Abstract
Quiescent cells play a predominant role in most organisms. Here, we identify RNA interference (RNAi) as a major requirement for quiescence (G0) in Schizosaccharomyces pombe. RNAi mutants lose viability at G0-entry and are unable to maintain long-term quiescence. We obtained dcr1Δ G0 suppressors, which mapped to genes involved in chromosome segregation, RNA polymerase-associated factors, and heterochromatin formation. We propose a model in which RNAi promotes RNA polymerase release in cycling and quiescent cells: (i) RNA pol II release mediates heterochromatin formation at centromeres allowing proper chromosome segregation during mitotic growth and G0-entry, and (ii) RNA pol I release prevents heterochromatin formation at rDNA during quiescence maintenance. Our model may account for the co-dependency of RNAi and H3K9 methylation throughout eukaryotic evolution.
Main Text
In nature, most cells exist in a non-dividing state (the ‘G0’ phase of the cell cycle). This state is commonly referred to as ‘quiescent’ when cells are still metabolically active and able to re-enter the cell cycle given the appropriate signal. Examples of quiescent cells include stem cell niches (1), neuronal progenitor cells (2) and memory T cells (3). Yeasts and other micro-organisms are also able to enter a reversible quiescent state in response to environmental cues (4), and in several pathogenic species this state allows persistence of infection in the host (5). Despite their tremendous importance, the molecular pathways involved in quiescence entry, maintenance, and exit, are not well understood. Given that long-term stability and reversibility are hallmarks of quiescence, epigenetic mechanisms are likely involved, and we are studying their contribution to quiescence, using the fission yeast Schizosaccharomyces pombe as a model.
In S. pombe, synchronized entry into the reversible quiescent state is induced upon nitrogen starvation in the absence of a mating partner. Under these conditions the rod-shaped cells divide twice without growth and differentiate into quiescent G0 cells, which retain viability and the ability to re-enter the cell cycle for more than 1 month (4). G0 cells have a characteristic transcriptome (6, 7), morphology and metabolic response (4, 8, 9). RNAi-guided heterochromatic silencing is a major epigenetic pathway in S. pombe, which has one copy of each of the key enzymes involved: Dicer (Dcr1), Argonaute (Ago1) and RNA-dependent RNA polymerase (Rdp1). Transcription of pericentromeric repeats on both strands by RNA polymerase II is thought to generate partially double-stranded RNAs, which are cleaved by Dcr1 to small RNAs (21–23nt). Small RNAs bound to Ago1 are targeted to homologous transcripts, and lead to the recruitment of Rdp1 to generate more template dsRNA. Subsequent engagement of the CLRC/Rik1 silencing complex, which includes the histone H3 lysine 9 (H3K9) methyltransferase Clr4, and the H3K4 demethylase Lid2, silences heterochromatic transcripts and reporter genes. This process is tightly regulated during S-phase in order to ensure heterochromatin inheritance (10, 11), and is coupled to the DNA replication fork, which promotes heterochromatin spreading (12). RNAi mutants are viable but lose silencing of the pericentromeric repeats (13) resulting in chromosome segregation defects (14).
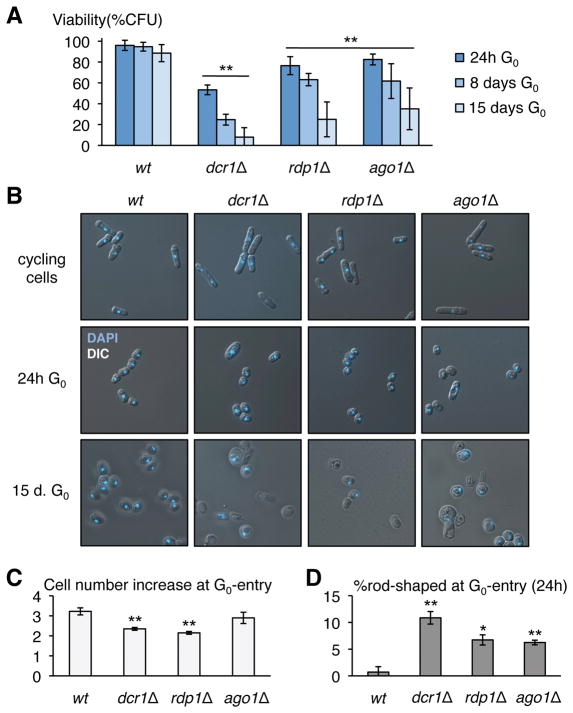
We induced prototrophic wild-type, dcr1Δ, ago1Δ and rdp1Δ strains into G0 by nitrogen starvation, and determined viability at different time-points up to 15 days (15). While the wild-type strain retained 88.5% viability after 15 days of G0, dcr1Δ viability was reduced to 53.3% at 24h and then gradually decreased to 7.9% at 15 days. rdp1Δ and ago1Δ cells had comparable, though less severe viability defects (Fig. 1A; Fig. S1; Fig. S2). We distinguished two different phases in the loss of viability of RNAi mutant strains: initial loss, which is characterized either by cells failing to enter G0 or by cells unable to exit G0, and subsequent gradual loss of viability over time, indicative of a defect in quiescence maintenance. In accordance with a major defect in G0-entry, there was an increase in the number of cells retaining a ‘rod-like’ shape (Fig. 1BD), resulting in a lower number of cell divisions after nitrogen-starvation (Fig. 1C). Mis-segregated DNA at the first division was readily detected by DAPI staining. Furthermore, while wild-type G0-exiting cells displayed a ‘schmoo’ morphology at the first S-phase (4), this was only observed for viable RNAi mutant cells (that form colonies when micro-isolated on rich medium) and not for the majority of inviable cells, supporting the idea that initial loss of viability reflects an inability to properly enter the quiescent state. Over time, refraction-negative (dead) and mis-shaped cells constituted an increasing proportion of RNAi defective G0 cells (Fig. 1B). Interestingly, the dead cells were morphologically heterogeneous, by comparison with, for example, tdp1Δ mutants in DNA repair (16), indicating that the loss of viability may be a complex effect.
Figure 1. RNAi mutants lose viability in G0.
(A) Loss of viability at both G0-entry and during quiescence maintenance in prototroph dcr1Δ, rdp1Δ and ago1Δ mutants (n=5 biological replicates for wild-type, n=6 for dcr1Δ and rdp1Δ, n=7 for ago1Δ). ** indicates a statistically significant difference (p<0.01, t-test) between each mutant and wt for all time-points, and between time-points 24h and 15 days (B) Microscopic observation of DAPI-stained RNAi mutants reveals an increased proportion of cells retaining a rod-shape in early G0 (24h). After 15 days, while wild-type G0 cells look uniform, RNAi mutants display a variety of morphological defects and many dead refraction-negative and/or DAPI-negative cells. Upon G0-entry (C) RNAi mutants have a reduction in the increase in cell number found in wild type cells during the initial two divisions (24h G0; n=2; ** p<0.01), and (D) an increase in the number of cells staying rod-shaped (24h G0; n=2; * p<0.05, ** p<0.01). All error bars indicate standard deviation; n indicates biological replicates.
To assess whether the G0 phenotype of RNAi mutants was due to the loss of pericentromeric heterochromatin, we assayed swi6HP1Δ, chp2HP1Δ, and clr4Δ for viability in G0. We observed that chp2Δ had a wild-type G0 phenotype, and that swi6Δ and clr4Δ strains were affected only during G0-entry and subsequently viable (Fig. S3ABC). In parallel, we treated the wild-type strain with trichostatin A (TSA) for 24 hours before G0-induction; this treatment results in loss of heterochromatin, but only results in a minor loss of viability in G0 (Fig. S3DE). These results indicate that whereas the G0-entry loss of viability was common to RNAi and heterochromatin mutants, the loss of viability in quiescence maintenance of RNAi mutants was not due to loss of pericentromeric heterochromatin. The two distinct phases of viability loss therefore rely on two different mechanisms.
To further explore the function of RNAi during quiescence maintenance, we established a suppressor screen based on the sharp decrease in viability of dcr1Δ in G0, as follows: a population of dcr1Δ cells was induced into G0 by nitrogen-starvation (EMM-N media) for 1–3 days, and was then put back into the cell cycle in rich medium (YES media) for ~4–7 generations. The population was then re-induced into G0, and the process repeated, alternating between quiescence and growth for up to 20 cycles (Fig. 2A). Cells carrying a spontaneous suppressor mutation have a fitness advantage at every cycle, and are progressively enriched in the culture. We tested the protocol for proof-of-concept using a known suppressor (15). We subsequently performed the suppressor screen on >50 independent populations, and we identified 13 dcr1Δ independent suppressors by whole genome sequencing (17) and validation in backcrosses (15) (Fig. S4; Fig. S5). The suppressors fell into three main classes (Fig. 2B):
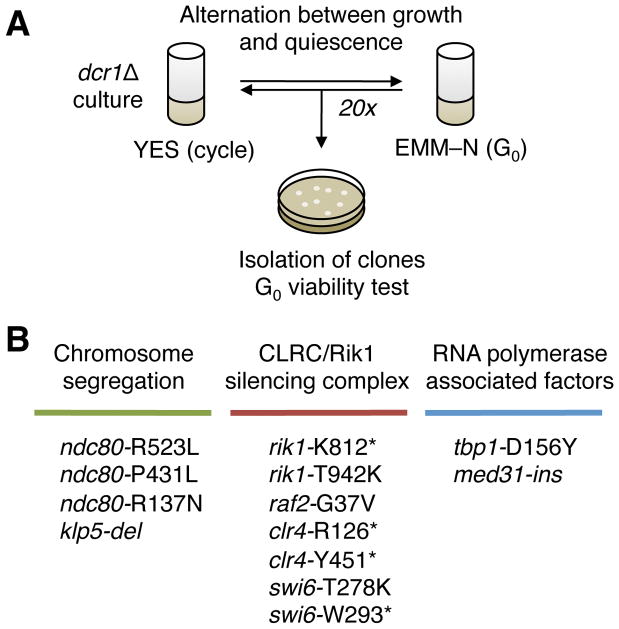
Figure 2. Design of a G0 suppressor screen.
(A) Experimental design of the suppressor screen: a dcr1Δ population is alternated between the cell cycle (in rich medium, YES) and quiescence (in nitrogen-deprived medium, EMM-N) every 1 to 3 days. Spontaneous suppressors, by their relative fitness advantage compared to the parental strain, get enriched during the alternations. After up to 20 alternations, individual clones are isolated and re-assayed in G0 to check for suppression. (B) Summary of mutations present in 13 isolated dcr1Δ G0 suppressors showing three classes of mutants, in chromosome segregation, the CLRC/Rik1 complex and Swi6HP1, and RNA polymerase-associated factors. Each mutation in the list arose as an independent suppressor.
Mutants in chromosome segregation and kinetochore assembly: ndc80-R523L, ndc80-P431L, ndc80-R137N, klp5-del. The Ndc80 mutants affect each of the functional domains of the protein: R137 is in the globular domain, P431 in a Dis1-interaction site (18), R523 is in the second long tail of unknown function. The klp5-del allele consists of a short deletion with a frameshift at codon 772, resulting in a different 772–814 C-terminus.
Mutants in the CLRC/Rik1 complex and HP1: rik1-K812*, raf2-G37V, swi6-W293*, swi6-T278K, rik1-T942K, clr4-R126*, clr4-Y451*.
Mutants in RNA polymerase-associated factors: tbp1-D156Y, med31-ins. The insertion in med31-ins consists of the replacement of Phe23 by the sequence LVRIC, without frameshift. This phenylalanine is highly conserved from budding yeast Soh1pMED31 to human MED31p (19).
Class (i) suppressors fell in the essential kinetochore Ndc80 subunit and the kinesin-8 ortholog Klp5, which promotes microtubule catastrophe (20). These mutants might be expected to suppress chromosome mis-segregation defects of RNAi mutants during the two accelerated divisions that S. pombe cells undergo at the entry of quiescence (Fig. 1B), as well as in mitosis (14) and in meiosis (21). Consistent with this hypothesis, we found that ndc80-R523L, ndc80-P431L, ndc80-R137N and klp5-del are TBZ-resistant, similarly to klp5Δ (20), and these strains have less rod-shaped cells after 24h in G0 in a dcr1Δ background than the single-mutant dcr1Δ (Fig. S6AB). Consistently, the lower suppression by ndc80-R137N may indicate that the Ndc80 loop may play a more important role in the suppression, potentially via Dis1 by stabilizing the spindle-microtubule attachment (18). We were additionally able to phenocopy the mis-segregation phenotype in wild-type cells by treatment with TBZ during G0-entry (24h) (Fig. S6E). Chromosome mis-segregation in RNAi mutants is caused by loss of pericentromeric heterochromatin, and we predicted that restoring pericentromeric heterochromatin in RNAi mutants (22–24) should rescue the G0-entry phenotype. Mutations in the H3K14-acetyltransferase Mst2 complex reduce pericentromeric transcription, allowing heterochromatin to be maintained in a RNAi-independent manner (22) and mst2Δ indeed strongly rescued the G0-entry phenotype of RNAi mutants (Fig. S6F).
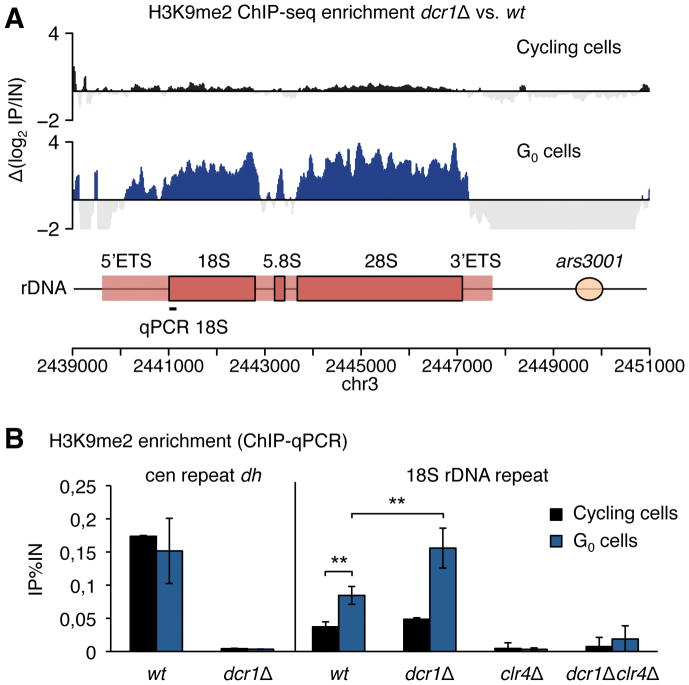
Class (ii) suppressors suppress the quiescence maintenance defect of dcr1Δ (Fig. S5). All suppressors in the CLRC/Rik1 complex lose silencing of the pericentromeric dg/dh repeats, and are therefore defective in heterochromatin formation; consistently raf2-G37V is mutated in the RFTS (Replication Foci Targeting Sequence) (25) while swi6-T278K and swi6-W293* are mutated in the chromoshadow domain. We subsequently found that clr4Δ and swi6Δ suppressed dcr1Δ, ago1Δ and rdp1Δ, as did rik1Δ (Fig. S7) indicating that H3K9-methylation may be responsible for the progressive loss of viability during G0 in the absence of RNAi. To investigate this further, we performed ChIP-seq of H3K9me2 in G0 wild-type and dcr1Δ cells (Fig. 3A; Fig. S8; Fig. S9). H3K9me2 localization in wild-type was prevalent at the centromere and subtelomeres, but not at the recently described transient heterochromatic islands found along chromosome arms (Fig. S9), consistent with the observation that these islands disappear following nitrogen-starvation (26). The dcr1Δ strain showed near-complete loss of centromeric H3K9me2 (Fig. S8C), as well as at subtelomeric tlh loci (Fig. S8B); however, a striking accumulation of H3K9me2 was observed at the rDNA locus (Fig. 3A; Fig. S8A). Intriguingly, H3K9me2 accumulation matched the transcribed region of rDNA (Fig. 3A). H3K9me2 accumulation at the rDNA was also found in wild-type G0 cells at lower levels and depends on the H3K9 methyltransferase Clr4 (Fig. 3B). This H3K9me2 accumulation continues at longer G0 times in both wild-type and dcr1Δ cells, further increasing the difference between both genotypes as cells enter deeper into quiescence.
Figure 3. Quiescence maintenance of dcr1Δ results in rDNA heterochromatinization by H3K9me.
(A) H3K9me2 ChIP-seq enrichment in dcr1Δ vs. wt cells. In G0 cells, but not cycling cells, dcr1Δ strongly accumulates H3K9me2 at the rDNA locus (n=2 biological replicates, cycling cell data from (31)). (B) Validation by H3K9me2 ChIP-qPCR in wild-type and dcr1Δ cycling and G0 cells shows a stronger rDNA H3K9me2 increase in G0 cells in dcr1Δ as compared to wild-type G0 cells (n≥3 biological replicates; ** p<0.01, t-test).
To ascertain that the suppression effect of CLRC/Rik1 mutants is mediated by loss of H3K9me2 and not through a potential new target of the complex, we created a set of prototroph strains to assay histone H3 mutants in G0 (hht3Δ hht1-K9R hht2-K9R), keeping viability for over a week (15) (Fig. S10A). The dcr1Δhht3Δ hht1-K9R hht2-K9R strains rescued G0 viability (Fig. S10B); and interestingly, the reduction of H3K9me2 in dcr1Δhht3Δ hht1-K9R and in dcr1Δhht3Δ hht2-K9R strains alone was also sufficient to recover G0 viability (Fig. S10BC). These results confirm that H3K9me2 is central to the quiescence maintenance defect of dcr1Δ cells. We then attempted to increase the levels of H3K9me at the rDNA to see if the defect would be enhanced. As the recruitment factor for Clr4 at rDNA in G0 is unknown, we decided instead to increase global Clr4 levels in G0. We replaced the promoter of clr4 at its endogenous location by the promoter of urg1 (Fig. S11A), which is induced not only by uracil but also by nitrogen-starvation (15, 27). The resulting purg1-clr4 strain entered G0 with wild-type viability but over-accumulated H3K9me2 at rDNA and quickly lost viability specifically in quiescence maintenance (Fig. S11BC); in the dcr1Δ purg1-clr4 background, the H3K9me2 rDNA accumulation was increased even further, resulting in very low viability, almost complete after a week in G0 (Fig. S11BC). However as the accumulation of H3K9me2 is not specific to the rDNA in these strains, we cannot exclude that other genomic locations participate in the loss of viability in this assay, via silencing of essential genes.
The class (iii) suppressors also suppressed the quiescence maintenance defect (Fig. S5). These suppressor mutations lay in RNA polymerase factors: Tbp1TBP (in RNA pol I, II and III) (28) and the Mediator complex, a co-regulator of RNA pol II, which may also play a role in transcription by other RNA polymerases (29). These genes act upstream of rDNA heterochromatinization, as H3K9me2 at rDNA is reduced in dcr1Δ med31-ins and dcr1Δ tbp1-D156Y G0 cells (Fig. 4H). Interestingly, the strong suppressor med31-ins also suppresses G0-entry and TBZ-sensitivity (Fig. S5), and might therefore additionally act at centromeric heterochromatin, similarly to mst2Δ and the Mediator subunit pmc2Δ (22). In support of this idea, distinct subunits of Mediator have been shown to play a role in silencing (30) and anti-silencing (22). In dividing cells, Dicer is required for efficient RNA polymerase II termination at various genomic locations including centromeres, tDNAs, and rDNA, and stalled RNA pol II accumulates in dcr1Δ at these loci (31) (Fig. S12). We performed ChIP-seq using an antibody directed against RNA pol II (unphosphorylated CTD) in wild-type and dcr1Δ G0 cells, but in contrast to the situation in cycling cells (31), no significant increase was detected at the rDNA in dcr1Δ G0 cells (Fig. S12). ChIP-qPCR using antibodies against the phosphorylated CTD pol II (S2P, S5P) did not show significant differences either. Small RNA sequencing revealed that, in G0 cells, centromeric small RNAs were strongly reduced at pericentromeric regions (Fig. S14AB), as well as other siRNAs, consistent with their production in the G1- and S-phases of the cell cycle (32). Furthermore, in contrast to the situation in cycling cells (31), no small RNA antisense to the rRNA were found (Fig. S14C), and we did not detect any new loci of Dicer-dependent small RNA production in G0 cells (15).
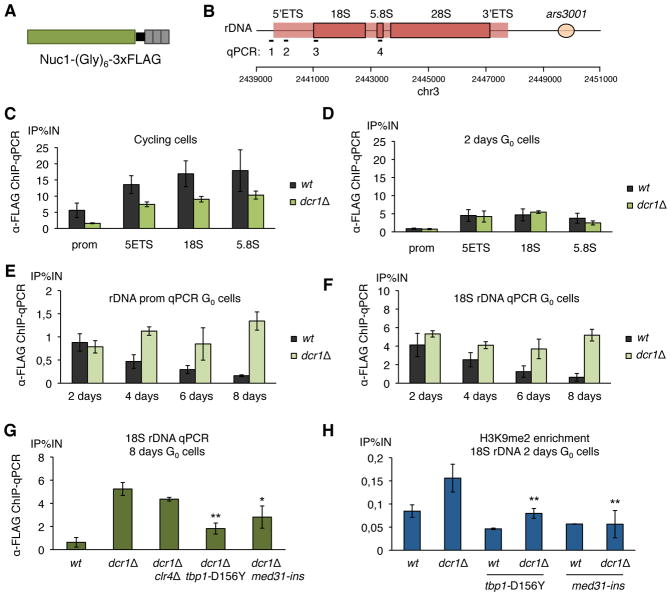
Figure 4. dcr1Δ mutants are defective in RNA polymerase I release in quiescence maintenance.
(A) The main subunit of RNA polymerase I Nuc1RPA190 was tagged with a (Gly)6 linker and 3xFLAG. (B) Probe location for ChIP-qPCRs: 1: rDNA promoter, 2: 5′ETS, 3: 18S, 4: 3′ETS. (C) RNA polymerase I enrichment at the rDNA was assayed by ChIP-qPCR using anti-FLAG antibody in the nuc1-(Gly)6-FLAG background, showing an higher occupancy in wild-type than dcr1Δ mutants in cycling cells, due to the reduced growth rate of dcr1Δ, and (D) a similar occupancy in early G0 cells (n=2 biological replicates). (E, F) RNA polymerase I enrichment at 18S rDNA over time spent in G0 (2 to 8 days) shows a reduction in wild-type cells, but dcr1Δ cells fail to release RNA pol I (n≥2 biological replicates). (G) The class (iii) suppressors, dcr1Δ tbp1-D156Y and dcr1Δmed31-ins, but not the class (ii) suppressor dcr1Δclr4Δ, significantly decrease RNA polymerase I occupancy at rDNA in 8 days G0 cells. (n=2 biological replicates, * p<0.05, ** p<0.01, t-test) (H) The accumulation of H3K9me2 at rDNA repeats, assayed by ChIP-qPCR, is significantly reduced in class (iii) suppressors (n≥3 biological replicates, ** p<0.01, t-test).
We therefore decided to assay RNA polymerase I, which is responsible for rDNA transcription. RNA pol I occupancy at rDNA, assayed by ChIP-qPCR of tagged Nuc1RPA190 (15) (Fig. 4A), was lower than wild type in cycling dcr1Δ cells, likely due to the slightly reduced growth rate of dcr1Δ due to accumulation of DNA damage (10, 31) (Fig. 4C; Fig. S15B). There was no significant difference in RNA pol I occupancy between wild-type and dcr1Δ initially in G0, but strikingly we saw that RNA pol I occupancy is further reduced as cells enter into deeper quiescence, and that dcr1Δ cells do not have this reduction, indicative of a failure to release RNA pol I (Fig. 4DEF). This accumulation is detected at the 5′ETS, 18S, 5.8S, 28S repeats, as well as at the very end of the 3′ETS next to the termination site Ter3. Strikingly, the accumulation of RNA pol I was not reduced in dcr1Δclr4Δ, suggesting that failure to release RNA pol I lies upstream of H3K9me accumulation. In contrast, RNA pol I occupancy was strongly reduced in dcr1Δtbp1-D156Y, and slightly reduced in dcr1Δmed31-ins, providing an explanation for their suppression effect, as well as for their reduced accumulation of H3K9me2 (Fig. 4GH).
The failure to release RNA pol I at rDNA in G0 may result in DNA damage, similar to the failure to release RNA pol II at centromeres and rDNA in the cell cycle (31). As G0 cells do not rely on HR for DNA repair, rad22-YFP foci are not formed in quiescent 1c cells (8), although we observed that they strongly accumulate upon G0-exit of dcr1Δ (Fig. S13BF). We therefore used H2A and H2A-S129phos (equivalent to γH2AX) antibodies to assay DNA damage directly in G0 cells by both ChIP-seq and ChIP-qPCR in wt and dcr1Δ, and observed an increase in rDNA in dcr1Δ cells (Fig. S13AC). The H2A-S129phos/H2A ratio was reduced in suppressors rescuing the RNA pol I defect, dcr1Δtbp1-D156Y and dcr1Δmed31-ins (Fig. S13C); the absence of reduction in dcr1Δclr4Δ suggests that DNA damage accumulation is correlated to stalled RNA pol I at the rDNA, but that the primary cause of death is not DNA damage itself but rather the accumulation of H3K9me2.
We further explored the distinction between RNA pol II release during S-phase and RNA pol I release during G0 by constructing a dcr1Δreb1Δ double mutant strain: Reb1TTFI is a factor that participates in RNA pol I termination and forces the direction of replication to occur in the same direction as transcription at the rDNA, thus avoiding collisions between DNA and RNA polymerase (33). While reb1Δ cells are viable, the accumulation of RNA pol II in dcr1Δ may increase the need for Reb1 in order to avoid polymerase collisions and subsequent DNA damage at the rDNA. In accordance with this hypothesis, dcr1Δreb1Δ strains show a strong negative epistatic interaction in dividing cells resulting in extremely poor growth (Fig. S15B); however, their viability is unaffected in G0, and is similar to the dcr1Δ single-mutant (Fig. S15A).
If RNA polymerase I is responsible for the quiescence maintenance defect of dcr1Δ, we reasoned that deletion of non-essential subunits of RNA pol I may destabilize it, and therefore, similarly to class (iii) suppressors, would allow RNAi-independent release. We therefore assayed rpa12Δ mutants in G0. Rpa12 is a non-essential subunit of RNA pol I required for polymerase termination (34), and strikingly, we found that dcr1Δrpa12Δ suppressed the quiescence maintenance defect (Fig. S15C), although it does not affect H3K9me2 levels at the rDNA. We were unfortunately unable to assay RNA pol I levels in these cells as nuc1-FLAG rpa12Δ double-mutants are inviable, most likely because RNA pol I becomes too unstable in this background. Thus, Rpa12 can be considered a “class iv” suppressor. As the suppressor screen is not saturated yet, we predict that sequencing many more of the >50 suppressors we obtained will yield interesting alleles in RNA polymerase-associated factors in both class iii (general transcription) and iv (RNA pol I). We propose in particular that the specific recruitment factor responsible for rDNA H3K9me2 increase in G0 could be a subunit of RNA pol I itself, providing a potential explanation as to why it would be difficult to recover such a suppressor.
We have identified RNAi as a novel essential regulator of quiescence in S. pombe. RNAi both promotes heterochromatin formation at centromeres allowing proper chromosome segregation during G0-entry, and prevents heterochromatin formation at the rDNA locus during quiescence maintenance (Fig. S16). At G0-entry, mis-segregation results in cell death, and can be suppressed by restoring heterochromatin or by strengthening segregation. In dividing cells RNAi is required to release RNA pol II from rDNA and from pericentromeric heterochromatin, and in RNAi mutants pol II must be removed from stalled replication forks by homologous recombination repair (HR) (10), resulting in a severe reduction in rDNA copy number (31). G0 cells possess a 1c DNA content and therefore use non-homologous end-joining (NHEJ) instead of HR for repair (8). Consistently, rDNA copies are not lost in RNAi mutants during quiescence. During quiescence maintenance, RNAi is required to release RNA pol I from rDNA. The failure to release RNA pol I results in over-recruitment of rDNA silencing factors, possibly by RNA pol I itself; in mammalian cells (35), rRNA interacts with SUV39H1 via NML (36) and RNA pol I interacts with the G9a methyltransferase (37). Stalled RNA pol I results in an over-accumulation of H3K9me2 at rDNA, and DNA damage, resulting in loss of viability of these cells, similarly to when essential genes are silenced by H3K9me2 (38). These defects can be suppressed by mutants in the silencing CLRC/Rik1 complex, the effector Swi6HP1, and specific mutants in RNA pol I non-essential subunits. Both RNA pol II and RNA pol I defects can be suppressed by specific alleles in the key transcription TATA Binding Protein (TBP) and Mediator (Med) complexes.
Thus each class of suppressors illuminates key roles of RNAi in quiescent cells: in chromosome segregation, in heterochromatin formation and spreading, and in transcription. As spontaneous mutations in essential genes are much more rare, we obtained relatively few class (iii) suppressors, and none from class (iv). We expect that saturation of the G0 screen may uncover the precise components of RNA polymerase I, or rDNA-associated factors, involved in recruiting the CLRC/Rik1 complex for G0 rDNA silencing, and more ‘master regulators’ genes which, like Dicer and TBP, regulate transcription by all RNA polymerases.
The rDNA locus plays a central role in ageing (39) and H3K9me is a hallmark of rDNA silencing in plants (40) as well as in mammalian cells (36). The role of RNAi at rDNA may also be evolutionarily conserved; in mammalian cells Dicer physically associates with rDNA (41), while in Candida albicans Dicer is involved in pre-rRNA processing by cleaving the 3′ETS (42). In Neurospora crassa quelling (RNAi) targets rDNA (43), and qiRNAs are generated at the rDNA locus by Dicer when cells are treated with alkylating agents or with hydroxyurea (44). Furthermore, there is evidence that during evolution RNAi and heterochromatin proteins (H3K9me, HP1) have been lost together, a loss which has happened independently in distinct fungal lineages such as budding yeasts of the subphylum Saccharomycotina (45), Ustilago maydis (Basidiomycota, subphylum Ustilagomycotina) (46), and the human pathogen Pneumocystis jirovecii (Ascomycota, subphylum Taphrinomycotina) (47). In budding yeasts, the loss of RNAi is correlated with the ability to acquire Killer RNA viruses (48). Our results provide a possible explanation for a strong selective pressure to lose H3K9me-based heterochromatin upon RNAi loss, accounting for their co-dependency in eukaryotic evolution.
Our finding sheds light on the anecdotal observation that some mutants in S. pombe, like dcr1Δ, do not keep well in long-term storage at low temperatures. We have found that repeated thawing/freezing cycles can select for spontaneous suppressors, which are quite widespread in laboratory strains of RNAi mutants. RNAi mutants also fail to differentiate into dormancy in asexual and sexual spores in Cryptococcus neoformans, an important human basidiomycete pathogen (49), and in the zygomycete Mucor circinelloides (50). In metazoans, such as Cænorhabditis elegans (51) and Drosophila melanogaster (52), Dicer mutants typically affect germ cells, which also spend long periods in quiescence. Given the importance of quiescence in the life cycle of unicellular as well as multicellular organisms, it is likely that epigenetic pathways will be found to be essential in neurons, germ cells and cancer stem cells, which can spend many years in a quiescent state. Uncovering the mechanisms underlying the epigenetic regulation of quiescence thus opens a promising new field of study.
Supplementary Material
Acknowledgments
This work was funded by NIH grant R01 GM076396-08, by the Howard Hughes Medical Institute and Gordon & Betty Moore Foundation Plant Biology Investigator Program [to R.M.], and by the Agence Nationale de la Recherche ANR-13-BSV8-0018 [to B.A.]. The authors acknowledge support from the Chaire Blaise Pascal (Fondation de l’École Normale Supérieure, France). We would like to thank the Cold Spring Harbor Laboratory Woodbury Sequencing Facility. We would like to thank everyone at the fission yeast database PomBase, as its resources have been extremely helpful for research in our lab. We would also like to thank the reviewers for their helpful comments and suggestions. The raw data of the next-generation sequencing libraries reported in this paper are available at the Sequence Read Archive (SRA) database, accession number SRP087488 (BioProject PRJNA341984). The authors report no conflicts of interest.
Footnotes
References and Notes
- 1.Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Codega P, et al. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okhrimenko A, et al. Human memory T cells from the bone marrow are resting and maintain long-lasting systemic memory. Proc Nat Acad Sci USA. 2014;111:9229–9234. doi: 10.1073/pnas.1318731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su SS, Tanaka Y, Samejima I, Tanaka K, Yanagida M. A nitrogen starvation-induced dormant G0 state in fission yeast: the establishment from uncommitted G1 state and its delay for return to proliferation. J Cell Sci. 1996;109:1347–1357. doi: 10.1242/jcs.109.6.1347. [DOI] [PubMed] [Google Scholar]
- 5.Ritterhaus ES, Baek SH, Sassetti CM. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe. 2013;13:643–651. doi: 10.1016/j.chom.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimanuki M, et al. Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells. 2007;12:677–692. doi: 10.1111/j.1365-2443.2007.01079.x. [DOI] [PubMed] [Google Scholar]
- 7.Marguerat S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mochida S, Yanagida M. Distinct modes of DNA damage response in S. pombe G0 and vegetative cells. Genes Cells. 2006;11:13–27. doi: 10.1111/j.1365-2443.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 9.Takeda K, et al. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc Nat Acad Sci USA. 2010;107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castel ES, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F, Martienssen RA, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 14.Volpe TA, et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supplementary materials on Science Online.
- 16.Ben Hassine S, Arcangioli B. Tdp1 protects against oxidative DNA damage in non-dividing fission yeast. EMBO J. 2009;28:632–640. doi: 10.1038/emboj.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irvine DV, et al. Mapping epigenetic mutations in fission yeast using whole-genome next-generation sequencing. Genome Res. 2009;19:1077–1083. doi: 10.1101/gr.089318.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linder T, Gustafsson CM. The Soh1/MED31 protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae Mediator. J Biol Chem. 2004;279:49455–49459. doi: 10.1074/jbc.M409046200. [DOI] [PubMed] [Google Scholar]
- 20.West RR, Malmstrom T, Troxell CL, McIntosh JR. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol Biol Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Nat Acad Sci USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy BD, et al. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 2011;25:214–219. doi: 10.1101/gad.1993611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol. 2011;18:1132–1138. doi: 10.1038/nsmb.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tadeo, et al. Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev. 2013;27:2489–2499. doi: 10.1101/gad.226118.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White SA, et al. The RFTS domain of Raf2 is required for Cul4 interaction and heterochromatin integrity in fission yeast. PLoS One. 2014;9:e104161. doi: 10.1371/journal.pone.0104161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zofall M, et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watt S, et al. urg1: a uracil-regulatable promoter system for fission yeast with short induction and repression times. PLoS One. 2008;3:e1428. doi: 10.1371/journal.pone.0001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamada M, Huang Y, Lowe TM, Maraia RJ. Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast. Mol Cell Biol. 2001;21:6870–6881. doi: 10.1128/MCB.21.20.6870-6881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlsten JO, Zhu X, López MD, Samuelsson T, Gustafsson CM. Loss of the Mediator subunit Med20 affects transcription of tRNA and other non-coding RNA genes in fission yeast. Biochim Biophys Acta. 2016;1859:339–347. doi: 10.1016/j.bbagrm.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Thorsen M, Hansen H, Venturi M, Homlberg S, Thon G. Mediator regulates non-coding RNA transcription at fission yeast centromeres. Epigenetics Chromatin. 2012;5:19. doi: 10.1186/1756-8935-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castel SE, et al. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell. 2014;159:572–583. doi: 10.1016/j.cell.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kloc A, Zaratiegui M, Nora E, Martienssen RA. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez-Gorostiaga A, López-Estraño C, Krimer DB, Schvartzman JB, Hernández P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol. 24:398–406. doi: 10.1128/MCB.24.1.398-406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prescott EM, et al. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc Nat Acad Sci USA. 2004;101(16):6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voit R, Grummt I. The RNA polymerase I transcription machinery. In: Olson M, editor. The Nucleolus vol 15 of Springer Protein Reviews. 2011. [Google Scholar]
- 36.Murayama A, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 37.Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Reddy BD, Jia S. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. Elife. 2015;4 doi: 10.7554/eLife.06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganley AR, Kobayashi T. Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 2014;14:49–59. doi: 10.1111/1567-1364.12133. [DOI] [PubMed] [Google Scholar]
- 40.Pontvianne F, et al. Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 2012;26:945–957. doi: 10.1101/gad.182865.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS One. 2010;5:e12175. doi: 10.1371/journal.pone.0012175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernstein DA, et al. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. Proc Nat Acad Sci USA. 2012;109:523–528. doi: 10.1073/pnas.1118859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cecere G, Cogoni C. Quelling targets the rDNA locus and functions in rDNA copy number control. BMC Microbiol. 2009;9:44. doi: 10.1186/1471-2180-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z, et al. Homologous recombination as a mechanism to recognize repetitive DNA sequences in an RNAi pathway. Genes Dev. 2013;27:145–150. doi: 10.1101/gad.209494.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drinnenberg IA, et al. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laurie JD, et al. Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell. 2012;24:1733–1745. doi: 10.1105/tpc.112.097261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cissé OH, Pagni M, Hauser PM. Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol Evol. 2014;6:1938–1948. doi: 10.1093/gbe/evu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, et al. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 2010;24:2566–2582. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Haro JP, et al. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryot Cell. 2009;8:1486–1497. doi: 10.1128/EC.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ketting RF, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012 arXiv:1207.3907v2. [Google Scholar]
- 56.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 59.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.