
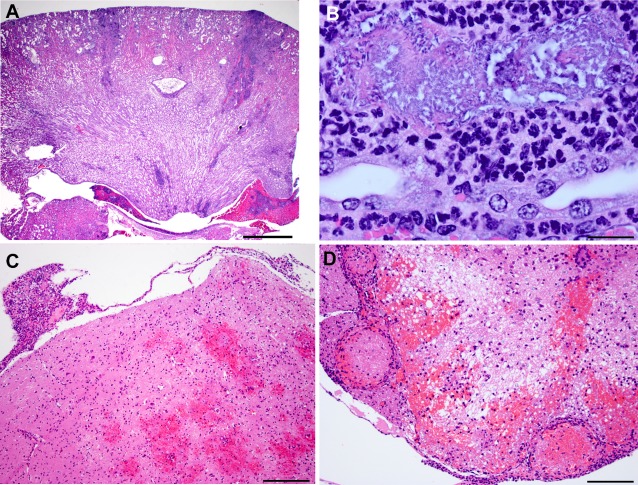
In 5 out of 12 female mice, the renal architecture was multifocally disrupted by necrosis and inflammation. The renal pelvis was dilated and contained hemorrhage and inflammatory cells (Fig 7A). The affected renal tubules were lined by attenuated epithelium and occasionally the tubular epithelial cells with intact basement membrane were lost. The tubular lumen contained cellular debris and abundant bacteria. The tubular interstitium was multifocally infiltrated by granulocytes and mononuclear cells (Fig 7B).
Fig 7.
Histopathological lesions induced by pks+ E. coli (A) Moderate multifocal subacute suppurative pyelonephritis with tubular necrosis and intraluminal bacterial colonies. The renal architecture is multifocally disrupted by pyelonephritis that discretely extends from the capsular surface deep in to the medulla. The ectatic renal pelvis contains hemorrhage admixed with inflammatory cells. H&E Scale: 1mm. (B) Higher magnification of the affected renal tubule. Focally, ectatic tubule contained large numbers of bacteria admixed with cellular debris. The epithelial lining is completely lost with intact basement membrane. The adjacent tubular interstitium is infiltrated by granulocytes and mononuclear cells. H&E Scale: 20μm. (C) Mild focally extensive subacute necrohemorrhagic meningoencephalitis. The brain parenchyma and meninges are multifocally disrupted by hemorrhage and inflammatory infiltrates. H&E Scale: 200μm. (D) Lumbar spinal cord: Moderate multifocal subacute hemorrhagic meningiomyelitis with vasculitis and fibrin thrombi. The gray and white matter are disrupted by hemorrhage, vacuolations, granulocytes, and mononuclear cells. The meningeal blood vessels are multifocally affected by vasculitis and fibrin thrombi.
In 4 out of 12 female mice, the brain and spinal cord were variably affected by necrohemorrhagic meningoencephalomyelitis. The affected brain and spinal cord parenchyma were disrupted by minimal amounts of hemorrhage and vacuolations. These affected areas were infiltrated by granulocytes and mononuclear infiltrates and the meningeal blood vessels were disrupted by necrotizing vasculitis and fibrin thrombi (Fig 7C and 7D). Other significant findings in these affected mice included interstitial pneumonia, rhinitis, necrotizing granulocytic hepatitis, and mild granulocytic ureteritis and cystitis.
Since all of the above mentioned organs showed significant findings and signs of disease, we hypothesized that these tissues may have been infected with E. coli. Thus, representative tissue sections from the uterus, kidneys, urogenital tract, heart, and ear were subjected to E. coli specific PNA-FISH assay [16]. The PNA-FISH assay revealed E. coli positivity by the presence of multifocal and discrete clumps of intra luminal bacilli in the tissue sections from the above mentioned organs (Fig 6A).
Further, four rederived mice (3 males and 1 female), confirmed negative for pks+ E. coli, were analyzed by a detailed gross necropsy followed by histopathological evaluation of the liver, spleen, pancreas, mesenteric lymph node, kidneys, heart, lungs, urinary bladder, reproductive organs (testes, epididymis, prostate or uterus, ovaries, vagina and vulva). No significant pathological findings were noted other than incidental findings of sparse to minimal perivascular interstitial lympho-plasmacytic inflammatory aggregates in the all the renal medulla and/or pelvis of all animals. The female mouse had a gravid uterus.
Discussion
Although E. coli infections in laboratory mice have been previously reported [10,13,25–29], a description of spontaneous disease which includes biochemical, genetic, and phenotypic characterization of pks+ E. coli isolates has not been undertaken. In one of the previously reported studies, natural infection with uncharacterized E. coli was associated with urogenital lesions, pneumonia, and septicemia in mice [26]. Characterization of pks+ E. coli NC101 in experimentally infected IL10-/- mice and the natural and experimental characterization of O7:H7:K1 in IL10-/- mice indicated the pathogenic potential of Clb-encoding E. coli in experimentally infected susceptible animals [10,11,13,30]. The current study characterized pks+ E. coli in clinically affected mice with genitourinary infection, septicemia, and meningitis. The mouse E. coli isolates encoding two Clb genes (clbQ and clbA), were cytotoxic to cells in vitro. The isolates in this study were serotype O2:H6, which is the same serotype as that of NC101 [12]. Serotype O2 in humans is also commonly associated with UTIs, septicemia, and meningitis [31]. Importantly, barrier-maintained rederived mice without pks+ E. coli in their intestinal tracts have produced multiple litters without adverse health effects, nor have lesions noted in the original outbreak observed.
The isolates in the current study were cytotoxic to cells in vitro suggesting that mouse Clb-encoding E. coli have the potential to induce clinical disease in laboratory animals, as noted in this study. In a rodent model of septicemia, 8–9 week old female C57BL/6J mice were injected subcutaneously with a pks+ E. coli colibactin producing 018:K1:H7, ExPEC strain S15 isolated from a meningitis case in a newborn infant [14]. The E. coli induced a profound lymphopenia in septicemic mice, which was attenuated when an isogenic mutant lacking colibactin genotoxin activity was injected into the mice [14]. The authors argued that the production of colibactin by E. coli exacerbates lymphopenia associated with septicemia, reducing the chances that the mice, and by association humans, would survive pks+ E. coli induced sepsis [14]. The prolonged immunosuppression often referred to as immunoparalysis, in sepsis cases, predisposes the patients to nosocomial infections and reactivation of latent viruses [32,33]. Isolation of commensals such as Enterococcus spp. and S. xylosus along with pks+ E. coli from these affected mice in our study may be attributed to immunosuppression. E. faecalis has been shown to exacerbate pathogenic effects induced by Gram-negative bacterial infection [34]. However, E. coli was isolated in 93% of cases in this study either with or without concurrent isolation of Enterococcus spp. and S. xylosus. Recent studies have demonstrated the importance of colibactin in intestinal colonization and invasion of other organs by pks+ E. coli that leads to systemic infection in neonates [15]. Hence, there is a distinct possibility that pks+ E. coli infection facilitates colonization and/or co-infection by other commensal bacteria such as Enterococcus spp. and S. xylosus. Furthermore, our findings of pks+ E. coli strains responsible for urosepsis in C57BL/6 Rag2 and Il2rγ knockout Wsh/Wsh mice reinforce the susceptibility of immuno-compromised mice to opportunistic pathogens.
The breeding colony of Rag2 and Il2rγ knockout Wsh/Wsh mice also presented with a variable incidence of vaginal septa. The vaginal septal defect apparently has not been noted to interfere in the overall breeding performance in this colony. Our results however, suggest that the vaginal defect may have predisposed the female mice to pks+ E. coli urosepsis. The congenital vaginal septal defect has been reported previously and has been associated with compromising successful breeding and parturition, in both mice and rats [35,36]. The vaginal defect has been linked to metritis in rats used on reproductive toxicology studies [36]. Although rats with vaginal septal defects were screened for rat microbial pathogens, none were noted. However, the authors mention that bacterial cultures obtained from abnormal uterine contents identified the presence of abundant E. coli. Unfortunately, further molecular identification of putative virulence factors was not undertaken [36].
Investigators have also capitalized on the use of neonatal rats to explore the pathogenicity of colibactin expressing E. coli. Two day old Wistar rats were orally dosed with pks+ E. coli which efficiently colonized the intestinal tract and translocated across the immature GI tract resulting in sepsis [15]. Inactivation of the clbA and clbP genes responsible for colibactin production in pks+ E. coli strains, significantly reduced the capacity of the E. coli strain A192PP to colonize the intestine, translocate, induce septicemia, and cause the death of the neonatal rats [15]. This neonatal rat model replicates the age dependency seen in newborn children where ExPEC B2 strains of pks+ E. coli are noted to express colibactin and cause significant disease [37]. Indeed, ExPEC infections are associated with urosepsis, bacteremia, and neonatal meningitis in infants [15,38,39]. E. coli isolates from human prostatitis cases also have been characterized as ExPEC and some encode Clb [40].
Previously, we reported the draft genomes of two E. coli isolates from this study that were recovered from the blood of mice (accession numbers 1512290008 and 1512290026)[24]. Both genomes contained a complete ~54-kb PKS pathogenicity island required for colibactin synthesis[24]. Cytolethal distending toxin (cdt), cytotoxic necrotizing factor (cnf), and cycle inhibiting factor (cif) genes known to cause megalocytosis were not present in either of these genomes, which substantiated our PCR results, suggesting the megalocytic cytotoxicity observed in our in vitro infection experiments was the result of colibactin activity. Aside from PKS, both of our novel pks+ E. coli isolate genomes also contained homologous virulence genes that regulate colonization, adherence, immune evasion, cytotoxicity, and iron acquisition at intestinal, vascular, and renal sites by EPEC and ExPEC strains. These genes included enterobactin siderophore receptor protein[41,42], s-fimbriae minor subunit[43], glutamate decarboxylase[44,45], per-activated serine protease autotransporter enterotoxin[46–48], and iss/bor protein precursor[49,50]. The genomic presence of pks and other virulence genes supports our hypothesis that these novel mouse E. coli isolates have the pathogenic potential to infect and induce clinical diseases in susceptible hosts.
The genetic profile of the Rag2 and Il2rγ knockout Wsh/Wsh mice predisposes them to progressive kidney disease. The underlying mutation in the Wsh/Wsh strains is an inversion mutation in the upstream transcriptional regulatory elements of the c-kit gene that also disrupts the corin gene [51–53]. Corin is a trypsin-like transmembrane serine protease that activates atrial natriuretic peptide, which regulates salt-water balance and blood pressure by promoting renal natriuresis. In humans, abnormalities in corin expression result in hypertension [54], heart failure [55], and chronic kidney disease [56]. In our study, the tubular lesions noted in the mice closely resemble obstructive nephropathy. However, there was no evidence for obstruction of renal tubules, urinary bladder, and urethra by crystals, proteinaceous plugs, and associated inflammation. The observation from our study and findings reported in previous studies suggests decreased expression of c-kit and corin could have resulted in the chronic progressive nephropathy observed in these mice due to impaired sodium excretion. However, the exact mechanism by which c-kit and corin exert this detrimental effect on murine renal tubular epithelial cells and whether this defect predisposes the mice to urosepsis remains to be elucidated.
In summary, the pks pathogenicity island is present in a high percentage of extraintestinal pathogenic B2 strains of E. coli and exhibits an increased predilection of causing bacteremia [3]. This current finding of pks+ E. coli strains in cases of urosepsis, as well as pks+ E. coli in the feces of clinically normal Rag2 and Il2rγ-/- mice, and our recent characterization of E. coli strains from clinically normal and clinically affected laboratory mice indicates that mice can be colonized with Clb-encoding cytotoxic and pathogenic E. coli [16]. Phylogenetic B2 pks+ E. coli strains have a high likelihood of persistently colonizing infants and are commonly isolated in adults. Given that pks+ E. coli B2 isolates are present within 6-12-month-old mice obtained from a mouse colony endemically infected with pks+ E. coli, this supports the hypothesis that mice can also be persistently infected with this colibactin-producing E. coli [16]. The epidemiology and virulence determinants of mouse pks+ E. coli strains should be investigated further. Also, studies examining their impact in mouse models of inflammation, both intestinal and extraintestinal should be conducted. Importantly, our findings of urosepsis in immunocompromised mice caused by pks+ E. coli strain O2:H6 represents an opportunity to develop experimental mouse models to investigate the pathogenic potential of pks+ E. coli and cytotoxin-associated induction of urogenital disease and septicemia noted in humans.
Many murine models used in human research are genetically engineered to be immune-compromised in order to test human cell transplantations and viral gene therapies for translational research. This report is the first to describe acute morbidity and mortality associated with pks+ E. coli urosepsis and meningitis in immunocompromised mice. As E. coli strains have been frequently isolated from rodents, but are not routinely or completely characterized, our study highlights the importance of routine and complete characterization of pks+ E. coli. Also, health surveillance strategies to ensure that these E. coli strains are excluded from specific pathogen-free mouse colonies should be instituted.
Supporting information
Top row: clbA gene, bottom row: clbQ gene. Lane 1 to lane 17, 17 E.coli isolates from mice fecal samples; line 18, NC101 (positive control); line 19, no DNA control; line 20, 1 Kb plus molecular marker.
(PDF)
Lane 1 to lane 17, 17 E.coli isolates from mice fecal samples; line 18, NC101 (B2 group positive control); line 19, no DNA control; line 20, 1 Kb plus molecular marker.
(PDF)
Acknowledgments
We wish to thank the DCM and Whitehead staff for their assistance, as well as Alyssa Pappa for her assistance with manuscript preparation.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Institutes of Health under the following grants: T32OD010978; P30ES002109; R01OD011141 (all to J.G.F.). NIH/NCI F32CA196065 (K.J.W.) NIH grants HD045022, R37-CA084198, HG002668 (RJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cohen MA, Wert KJ, Goldmann J, Markoulaki S, Buganim Y, Fu D, et al. Human neural crest cells contribute to coat pigmentation in interspecies chimeras after in utero injection into mouse embryos. 2016; Proc Natl Acad Sci U S A 113: 1570–1575. doi: 10.1073/pnas.1525518113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. 1999; Infect Immun 67: 4499–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. 2008; J Clin Microbiol 46: 3906–3911. doi: 10.1128/JCM.00949-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen J. The immunopathogenesis of sepsis. 2002; Nature 420: 885–891. doi: 10.1038/nature01326 [DOI] [PubMed] [Google Scholar]
- 5.Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. 2006; Science 313: 848–851. doi: 10.1126/science.1127059 [DOI] [PubMed] [Google Scholar]
- 6.Brotherton CA, Wilson M, Byrd G, Balskus EP. Isolation of a metabolite from the pks island provides insights into colibactin biosynthesis and activity. 2015; Org Lett 17: 1545–1548. doi: 10.1021/acs.orglett.5b00432 [DOI] [PubMed] [Google Scholar]
- 7.Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. 2009; Infect Immun 77: 4696–4703. doi: 10.1128/IAI.00522-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossuet-Greif N, Dubois D, Petit C, Tronnet S, Martin P, Bonnet R, et al. Escherichia coli ClbS is a colibactin resistance protein. 2016; Mol Microbiol 99: 897–908. doi: 10.1111/mmi.13272 [DOI] [PubMed] [Google Scholar]
- 9.Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. 2015; Nat Chem 7: 411–417. doi: 10.1038/nchem.2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wohlgemuth S, Haller D, Blaut M, Loh G. Reduced microbial diversity and high numbers of one single Escherichia coli strain in the intestine of colitic mice. 2009; Environ Microbiol 11: 1562–1571. doi: 10.1111/j.1462-2920.2009.01883.x [DOI] [PubMed] [Google Scholar]
- 11.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. 2012; Science 338: 120–123. doi: 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Hansen JJ, Holt LC, Kim SC, Dogan B, Simpson KW, et al. W1183 Differential In Vitro Epithelial Translocation and Resistance to Phagocyte Uptake and Killing By Escherichia coli Strains Correlate with Their Ability to Induce Colitis in Monoassociated IL-10-/- Mice. 2008; Gastroenterology 134: A-650. [Google Scholar]
- 13.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. 2005; Gastroenterology 128: 891–906. [DOI] [PubMed] [Google Scholar]
- 14.Marcq I, Martin P, Payros D, Cuevas-Ramos G, Boury M, Watrin C, et al. The genotoxin colibactin exacerbates lymphopenia and decreases survival rate in mice infected with septicemic Escherichia coli. 2014; J Infect Dis 210: 285–294. doi: 10.1093/infdis/jiu071 [DOI] [PubMed] [Google Scholar]
- 15.McCarthy AJ, Martin P, Cloup E, Stabler RA, Oswald E, Taylor PW. The Genotoxin Colibactin Is a Determinant of Virulence in Escherichia coli K1 Experimental Neonatal Systemic Infection. 2015; Infect Immun 83: 3704–3711. doi: 10.1128/IAI.00716-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia A, Mannion A, Madden CM, Bakthavatchalu V, Shen Z, Ge Z, et al. Cytotoxic Escherichia coli strains encoding colibactin colonize laboratory mice. 2016; Microbes Infect 18: 777–786. doi: 10.1016/j.micinf.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raisch J, Buc E, Bonnet M, Sauvanet P, Vazeille E, de Vallee A, et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. 2014; World J Gastroenterol 20: 6560–6572. doi: 10.3748/wjg.v20.i21.6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. 2013; Science 339: 819–823. doi: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HY, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. 2013; Cell 153: 910–918. doi: 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Wang HY, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. 2014; Nature Protocols 9: 1956–1968. doi: 10.1038/nprot.2014.134 [DOI] [PubMed] [Google Scholar]
- 21.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A Rapid and General Assay for Monitoring Endogenous Gene Modification. 2010; Engineered Zinc Finger Proteins: Methods and Protocols 649: 247–256. [DOI] [PubMed] [Google Scholar]
- 22.Shibue T, Brooks MW, Inan MF, Reinhardt F, Weinberg RA. The Outgrowth of Micrometastases Is Enabled by the Formation of Filopodium-like Protrusions. 2012; Cancer Discovery 2: 706–721. doi: 10.1158/2159-8290.CD-11-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. 2013; PLoS One 8: e56964 doi: 10.1371/journal.pone.0056964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mannion A, Shen Z, Feng Y, García A, Fox JG. Draft genome sequences of five novel polyketide synthetase -containing mouse Escherichia coli strains 2016; Genome Announcements 4: e01082–01016. doi: 10.1128/genomeA.01082-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goo JS, Jang MK, Shim SB, Jee SW, Lee SH, Bae CJ, et al. Monitoring of antibiotic resistance in bacteria isolated from laboratory animals. 2012; Lab Anim Res 28: 141–145. doi: 10.5625/lar.2012.28.2.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benga L, Benten WP, Engelhardt E, Gougoula C, Sager M. Spontaneous bacterial and fungal infections in genetically engineered mice: Is Escherichia coli an emerging pathogen in laboratory mouse? 2015; Berl Munch Tierarztl Wochenschr 128: 278–284. [PubMed] [Google Scholar]
- 27.Nakagawa M, Sakazaki R, Muto T, Saito M, Hagiwara T. Infectious megaenteron of mice. II. Detection of coliform organisms of an unusual biotype as the primary cause. 1969; Jpn J Med Sci Biol 22: 375–382. [PubMed] [Google Scholar]
- 28.Schiff LJ, Barbera PW, Port CD, Yamashiroya HM, Shefner AM, Poiley SM. Enteropathogenic Escherichia coli infections: increasing awareness of a problem in laboratory animals. 1972; Lab Anim Sci 22: 705–708. [PubMed] [Google Scholar]
- 29.Waggie KS, Hansen CT, Moore TD, Bukowski MA, Allen AM. Cecocolitis in immunodeficient mice associated with an enteroinvasive lactose negative E. coli. 1988; Lab Anim Sci 38: 389–393. [PubMed] [Google Scholar]
- 30.Patwa LG, Fan TJ, Tchaptchet S, Liu Y, Lussier YA, Sartor RB, et al. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. 2011; Gastroenterology 141: 1842–1851 e1841-1810. doi: 10.1053/j.gastro.2011.06.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poolman JT, Wacker M. Extraintestinal Pathogenic Escherichia coli, a Common Human Pathogen: Challenges for Vaccine Development and Progress in the Field. 2016; J Infect Dis 213: 6–13. doi: 10.1093/infdis/jiv429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawkins CA, Collignon P, Adams DN, Bowden FJ, Cook MC. Profound lymphopenia and bacteraemia. 2006; Intern Med J 36: 385–388. doi: 10.1111/j.1445-5994.2006.01076.x [DOI] [PubMed] [Google Scholar]
- 33.Hotchkiss RS, Coopersmith CM, McDunn JE, Ferguson TA. The sepsis seesaw: tilting toward immunosuppression. 2009; Nat Med 15: 496–497. doi: 10.1038/nm0509-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchimori N, Hayashi R, Shino A, Yamazaki T, Okonogi K. Enterococcus faecalis aggravates pyelonephritis caused by Pseudomonas aeruginosa in experimental ascending mixed urinary tract infection in mice. 1994; Infect Immun 62: 4534–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang TK, Ho P, Liang CT, Yu CK. Effects of vaginal septa on the reproductive performance of BALB/cByJNarl mice. 2013; J Am Assoc Lab Anim Sci 52: 520–523. [PMC free article] [PubMed] [Google Scholar]
- 36.Lezmi S, Thibault-Duprey K, Bidaut A, Hardy P, Pino M, Macary GS, et al. Spontaneous metritis related to the presence of vaginal septum in pregnant Sprague Dawley Crl:CD(SD) rats: impact on reproductive toxicity studies. 2011; Vet Pathol 48: 964–969. doi: 10.1177/0300985810391113 [DOI] [PubMed] [Google Scholar]
- 37.Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. 2003; Microbes Infect 5: 449–456. [DOI] [PubMed] [Google Scholar]
- 38.Dubois D, Delmas J, Cady A, Robin F, Sivignon A, Oswald E, et al. Cyclomodulins in urosepsis strains of Escherichia coli. 2010; J Clin Microbiol 48: 2122–2129. doi: 10.1128/JCM.02365-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. 2000; J Infect Dis 181: 261–272. doi: 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- 40.Krieger JN, Dobrindt U, Riley DE, Oswald E. Acute Escherichia coli prostatitis in previously health young men: bacterial virulence factors, antimicrobial resistance, and clinical outcomes. 2011; Urology 77: 1420–1425. doi: 10.1016/j.urology.2010.12.059 [DOI] [PubMed] [Google Scholar]
- 41.Russo TA, Carlino UB, Mong A, Jodush ST. Identification of Genes in an Extraintestinal Isolate of Escherichia coli with Increased Expression after Exposure to Human Urine. 1999; Infect Immun 67: 5306–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo TA, McFadden CD, Carlino-MacDonald UB, Beanan JM, Barnard TJ, Johnson JR. IroN Functions as a Siderophore Receptor and Is a Urovirulence Factor in an Extraintestinal Pathogenic Isolate of Escherichia coli. 2002; Infect Immun 70: 7156–7160. doi: 10.1128/IAI.70.12.7156-7160.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marre R, Kreft B, Hacker J. Genetically engineered S and F1C fimbriae differ in their contribution to adherence of Escherichia coli to cultured renal tubular cells. 1990; Infect Immun 58: 3434–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant MA, Weagant SD, Feng P. Glutamate Decarboxylase Genes as a Prescreening Marker for Detection of Pathogenic Escherichia coli Groups. 2001; Applied and Environmental Microbiology 67: 3110–3114. doi: 10.1128/AEM.67.7.3110-3114.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergholz TM, Tarr CL, Christensen LM, Betting DJ, Whittam TS. Recent gene conversions between duplicated glutamate decarboxylase genes (gadA and gadB) in pathogenic Escherichia coli. 2007; Mol Biol Evol 24: 2323–2333. doi: 10.1093/molbev/msm163 [DOI] [PubMed] [Google Scholar]
- 46.Mellies JL, Navarro-Garcia F, Okeke I, Frederickson J, Nataro JP, Kaper JB. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. 2001; Infect Immun 69: 315–324. doi: 10.1128/IAI.69.1.315-324.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elisa Drago-Serrano M, Gavilanes Parra S, Angel Manjarrez-Hernández H. EspC, an autotransporter protein secreted by enteropathogenic Escherichia coli (EPEC), displays protease activity on human hemoglobin. 2006; FEMS Microbiology Letters 265: 35–40. doi: 10.1111/j.1574-6968.2006.00463.x [DOI] [PubMed] [Google Scholar]
- 48.Guignot J, Segura A, Tran Van Nhieu G. The Serine Protease EspC from Enteropathogenic Escherichia coli Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System. 2015; PLoS Pathogens 11: e1005013 doi: 10.1371/journal.ppat.1005013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lynne AM, Skyberg JA, Logue CM, Nolan LK. Detection of Iss and Bor on the surface of Escherichia coli. 2007; J Appl Microbiol 102: 660–666. doi: 10.1111/j.1365-2672.2006.03133.x [DOI] [PubMed] [Google Scholar]
- 50.Johnson TJ, Wannemuehler YM, Nolan LK. Evolution of the iss gene in Escherichia coli. 2008; Appl Environ Microbiol 74: 2360–2369. doi: 10.1128/AEM.02634-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mallen-St Clair J, Pham CT, Villalta SA, Caughey GH, Wolters PJ. Mast cell dipeptidyl peptidase I mediates survival from sepsis. 2004; J Clin Invest 113: 628–634. doi: 10.1172/JCI19062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimbaldeston MA, Chen CC, Piliponsky AM, Tsai M, Tam SY, Galli SJ. Mast cell-deficient W-sash c-kit mutant Kit W-sh/W-sh mice as a model for investigating mast cell biology in vivo. 2005; Am J Pathol 167: 835–848. doi: 10.1016/S0002-9440(10)62055-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nigrovic PA, Gray DH, Jones T, Hallgren J, Kuo FC, Chaletzky B, et al. Genetic inversion in mast cell-deficient (Wsh) mice interrupts corin and manifests as hematopoietic and cardiac aberrancy. 2008; Am J Pathol 173: 1693–1701. doi: 10.2353/ajpath.2008.080407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Liao X, Fukuda K, Knappe S, Wu F, Dries DL, et al. Corin variant associated with hypertension and cardiac hypertrophy exhibits impaired zymogen activation and natriuretic peptide processing activity. 2008; Circ Res 103: 502–508. doi: 10.1161/CIRCRESAHA.108.177352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong N, Chen S, Yang J, He L, Liu P, Zheng D, et al. Plasma soluble corin in patients with heart failure. 2010; Circ Heart Fail 3: 207–211. doi: 10.1161/CIRCHEARTFAILURE.109.903849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang C, Shen L, Dong L, Liu M, Shi S, Dong N, et al. Reduced urinary corin levels in patients with chronic kidney disease. 2013; Clin Sci (Lond) 124: 709–717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top row: clbA gene, bottom row: clbQ gene. Lane 1 to lane 17, 17 E.coli isolates from mice fecal samples; line 18, NC101 (positive control); line 19, no DNA control; line 20, 1 Kb plus molecular marker.
(PDF)
Lane 1 to lane 17, 17 E.coli isolates from mice fecal samples; line 18, NC101 (B2 group positive control); line 19, no DNA control; line 20, 1 Kb plus molecular marker.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.