Abstract
Germ cell tumors (GCTs) are a heterogeneous group of tumors occurring in gonadal and extragonadal locations. GCTs are hypothesized to arise from primordial germ cells (PGCs), which fail to differentiate. One recently identified susceptibility loci for human GCT is PR (PRDI-BF1 and RIZ) domain proteins 14 (PRDM14). PRDM14 is expressed in early primate PGCs and is repressed as PGCs differentiate. To examine PRDM14 in human GCTs we profiled human GCT cell lines and patient samples and discovered that PRDM14 is expressed in embryonal carcinoma cell lines, embryonal carcinomas, seminomas, intracranial germinomas and yolk sac tumors, but is not expressed in teratomas. To model constitutive overexpression in human PGCs, we generated PGC-like cells (PGCLCs) from human pluripotent stem cells (PSCs) and discovered that elevated expression of PRDM14 does not block early PGC formation. Instead, we show that elevated PRDM14 in PGCLCs causes proliferation and differentiation defects in the germline.
Keywords: Germ cell tumor, PRDM14, Cell differentiation, Primordial germ cell, Proliferation
1. Introduction
Germ cell tumors (GCTs) make up a heterogeneous group of tumors, encompassing five histologic subtypes. These are, germinomas, embryonal carcinoma, yolks sac, choriocarcinoma and teratomas. GCTS occur in gonadal and extragonadal locations, with extragonadal occurring primarily along the midline in the sacrum, mediastinum and pineal and/or suprasellar region of the brain. GCTs occur in a bimodal fashion with pediatric patients being most affected from birth to 3 years of age, and then again in adolescences into young adulthood. Although considered rare, GCTs account for 15% of malignancies in the adolescent to young adult population (15–40 years), with testicular GCTs being the most common malignancy of males in this age group (Calaminus and Joffe, 2016).
GCTs are hypothesized to arise from a common cell of origin, the embryonic progenitors of gametes called primordial germ cells (PGCs). Extragonadal GCTs are hypothesized to arise from PGCs that migrated inappropriately (Schmoll, 2002). PGCs are specified very early in the peri-implantation human embryo, between 2 and 3 weeks post-fertilization (De Felici, 2013). After specification, PGCs migrate through the hindgut, into the dorsal mesentery and begin colonizing the genital ridges starting at week 4–5 post-fertilization (De Felici, 2013). Specified human PGCs that are negative for the gonadal-stage germline markers VASA and deleted in azoospermia like (DAZL) are referred to as “early PGCs”, whereas DAZL and VASA positive PGCs that have migrated into the dorsal mesentery and genital ridges are called “late PGCs” (Gkountela et al., 2015; Gkountela et al., 2013; Guo et al., 2015; Irie et al., 2015; Chen et al., 2017a). PGCs in the gonads are classically referred to as gonocytes. In humans, the PGC stage of germline development ends at around 8–10 weeks post-fertilization (De Felici, 2013). After this, PGCs in the gonad start to advance in differentiation, and transition into oogonia or pro-spermatogonia which ultimately become female and male gametes, respectively.
Much of what is known about mammalian PGC development comes from work in mouse models. However, recent studies revealed critical species-specific differences between mouse and human PGC development, including differences in the transcription factor network (Irie et al., 2015; Tang et al., 2016), and most importantly many of the GCT types found in humans, such as seminomas, yolk sac tumors, and intracranial germ cell tumors, do not occur in mouse models of the disease (Heaney et al., 2012; Irie et al., 2014). Therefore, understanding the cell and molecular origins of GCTs in humans requires developing new human cell-based models. Given that PGCs are embryonic progenitors that differentiate into more mature germline cell types at the end of the first trimester, it is not possible to isolate these cells from children or young adults at risk for GCT formation. Instead, we hypothesize that the use of pluripotent stem cells (PSC), and the generation of human PGC-like cells (PGCLCs), will allow for further investigation into the mechanisms of GCT formation in humans.
In a recent genome-wide association study of testicular and intracranial GCTs, the transcription factor PRDM14 was identified as being a susceptibility locus for this disease (Ruark et al., 2013; Terashima et al., 2014). In mice, Prdm14 is critical for PGC formation, being highly expressed from the time of specification until the end of the PGC period, which is embryonic (E) day E13.5 in the mouse (Nakaki and Saitou, 2014; Yamaji et al., 2008; Kurimoto et al., 2008). A homozygous null mutation in Prdm14 in mice causes loss of PGCs by E12.5 due to a failure of Prdm14 mutant PGCs to undergo germline reprogramming (Yamaji et al., 2008). In humans, the function of PRDM14 in PGCs is unclear. RNA-Sequencing and immunofluorescence studies have found that human PGCs express low levels of PRDM14 (Gkountela et al., 2015; Guo et al., 2015; Irie et al., 2015; Tang et al., 2015), and a knockdown of PRDM14 has no effect on human PGCLC differentiation (Sugawa et al., 2015). Combined, these results suggest that the role of PRDM14 in human PGCs may be different from mice, with one hypothesis being the repression of PRDM14 is required for PGC differentiation.
In the current study, we used human GCT tissue samples, and the differentiation of PGCLCs from human PSC to address the hypothesis that PRDM14 is expressed in human GCTs, and that over expression of PRDM14 alters PGC differentiation.
2. Materials and methods
2.1. Cell lines and cell culture
Primed hESC lines were cultured on mitomycin C-inactivated mouse embryonic fibroblasts (MEFs) in hESC media, per Pastor et al. (2016) with the addition of 50 ng/mL primocin (InvivoGen, ant-pm-2). All hESC lines were split every 7 days with Collagenase type IV (GIBCO, 17104-019). All hESC lines used in this study are registered with the National Institute of Health Human Embryonic Stem Cell Registry and are available for research use with NIH funds. Specifically, the following hESC lines were used in this study: UCLA2 (46XY), UCLA6 (46XY). The derivation and basic characterization of UCLA2 and 6 were previously reported (Diaz Perez et al., 2012). Experiments were performed between passage 15–25, two passages were performed between thaw and use in experiments. Human embryonal carcinoma cell (ECC) lines, GCT27 and NTERA2 were cultured in media containing 10% fetal bovine serum (FBS) (EDM Millipore, TMS-013-B), 1× Penicillin-Streptomycin-Glutamine (PSG) (Gibco, 10378-016), 1× Non-essential amino acids (NEAA) (Gibco, 11140-050), 50 ng/mL primocin (IvivoGen, ant-pm-2) in DMEM High Glucose (Gibco, 11960-069). GCT27 cell line was donated from Dr. Martin Pera (derivation described in (Pera et al., 1987)), NTERA2 cl.D1 (NT2) line was obtained from America Type Culture Collection (ATCC) (ATCC CRL-1973). All ECC lines were grown to 80–90% confluence prior to split with 0.05% Trypsin-EDTA (Gibson, 25300-054). Experiments were performed between passages 20–30, one passage was used between thaw and use in experiments. Human embryonic kidney (HEK) 293T cells were cultured in 10% FBS (ThermoFisher, SH3007003), 1× PSG (Gibco, 10378-016), 1× NEAA (Gibco, 11140-050), 55 μM Sodium Pyruvate (Gibco, 21985-023), and 50 ng/mL primocin (InvivoGen, ant-pm-2) in KnockOut DMEM (Gibco, 10829-018). Cells were cultured to 80–90% confluency prior to split with 0.05% Trypsin-EDTA. Experiments were performed between passage 8–15, one passage was used between thaw and use in experiments. All cell lines used in these experiments were Mycoplasma negative. Mycoplasma testing was performed every 6–9 weeks, using MycoAlert Detection Kit (Lonza, LT07-418).
2.2. Induction of PGCLCs though iMeLCs from primed hESCs
PGCLCs were induced from primed hESCs as described in Sasaki et al. (2015), with some modifications (Chen et al., 2017b). Day 7 hESCs were dissociated into single cells with 0.05% Trypsin-EDTA and plated onto Human Plasma Fibronectin (Invitrogen, 33016-015)-coated 12-well-plate at the density of 200,000 cells/well in 2 mL/well of iMeLC media, which is composed of 15% KSR, 1× NEAA, 0.1 mM 2-Mercaptoethanol, 1× PSG (Gibco, 10378-016), 1 mM sodium pyruvate (Gibco, 11360-070), 50 ng/mL Activin A (Peprotech, AF-120-14E), 3 μM CHIR99021 (Stemgent, 04-0004), 10 μM of ROCKi (Y27632, Stemgent, 04-0012-10), and 50 ng/mL primocin in Glasgow’s MEM (GMEM) (Gibco, 11710-035). iMeLCs were dissociated into single cells with 0.05% Trypsin-EDTA after 24 h of incubation and plated into ultra-low cell attachment U-bottom 96-well plates (Corning, 7007) at the density of 3000 cells/well in 200 μL/well of PGCLC media, which is composed of 15% KSR, 1× NEAA, 0.1 mM 2-Mercaptoethanol, 1× PSG (Gibco, 10378-016), 1 mM sodium pyruvate (Gibco, 11360-070), 10 ng/mL human LIF (Millipore, LIF1005), 200 ng/mL human BMP4 (R&D systems, 314-BP), 50 ng/mL human EGF (R&D systems, 236-EG) 10 μM of ROCKi (Y27632, Stemgent, 04-0012-10), and 50 ng/mL primocin in Glasgow’s MEM (GMEM) (Gibco, 11710-035).
2.3. Fluorescence activated cell sorting
Day 4 aggregates were dissociated with 0.05% Trypsin-EDTA for 10 min at 37 °C. The dissociated cells were stained with conjugated antibodies, washed with FACS buffer (1% BSA in PBS) and resuspended in FACS buffer accompanying with 7-AAD (BD Pharmingen, 559925). The conjugated antibodies used in this study are: INTEGRINa6 conjugated with BV421 (BioLegend, 313624), EPCAM conjugated with 488 (BioLegend, 324210). PGCLCs were either sorted 1000 cells in RLT buffer (QIAGEN) for RNA extraction or media for culture.
2.4. PGCLC culture on transwell membrane
Sorted day 4 PGCLCs were cultured in primed hESC media (Pastor et al., 2016), naïve hESC media (Pastor et al., 2016; Theunissen et al., 2014) or 7-factor media. 7-factor media was based on the formulation of Farini et al. (2005), without the addition of retinoic acid. Therefore, 7-factor medium contains the following: 15% Hyclone FBS (ThermoFisher, SH3007003), DMEM high glucose (Gibco, 11960-069), 1× NEAA (Gibco, 11140-050), 0.1 mM 2-mercaptoethanol (Gibco, 21985-023), 0.25 mM sodium pyruvate (Gibco, 11360070), 1× PSG (Gibco, 10378-016), 50 ng/mL SCF (PeproTech, 250-03), 10 ng/mL bFGF (R&D System, 233-FB), 10 ng/mL SDF1 (R&D Systems, 350-NS), 25 ng/mL human BMP4 (R&D Systems, 314-BP), 500 U/mL LIF (Millipore, LIF1005), 5 μM forskolin (Sigma, D6886), and 1 mg/mL N-acetyl-L-cysteine (Sigma, A9165). 1000 sorted PGCLCs were plated directly on 0.4 μM PET membranes (BD Falcon) in 24-well plates and cultured at 37 °C with 5% CO2 with daily medium changes. For some downstream experiments, cultured PGCLCs were detached from the membrane with 0.05% trypsin for 5 min at 37 °C. Following trypsin cell numbers were counted with hemocytometer and trypan blue (Gibco, 15250061) for viability or placed in RLT buffer for RNA extraction (see below). For each experiment, three independent experiments were performed for each cell line UCLA2 and UCLA6.
2.5. Real time quantitative PCR
PGCLCs sorted or harvested from membranes were placed in 350 μL of RLT buffer (QIAGEN) and RNA was extracted using RNeasy micro kit (QIAGEN, 74004). cDNA was synthesized using SuperScript® II Reverse Transcriptase (Invitrogen, 18064-014). Real time quantitative PCR was performed using TaqMan® Universal PCR Master Mix (Applied Biosystems, 4304437) and the expression level of genes-of-interest were normalized to the expression of housekeeping gene GAPDH. The TaqMan probes used in this study include: GAPDH (Applied Biosystems, Hs99999905_m1), NANOS3 (Applied Biosystems, Hs00928455_s1), PRDM1 (Applied Biosystems, hs01068508_m1), TFAP2C (Applied Biosystems, Hs00231476_m1), POU5F1 (Applied Biosystems, Hs03005111_g1), SOX2 (Applied Biosystems, Hs01053049_s1), PRDM14 (Applied Biosystems, Hs01119056_m1).
2.6. Immunofluorescence, immunohistochemistry and microscopy
Immunostaining paraffin sections of aggregates and tumor sections were described previously (Gkountela et al., 2013). For cells cultured on chamber slides, cells were fixed in 4% paraformaldehyde in PBS for 10 min and washed with PBS containing 0.1% Tween 20 and permeabilized with PBS containing Triton X for 10 min. Slides were blocked with 10% donkey serum for 60 min before antibody incubation. The primary antibodies used for immunofluorescence in this study include: rabbit-anti-PRDM14 (Abcam, ab187881, 1:100), goat-anti-OCT4 (Santa Cruz Biotechnology, sc-8628, 1:100), goat-anti-VASA (R&D Systems, AF2030, 1:20), rabbit-anti cKIT (DAKO, A4502, 1:100), mouse-anti-PLAP (DAKO, IR779), mouse-anti-5mC (Aviva Bioscience, AMM99021), rabbit-anti-H3K27me3 (Millipore, 07-449), rabbit-anti-PRDM1 (Cell Signaling, 9115, 1:100), rabbit-anti-TFAP2C (Santa Cruz Biotechnology, sc-8977). The secondary antibodies used in this study are donkey anti-rabbit-488 (Jackson ImmunoResearch Laboratories, 711-545-152), donkey anti-goat-594 (Jackson ImmunoResearch Laboratories, 705-586-147), and donkey-anti-mouse-488 (Jackson ImmunoResearch Laboratories, 715-545-150). DAPI is counterstained to indicate nuclei. All slides were imaged with an LSM 780 confocal microscope (Zeiss) using ZEN 2011 software. Immunohistochemistry paraffin sections of tumor samples were deparafinized and re-hydrated followed by antigen retrieval in 10 mM Sodium Citrate, 0.05% Tween 20 at 95 °C for 40 min. Sections were washed in 20 mM Tris-HCl pH 7.4, 0.15 mM NaCl and 0.05% Tween 20, permeablized in 0.1% Triton X-100 in PBS. Quenching endogenous peroxidase activity was performed with application of Peroxidase Block for 5 min, provided in Dako EnVIsion + System-HRP-DAB, for Rabbit or Mouse primaries (Agilent Technologies, K4010 and K4006), and anti-Goat HRP-DAB (R&D systems HRP, CTS008). Slides were then blocked in 5% normal donkey serum, 0.05% Tween in PBS. Goat primaries were then incubated for 15 min with Avidin blocking reagent followed by Biotin blocking agent for 15 min (R&D system HRP, CTS008). Primary antibodies against rabbit-anit-PRDM14 (Abcam ab187881, 1:100), rabbit-anti-SOX2 (Abcam 97959, 1:1000), rabbit-anti-OCT4 (cell signaling 2840S, 1:100), mouse-anti-PLAP (DAKO IR779, 1:100), and goat-anti-SOX17 (Neuromics GT15094, 1:100) were incubated overnight. Slides were incubated with secondary-peroxidase labelled polymer-HRP (Dako EnVision + System-HRP or R&D systems HRP) per protocol recommendations. Slides were then washed for 5 min in PBS, containing 0.1% Tween 20. Substrate –chromogen (Dako EnVision + -HRP) was added for 10 min at room temperature. Slides were washed in distilled water and counterstained with hematoxylin, and mounted with VectaMount (Vector laboratories H-5000). Patient paraffin sections were banked specimens obtained from UCLA pathology and laboratory, samples were de-identified, IRB exempt.
2.7. Generation of human PRDM14 overexpression plasmid
Human PRDM14 cDNA (ThermoFisher, MHS6278-202833494) was used for hPRDM14 overexpression plasmid. The following primers were used: FWD-5′GAGCTAGCGAATTCGAATTTATGGCTC TACCCCGGCCAAG-3′, REV-5′-AGCGGCCGCGGATCCGATTTCGTAGTCTTCATGAAACTTCA-3′, to modify the hPRDM14 cDNA to be a compatible insert with 20 bp overhang on each end, for insertion into the bidirectional promoter lentivector pCDH-EF1-MCS-T2A-RFP(PGK-Puro) (System Biosciences, CD822A-1). 2× Phusion® High-Fidelity PCR master mix (New England BioLabs, M0531) was used per the product protocol, to generate the plasmid insert. The plasmid was ligated with restriction enzyme SwaI (New England BioLab, R0604), incubated at 25 °C for 60 min, followed by addition of Calf Intestine Alkaline Phosphatase (CIP) (New England BioLabs, M0290), then continued incubation at 37 °C for 15 min, followed by 65 °C for 15 min. The ligation product was then run on 1% agarose gel, and the resulting bands were extracted per QIAquik® gel extraction kit protocol (Qiagen, 28704). Gibson Assembly was utilized to generate the desired plasmid, 300 ng of insert and 100 ng of plasmid was added to Gibson Assembly master mix (New England BioLabs, E2611), and incubated at 50 °C for 60 min. The resulting product was transformed into One Shot® Stbl3 Chemically Competent E. coli (ThermoFisher Scientific, C737303), and incubated overnight on ampicillin LB agar plate. 5 colonies were selected, plasmid DNA extracted via QIAprep spin miniprep kit (Qiagen, 27104), and was sent for sequencing (Laragen, Culver City, CA) to verify insertion of hPRDM14. The resulting product was then transfected into HEK 293T cells (50,000 cells in a 6-well plate), using Lipofectamine™ 3000 reagent protocol (Invitrogen, L3000008) to verify functional protein.
2.8. Lentiviral production and transduction of iMeLC
HEK 293T cells were seeded in T175 flask (Corning, 10-126-13) and grown until 85–90% confluency. In tube A Opti-MEM I (Gibco, 31985070) and Lipofectamin 3000 (Invitrogen) were mixed (volumes per product recommendation), in tube B Opti-MEM I, 13.8 μg of expression plasmid, 5 μg pCMV-VSV-G (addgene, 8454), 12.5 μg of pCMV-dR8.91(addgene, 2221), and P3000 reagent. Contents of tube A was mixed with tube B to make DNA-lipid complex, and incubated for 20 min at room temperature. HEK 293T media was replaced with packaging medium consisting of Opti-MEM I, reduced serum, GlutaMAX (Gibco, 51985034), 5% FBS, 1 mM sodium pyruvate and DNA-lipid complex was added for 6 h. Fresh packing media was collected every 24 h for 72 h. The packaging media containing the virus was then filtered through 0.45 μm pore, PVDV membrane (EMD Millipore, SCHVU01RE). The filtrate was then centrifuged at 22,000 rpm for 90 min, and the pellet was resuspended in HEK 293T media. Lentivirus titration was then performed by plating 100,000 HEK 293T cells into 6 wells of a 24-well plate. The following day serial dilutions on virus is plated as follows, 4 μL undiluted virus in 400 μL media, 10−1 by adding 40 μL of undiluted in 360 μL media, this is repeated for 10−2, 10−3, 10−4, and no virus. Cells are incubated for 48 h, then detached with 0.05% trypsin for 5 min at 37 °C, neutralized, and spun down at 1600 rpm for 5 min. The pellet is then resuspended in 400 μL of FACs buffer and 7AAD is added for viability. Flow cytometry was then performed to evaluate percent of RFP positive cells. iMeLCs were transduced with the virus following 24 h of culture, as described above. iMeLCs were detached with 0.05% Trypsin and neutralized with MEF media. Cells were resuspended in 100 μL of PGCLC media. Virus was added, 30 μL of empty plasmid virus, 100 μL of hPRDM14 plasmid virus. Total volume was brought up to 200 μL with PGCLC media. Cells were incubated on the nutator with virus for 2 h. Cells were spun down at 1200 rpm following incubation, then resuspended in 500 μL of PGCLC media, and plated in 96 well, low adherence plate at 3000 cells per well.
2.9. Western blot analysis
Protein fractions (nuclear and cytoplasmic) were isolated using the QProteome Cell Compartment Kit (Qiagen, 37502) according to manufacturer’s instructions. Protein was quantified using the Pierce™ BCA Protein Assay Kit (ThermoFisher, 23227), 5 μg of protein was analyzed by electrophoreses on 12% NuPAGE Novex Bis-Tris gels (Invitrogen) and transferred to Hybond ECL Nitrocellulose Membrane (GE Healthcare) according to standard procedures. Membranes were divided to probe same blot with different species antibody. ECC western blot was performed as two separate blots from the same protein extracts. One blot was used for PRDM14 and beta-actin. Second blot was made for OCT3/4, given the proximity in size for OCT4 and beta-actin. One blot was used for PRDM14 and H3, in HEK 293T cells. Primary antibodies (1:1000) were rabbit-anti-PRDM14 (Abcam, ab187881), goat-anti-OCT3/4 (N-19) (Santa Cruz, sc-8628), mouse-anti-ACTIN (Santa Cruz, sc47778) and rabbit anti-H3 (Abcam ab1791). Secondary HRP-conjugate antibodies were from Abcam anti-rabbit and anti-goat, GE LifeScience anti-mouse, all used at 1: 5000. Blots were developed using Pierce ECL Western Blotting Substrate (ThermoFisher, 32106).
2.10. Edu analysis
PGCLCs cultured on membrane were evaluated with Edu Click-IT® EdU Alexa 488 imaging kit (ThermoFisher, C10337). PGCLCs we cultured on membrane in 7-factor as described above. EdU was added at 10 μM for 3 h. Cells were then fixed in 4% PFA, and processed per manufactures protocol. Transwell membrane was extracted with Nuclear counter staining performed with ProLong® Gold Antifade Mountant with DAPI. Cells were imaged with an LSM 780 confocal microscope (Zeiss) using ZEN 2011 software. EdU quantification was performed using Imaris microscope imaging analysis software (Bitplane).
2.11. Statistical analysis
qPCR data was analyzed using Prism 7.0 (GraphPad) software. 2way ANOVA tests with multiple comparisons were used for aggregates versus membrane comparisons of un-transduced PGCLCs, significance was denoted with p < 0.05. For PGCLCs transduced with hPDM14, ordinary one-way ANOVA, with multiple comparisons was utilized to evaluate hPRDM14 fold change between groups. Significance was denoted with p < 0.05. The remainder of genes for transduced PGCLCs were analyzed using 2way ANOVA test with multiple comparison, with significance denoted with p < 0.05. EdU analysis was performed with Prism 7.0, with unpaired t-test, significance denoted with a p < 0.05.
3. Results
3.1. Intracranial germinomas resemble late stage primordial germ cells
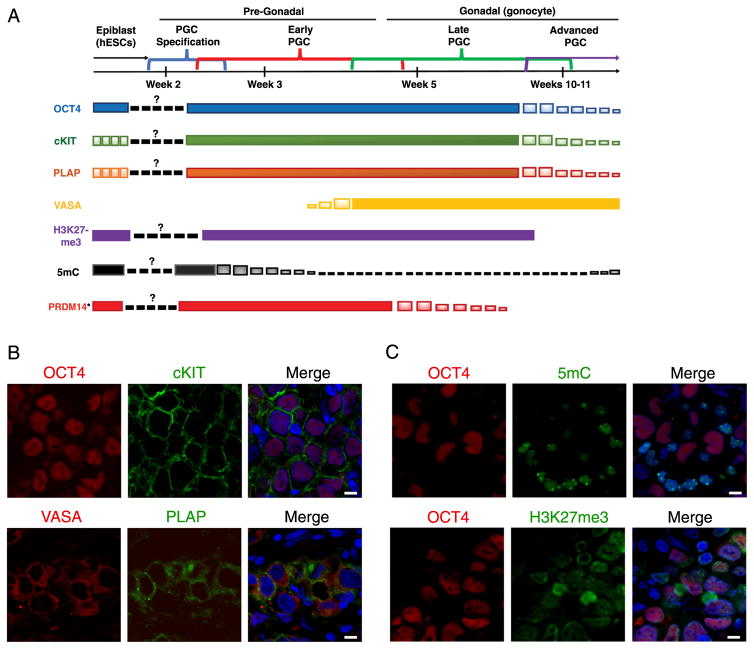
Extragonadal GCTs occur along the midline in the sacrum, mediastinum, and intracranial midline structures (pineal gland and suprasellar region). These, extragonadal GCTs histologically resemble gonadal GCTs, and are hypothesized to arise from PGCs that missed the genital ridge during migration (Schmoll, 2002). Using human fetal tissue consented to research and analysis of molecular markers at a single cell level, the molecular program for human PGC development is more precisely staged as “early PGCs”, “late PGCs” and “advanced PGCs” (Gkountela et al., 2015; Gkountela et al., 2013; Guo et al., 2015; Irie et al., 2015; Chen et al., 2017a). With late and advanced PGCs in the gonad referred to as gonocytes. We hypothesize that these markers provide an opportunity to determine whether extragonadal GCTs correspond to PGCs in one of the stages. Most notably, early PGCs are VASA−/5mC−/H3K27me3+, and late PGCs are VASA+/5mC−/H3K27me3+, and advanced PGCs are VASA+/5mC−/H3K27m3− (Fig. 1A) (Gkountela et al., 2013).
Fig. 1. Intracranial germinomas resemble late PGCs.
A. Timeline of germ cell development with expression patterns of germ line markers and epigenetic markers. *Early PGC expression pattern extrapolated from non-human primate data (Sasaki et al., 2016).
B. Immunofluorescence staining of intracranial germinomas for germ line markers, top panel: OCT4/cKIT/DAPI(merge), bottom panel: VASA/PLAP/DAPI(merge). Scale bars, 8 μm.
C. Immunofluorescence staining of intracranial germinomas for epigenetic markers, top panel: OCT4/5mC/DAPI (merge), bottom panel: OCT4/H3K27me3/DAPI(merge). Scale bars, 8 μm.
We began by examining intracranial germinomas for VASA, cKIT, 5mC and H3K27me3 together with two well-known GCT markers, OCT4 and PLAP. We discovered that OCT4 and PLAP positive germinoma cells, are positive for cKIT and VASA respectively (Fig. 1B). Indicating that the intracranial germinomas most likely originated from late PGCs during the 4–10th week of life. To confirm this, we stained for 5mC and H3K27me3 in the OCT4 positive germinomas (Fig. 1C). Our results show that intracranial germinomas are depleted of 5 mC and are enriched in H3K27me3 which rules out progression to advanced PGCs. Taken together, this data suggests that intracranial germ cell tumors most likely originate from late PGCs, equivalent to 4–10 weeks of development post-fertilization, that were unable to complete further differentiation, yet did not die in their extragonadal locations. Critically, this is the time when PRDM14 is normally repressed in primate PGCs (Sasaki et al., 2016).
3.2. PRDM14 is a novel marker of malignant germ cell tumors
Recently, two genome-wide association studies identified PRDM14 as a susceptibility locus for GCTs. However, it is not known whether PRDM14 is expressed in GCT cells. To address this, we first performed, a western blot analysis to examine PRDM14 expression in two human GCT embryonal carcinoma cell (ECC) lines, Ntera2 (NT2) and GCT27. NT2 cells differentiate into teratomas when transplanted into immunocompromised mice (Andrews et al., 1984), and GCT27 generates undifferentiated embryonal carcinomas and yolk sac tumors following transplant (Pera et al., 1987). In both ECC lines, we discovered that PRDM14 protein is enriched in the nucleus (Fig. 2A).
Fig. 2. PRDM14 expression in testicular and intracranial germ cell tumors.
A. Western blot of nuclear (5 μg) and cytoplasmic (5 μg) extracts from embryonal carcinoma cell (ECC) lines, Ntera2 (NT) and GCT27. Blots probed for PRDM14 (64 kD), OCT4 (45 kD), and beta-actin (42 kD).
B. Immunofluorescence of ECC line, GCT27, showing staining for PRDM14/OCT4, top panel. Immunofluorescence of paraffin section of a seminoma tumor sample, showing staining for PRDM14/OCT4, bottom panel. Scale bars, 15 μm.
C. Immunohistochemistry of paraffin section of tumor sample containing corresponding GCNIS portion of the tumor, staining for PRDM14, and GCNIS markers OCT4 and PLAP. Scale bars, 15 μm.
D. Immunohistochemistry of paraffin section of mixed germ cell tumor, corresponding to embryonal carcinoma, staining for PRDM14, SOX2 and SOX17, top panel. Magnified images of cells demarked in yellow box in top panel. Scale bars, 15 μm (top panel). 5 μm (bottom panel).
E. Immunohistochemistry staining of intracranial germinoma, yolk sac and teratoma, for PRDM14. Scale bars, 20 μm.
To examine expression of PRDM14 in testicular GCT tissue samples from patients, we performed immunofluorescence and immunocytochemistry on testicular germ cell neoplasm in situ (GCNIS), seminomas and testicular mixed GCTs. We chose these GCT types because like ECCs, these tumors express the pluripotency factor OCT4, however unlike ECCs, the seminomas and GCNIS express PGC markers including cKIT, VASA, and SOX17 (Rajpert-De Meyts et al., 2015; Leroy et al., 2002; Gillis et al., 2011). Using immunofluorescence, we confirmed that PRDM14 was expressed in ECC lines, demonstrating co-localization with the OCT4 positive cells (Fig. 2B). Likewise, PRDM14 was found to co-localize with the OCT4 positive cells in seminoma (Fig. 2B). We next evaluated GCNIS cells associated with seminomas. GCNIS is characterized by expression of PGC markers such as PLAP and OCT4 within tubules. Here we demonstrate that PRDM14 is also expressed in GCNIS cells as indicated by positive staining in serial sections to tubules containing OCT4 and PLAP positive GCNIS cells (Fig. 2C). In contrast, mixed GCTs were not uniformly positive for PRDM14. Specifically, in the mixed GCTs embryonal carcinoma, seminoma, and yolk sac, all showed positive nuclear staining, however, the non-malignant teratoma areas were negative for PRDM14 (quantified in Table 1). Fig. 2D represents an embryonal carcinoma component of a mixed GCT, in which we identified PRDM14 positive cells corresponding to SOX2 positive cells, a marker of embryonal carcinoma. We did not identify SOX17 positive cells in this mixed GCT section, which is a marker of seminomas, therefore PRDM14 in this case is specifically expressed in the embryonal carcinoma cells. Taken together, PRDM14 is expressed in ECC lines with a pluripotent identity, as well as testicular GCT tissue samples that exhibit both a PGC and pluripotent identity.
Table 1.
Testicular germ cell tumors histologic types evaluated with immunohistochemistry staining result for PRDM14. N = 12 seminomas, with N = 7 having GCNIS components. N = 5 mixed GCTs.
| Histology | PRDM14 present | PRDM14 absent |
|---|---|---|
| Testicular ITGCN | 7/7 | 0/7 |
| Testicular seminoma | 12/12 | 0/12 |
| Testicular mixed GCTa | ||
| Embryonal carcinoma | 4/4 | 0/4 |
| Yolk sac | 2/2 | 0/2 |
| Teratoma | 0/3 | 3/3 |
| Seminoma | 1/1 | 0/1 |
Denotes the various components present in tumor samples.
To determine whether PRDM14 is expressed in intracranial germ cell tumors (demographics in Table 2), we performed immunohistochemistry to detect PRDM14, and discovered positive nuclear staining in germinoma cells, as well as cells in the cells of the intracranial yolk sac tumors (Fig. 2E and quantified in Table 3). In contrast, intracranial teratomas had undetectable levels of PRDM14 protein (Fig. 2E and quantified in Table 3). Taken together, we show that PRDM14 is expressed in gonadal and extragonadal GCTs with a late PGC identity, as well as ECCs with a pluripotent identity. Therefore, we hypothesize that failure to repress PRDM14 during embryo development affects PGC differentiation.
Table 2.
Demographics by tissue histology types of intracranial germ cell tumors evaluated.
| Intracranial germ cell tumor demographics | ||
|---|---|---|
|
| ||
| Tumor type (N) | Mean age (age range) | Sex distribution |
| Germinoma (12) | 23 yo (9 yo–40 yo) | Male = 12, Female = 0 |
| Yolk sac (2) | 23 yo (20 yo–26 yo) | Male = 2, Female = 0 |
| Teratoma (3) | 22 yo (3 mo–25 yo) | Male = 2, Female = 1 |
Table 3.
Intracranial germ cell tumors histologic types evaluated with immunohistochemistry staining results for PRDM14. N = 12 germinomas, 2 yolk sacs and 3 teratomas.
| Histology | PRDM14 present | PRDM14 absent |
|---|---|---|
| Intracranial germinoma | 12/12 | 0/12 |
| Intracranial yolk sac | 2/2 | 0/2 |
| Intracranial teratoma | 0/3 | 3/3 |
3.3. Using human pluripotent stem cells to model human PGC differentiation in vitro
Given that PRDM14 positive intracranial GCTs (germinomas) express a signature that is reminiscent of human PGCs between 4 and 10 weeks of development, we were interested in addressing the hypothesis that elevated PRDM14 expression alters PGC differentiation. Given that PGCs do not exist in adult tissues, and PGCs cannot be maintained in culture as cell lines, we chose to address this hypothesis by differentiating PGCLCs from human PSCs.
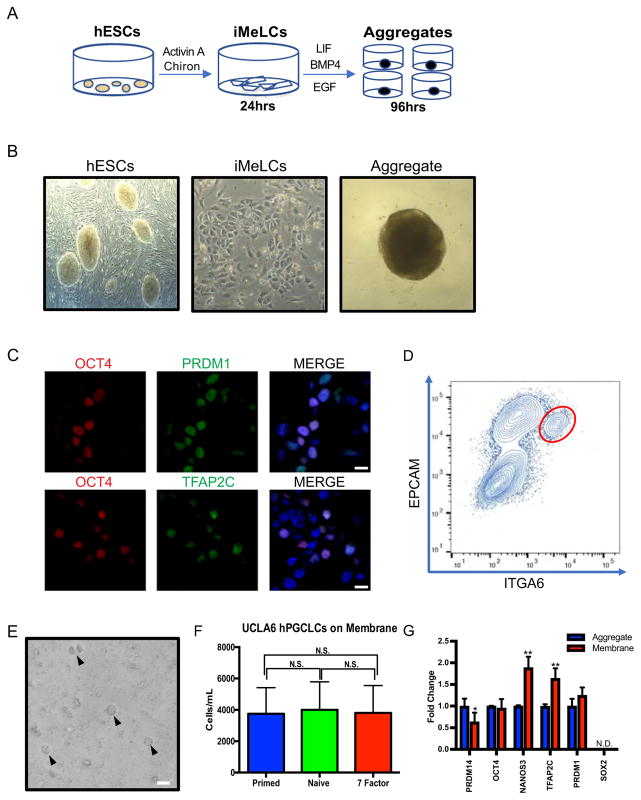
Using the directed differentiation approach published by Sasaki et al. (2015) and modified by (Chen et al., 2017b) (Fig. 3A), we differentiated a male human embryonic stem cell (hESC) line called UCLA6 into incipient mesoderm-like cells (iMeLCs), and used the iMeLC to generate three-dimensional aggregates (Fig. 3B). Using immunofluorescence, we discovered that the aggregates contained putative PGCLCs that were OCT4/PRDM1 and OCT4/TFAP2C double positive (Fig. 3C). Using Fluorescence activated cell sorting (FACS) we detected a population of Integrin alpha 6 (ITGA6) and Epithelial cell adhesion molecule (EPCAM) double positive PGCLCs at day 4 (Fig. 3D). Based upon the results of Sasaki et al., (2015) and Chen et al., (2017b), the ITGA6/EPCAM positive cells are the putative OCT4, PRDM1, TFAP2C positive PGCLCs.
Fig. 3. PGCLCs can be cultured on transwell membranes for at least four days.
A. Two-step differentiation model to generation PGCLCs from hESCs.
B. Bright filed images of hESCs, iMeLCs, and an aggregate generated through the two-step differentiation model.
C. IF of UCLA6 day 4 aggregates containing OCT4/PRDM1 (top) and OCT4/TFAP2C (bottom) positive PGCLCs. Scale bars, 15 μm.
D. FACS analysis of day 4 aggregates, red circle represents ITGA6/EPCAM double positive PGCLCs.
E. Bright field image of PGCLCs sorted according to D, followed by four additional days of culture on transwell membranes. Triangles denote individual PGCLCs. Scale bar, 20 μm.
F. Cell counts of PGCLCs isolated from UCLA6 day 4 aggregates by FACS according to D, followed by 4 days of additional culture on transwell membrane in medias indicated. Primed hESC media, naïve hESC, and 7-factor media, respectively. N.S. = not significant. Three independent experiments were performed.
G. Gene expression of PGCLCs sorted from UCLA6 aggregates at day 4 (blue) according to D, and after 4 days of additional culture on transwell membrane in 7-factor media (red). Three independent experiments were performed. **p < 0.0001, *p < 0.001.
Given that our goal was to determine the outcome of overexpressing PRDM14 in PGCLCs, we next determined whether ITGA6/EPCAM positive PGCLCs could be cultured on transwell membranes (Fig. 3E). In pilot tests, we cultured the ITGA6/EPCAM sorted PGCLCs in three types of media; primed hESC media (Amit et al., 2000), naïve hESC media (also called 5-inhibitor, LIF, ACTIVIN, FGF2 or 5iLAF) (Pastor et al., 2016; Theunissen et al., 2014) and 7-Factor media (Farini et al., 2005; Oliveros-Etter et al., 2015), and assayed PGCLC number four days later (Fig. 3F). After four days on the transwell membranes, the PGCLCs remained round, and well separated from each other (Fig. 3E). We also discovered that the media used for transwell culture did not have an affect PGCLC number (Fig. 3F), therefore we chose to use 7-Factor media for all future experiments given that 7-Factor media has been used to support the differentiation of PGCs sorted from mouse embryos (Oliveros-Etter et al., 2015).
To determine whether culturing PGCLCs on the transwell membranes leads to a change in PGCLC identity, we performed real time Polymerase Chain Reaction (RT-PCR) for PGC genes (OCT4, NANOS3, TFAP2C, PRDM1), as well as the pluripotency genes SOX2 and PRDM14. Our results show that OCT4 and PRDM1 are expressed in PGCLCs sorted from the aggregate at day 4 of differentiation, and expression of these genes does not change following an additional four days of transwell membrane culture (8 days total). Similarly, SOX2 RNA is not expressed in PGCLCs isolated from the aggregates at day 4, and SOX2 expression remains repressed after an additional 4 days of transwell culture. Interestingly, we discovered that the PGC genes NANOS3 and TFAP2C are further enriched with additional days on the transwell membrane, whereas PRDM14 expression levels are reduced (Fig. 3G). Taken together, this data shows that PGCLCs can be cultured in vitro on transwell membranes, and during this time the PGCLCs do not lose germline identity or revert to a pluripotent stem cell (they do not reactivate SOX2). Instead, we discovered that the PGCLCs significantly up-regulate the PGC genes, NANOS3 and TFAP2C, and repress PRDM14.
3.4. Constitutive overexpression of PRDM14 in PGCLCs does not affect PGCLC identity
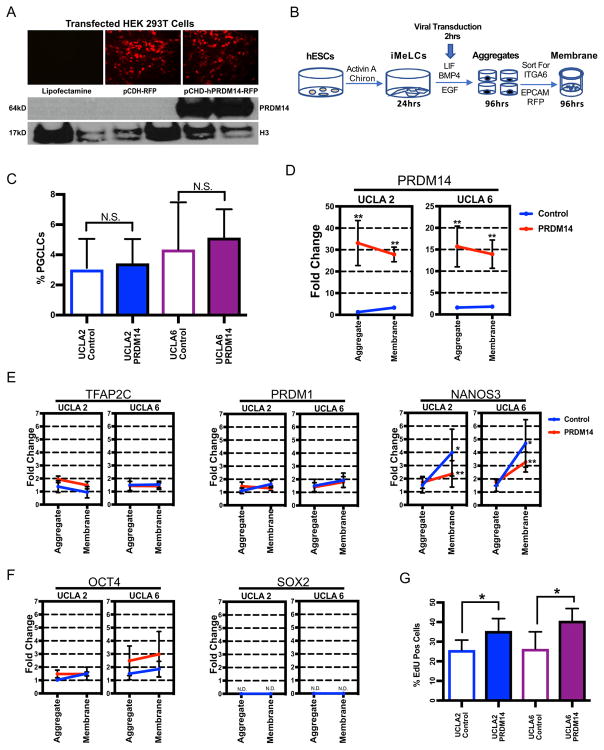
Using the model described above, we then sought to evaluate the effect of constitutive overexpression of PRDM14 on PGCLC differentiation, and proliferation using two different male hESC lines (UCLA2 and UCLA6). To achieve this, we generated a plasmid construct in which human PRDM14 (hPRDM14) was transcribed under the control of the elongation factor 1-alpha promoter with a T2A-RFP tag (pCDH-hPRDM14-RFP). The empty pCDH plasmid, called pCDH-RFP was used as a negative control. We first tested the constructs in HEK 293T cells by transfection and discovered that only cells transfected with pCDH-RFP plasmids express RFP (Fig. 4A), and using Western blot analysis, we discovered that only the pCDH-hPRDM14-RFP transduced cells expressed PRDM14 protein (Fig. 4A).
Fig. 4. PGCLCs constitutively overexpressing PRDM14 fail to increase NANOS3 and are more proliferative.
A. HEK 293T cells transfected with plasmid constructed to constitutively overexpress PRDM14. Images portraying no RFP signal in Lipofectamine control, positive RFP signal in pCDH-RFP (empty plasmid), pCDH-hPRDM14-RFP. Protein expression of PRDM14 in pCHD-hPRDM14-RFP confirmed with western blot. PRDM14 ~ 64 kD, H3 (loading control) ~ 17 kD. Two independent experiments were performed.
B. Experiment design for constitutive overexpression of PRDM14, using the two-step method of differentiation, day 4 PGCLCs were sorted by FACS by ITGA6/EPCAM and cultured for an additional 4 days on transwell membranes prior to analysis.
C. Percent of PGCLCs sorted from control and PRDM14 overexpressing aggregates, for both UCLA2 and UCLA6 hESC cell lines. Three independent experiments were performed for each cell line UCLA2 and UCLA6.
D. Gene expression level of PRDM14 in control (blue) and PRDM14 constitutive overexpression (red). Aggregate = PGCLCs isolated by FACS at day 4 for ITGA6/EPCAM. Membrane = ITGA6/EPCAM sorted PGCLCs cultured for four additional days on transwell membranes. ** = significance between control and PRDM14, p < 0.0001. Three independent experiments were performed for each cell line UCLA2 and UCLA6.
E. Gene expression levels of germ line genes, TFAP2C, PRDM1, NANOS3. Control (blue). PRDM14 overexpression (red). * = significance between aggregate and membrane, p < 0.0001. ** = significance between control and PRDM14 on membrane, p < 0.001. Aggregate = PGCLCs isolated by FACS at day 4 for ITGA6/EPCAM. Membrane = ITGA6/EPCAM sorted PGCLCs cultured for four more days on a transwell membrane. Three independent experiments were performed for each cell line UCLA2 and UCLA6.
F. Gene expression levels of pluripotency genes, OCT4 and SOX2 in control (blue) and PRDM14 overexpression (red). N.D. = none detected. Aggregate = PGCLCs isolated by FACS at day 4 for ITGA6/EPCAM. Membrane = ITGA6/EPCAM sorted PGCLCs cultured for four additional days on transwell membranes. Three independent experiments were performed for each cell line UCLA 2 and UCLA 6.
G. Percent of Edu positive PGCLCs on transwell membrane. * = significance between control and PRDM14, p < 0.05. PGCLCs for this experiment were differentiated for 4 days in aggregates before FACS using ITGA6/EPCAM to isolate PGCLCs. The sorted PGCLCs were cultured for an additional four days on transwell membranes. Three independent experiments were performed for each cell line UCLA2 and UCLA6.
Next, we used the plasmids to generate lentiviruses, and transduced the iMeLCs with pCDH-RFP (control) or pCDH-hPRDM14-RFP (PRDM14) lentivirus prior to aggregate formation (Fig. 4B). In our first experiment, we evaluated whether constitutive PRDM14 expression caused a change in the percentage of ITGA6/EPCAM PGCLCs sorted from the aggregates (Fig. 4C). Our results show that constitutive PRDM14 expression had no effect on the percentage of ITGA6/EPCAM PGCLCs sorted from the aggregates relative to control.
To determine whether constitutive overexpression of PRDM14 alters PGCLC identity, we examined PGC and pluripotency gene expression by Real-time PCR in RFP positive PGCLCs sorted directly from the aggregate by FACS. We also evaluated gene expression in the PGCLCs harvested from the transwell membrane after 4 additional days of culture. Our results show that PRDM14 levels are 15 to 30-fold higher in cells transduced with the PRDM14 expressing lentivirus compared to the control, and remain significantly elevated after 4 additional days of culture on the transwell membrane (Fig. 4D). Next, we evaluated germ cell identity using real time PCR to examine expression of TFAP2C, PRDM1, NANOS3 and OCT4 (Fig. 4E, F). Our results demonstrate that constitutive overexpression of PRDM14 does not alter PGC gene expression of TFAP2C, PRDM1, and OCT4. However, we did observe a significant decrease in NANOS3.
One of the hallmarks of human PGCs in the embryo, and PGCLC differentiation in vitro is the rapid and specific repression of SOX2 (Perrett et al., 2008). In mouse, Prdm14 promotes the expression of sox2 during PGC development (Yamaji et al., 2008). Therefore, it could be hypothesized that overexpression of PRDM14 in human PGCs results in maintenance of SOX2 expression. Instead, our data shows that SOX2 remains repressed in PGCLCs with constitutive PRDM14 (Fig. 4F). Therefore, PRDM14 does not cause reinstatement of pluripotency in PGCLCs.
Finally, we addressed whether constitutive overexpression of PRDM14 caused an increase in proliferation by monitoring uptake of 5-ethynyl-2′-deoxyuridine (EdU). PRDM14 has previously shown to increase proliferation in breast cancer cell lines overexpressing PRDM14 (Nishikawa et al., 2007). PGCLCs were cultured on membrane for 4 days, at which point PGCLCs were incubated with EdU for 3 h. Here we show that constitutive overexpression of PRDM14 in PGCLCs leads to a significant increase the percent of EdU positive cells (Fig. 4G), indicating that more PGCLCs are proliferating relative to control.
4. Discussion
In this study, we evaluated the expression of PRDM14 in human GCT ECC lines, as well as tissue samples of testicular and intracranial GCTs from patients. PRDM14 is expressed in early embryo and fetal development in humans while not being expressed in somatic cells after birth making it a desirable therapeutic target. We discovered that PRDM14 is expressed in OCT4 positive ECCs, testicular embryonal carcinoma, seminoma and GCNIS as well as intracranial germinomas and yolk sac tumors. Notably, we discovered that PRDM14 is not expressed in differentiated teratomas. This is in agreement with work in human pluripotent stem cells revealing that PRDM14 is a component of the core human pluripotency network together with OCT4, SOX2 and NANOG (Chia et al., 2010), and that loss of PRDM14 in human PSCs is associated with stem cell differentiation (Chia et al., 2010; Tsuneyoshi et al., 2008). Conversely, overexpression of PRDM14 prevents PSC differentiation (Tsuneyoshi et al., 2008). Therefore, PRDM14 may function in GCT cells to block differentiation.
In previous studies we, and others discovered that PRDM14 is repressed in late and advanced stage human PGCs (Gkountela et al., 2015; Irie et al., 2015), suggesting that PRDM14 repression may be required for PGC differentiation. To address this, in the current study we re-examined intracranial germinomas, which have a PGC identity, and a globally hypomethylated genome (Fukushima et al., 2017). Therefore, intracranial germinomas are consistent with the hypothesis that they originate from PGCs that survive and proliferate in an abnormal niche, yet fail to advance in differentiation. Our data is consistent with Fukushima et al. (2017), showing that intracranial germinomas are hypomethylated. However, we also discovered that the chromatin of intracranial germinomas is enriched in histone H3K27me3 indicating that intracranial germinomas most likely correspond to transformed PGCs that have not progressed on to the advanced stage where H3K27me3 is lost. Notably, PRDM14 is repressed in late PGCs in the embryo, whereas intracranial germinomas are positive for PRDM14.
Given that PGCs do not exist in adult tissues, and PGCs cannot be maintained in culture as cell lines, analysis of PGC transformation requires the differentiation of PGCLCs in vitro from PSCs together with the ability to maintain in culture. There are three alternate approaches for generating PGCLCs in vitro from PSCs. These include, spontaneous differentiation as embryoid bodies (Clark et al., 2004), directed differentiation from PSCs cultured in 4i media (Irie et al., 2015) and a two-step differentiation approach starting from PSCs cultured in primed media followed by differentiation through incipient mesoderm like cells (iMeLCs) prior to the differentiation of PGCLCs (Sugawa et al., 2015; Sasaki et al., 2015; Chen et al., 2017b). We chose to use the two-step PGCLC method of Sasaki et al., (2015), given that PGCLCs can be sorted from any PSC line following differentiation and FACS for ITGA6/EPCAM. We did not use the spontaneous differentiation method as it yields too few PGCLCs for analysis. PGCLCs generated by the two-step method creates PGCLCs in vitro that are equivalent to early PGCs found in the human embryo. Given that PGCLCs cannot be maintained in the aggregates for extended period, we invented a third step in the differentiation protocol by culturing the PGCLCs on transwell membranes for an additional four days leading to increased levels of the PGC-specific genes NANOS3 and TFAP2C. Notably this did not result in the reprogramming of PGCLCs to embryonic germ cells which are equivalent to PSCs that express SOX2 (Pashai et al., 2012). We discovered that constitutive expression of PRDM14 in PGCLCs leads to a significant decrease in NANOS3 expression after culture on the transwell membrane which may suggest complications with PGCLC differentiation. Previous studies using short hairpin RNA knockdown of NANOS3 in human PSCs leads to a reduction in germ cell numbers, and a decrease in the expression of germline genes (cKIT, PRDM1, and VASA) and meiotic initiation (STRA8, SCP3) (Julaton and Reijo Pera, 2011). Therefore, we speculate that constitutive PRDM14 expression alters PGCLC differentiation by first altering the levels of NANOS3 mRNA. However, our results do not favor the hypothesis that PRDM14 causes reprogramming of PGCs to a pluripotent state given that SOX2 remains repressed in PGCLCs with elevated PRDM14, and the PGCLCs do not form colonies on the transwell membranes.
Although PRDM14 is not expressed in adult cells in the human body, PRDM14 is also up-regulated in several somatic cancers, including lung, breast, and lymphoblastic leukemia (Nishikawa et al., 2007; Dettman et al., 2011; Zhang et al., 2013). In breast cancer, PRDM14 enhances cell growth, and reduce sensitivity to chemotherapy (Nishikawa et al., 2007). Similar to studies with breast cancer cell lines, we showed that constitutive overexpression of PRDM14, leads to a significant increase in the proportion of Edu positive PGCLCs suggestive of an increased proliferation rate.
5. Conclusions
In this study, we demonstrated that intracranial germinomas have a signature resembling late PGCs, a time point in human PGC development in which PRDM14 is normally repressed. By examining patient tumor samples, we found that PRDM14 is a marker of malignant GCTs, including those of testicular and intracranial origin. Utilizing a PSC model of generating human PGCLCs, we showed that constitutive over-expression of PRDM14 in PGCs leads to delayed differentiation and increased proliferation, as evident by the inability to upregulated NANOS3 and increased EdU positivity. Taken together, our work leads to a new model for GCT formation, particularly intracranial germinomas, in which constitutive overexpression of PRDM14 leads to increased PGC proliferation and alterations in PGC differentiation.
Acknowledgments
The authors would like to thank Jiaoti Juang, UCLA Translational Pathology Core Laboratory (TPCL), UCLA Brain Tissue Translational Resource (BTTR) laboratory, for providing tumor tissue samples and pathology review. The authors would also like to thank Jessica Scholes and Felicia Codrea of the UCLA BSCRC FACS core and Jinghua Tang of the UCLA BSCRC Stem Cell Banking core. The authors would also like to give special thanks to Isabel Monroy and Alanna Richman for assisting with experiments.
This work was supported by a K12 HD034610 UCLA Child Health Research Career Development Award awarded to JJG, T32 CA009056 UCLA Tumor Cell Biology Training Grant awarded to JJG, a UCLA BSCRC Postdoctoral Training grant awarded to DC, a BSCRC Clinical Fellow Training Grant awarded to JJG, an R01 HD079546 from NICHD awarded to ATC, and a Concern Foundation Award to ATC.
Footnotes
Conflicts of interest
The authors declare no potential conflicts of interest.
References
- Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- Andrews PW, Damjanov I, Simon D, Banting GS, Carlin C, Dracopoli NC, Fogh J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Investig. 1984;50:147–162. [PubMed] [Google Scholar]
- Calaminus G, Joffe J. Germ cell tumors in adolescents and young adults. Prog Tumor Res. 2016;43:115–127. doi: 10.1159/000447081. [DOI] [PubMed] [Google Scholar]
- Chen D, Gell JJ, Tao Y, Sosa E, Clark AT. Modeling human infertility with pluripotent stem cells. Stem Cell Res. 2017a;21:187–192. doi: 10.1016/j.scr.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Liu W, Lukianchikov A, Hancock G, Zimmerman J, Lowe M, Kim R, Galic Z, Irie N, Surani MA, Jacobsen SE, Clark AT. Germline competency of human embryonic stem cells depends on EOMESODERMIN. Biol Reprod. 2017b doi: 10.1093/biolre/iox138. https://doi.org/10.1093/biolre/iox138. [DOI] [PMC free article] [PubMed]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, Huss M, Soh BS, Kraus P, Li P, Lufkin T, Lim B, Clarke ND, Bard F, Ng HH. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Clark AT, Bodnar MS, Fox M, Rodriquez RT, Abeyta MJ, Firpo MT, Pera RA. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- De Felici M. Origin, migration, and proliferation of human primordial germ cells. In: Coticchio G, Albertini DF, De Santis L, editors. Oogenesis. Springer; London, New York: 2013. pp. 19–37. [Google Scholar]
- Dettman EJ, Simko SJ, Ayanga B, Carofino BL, Margolin JF, Morse HC, 3rd, Justice MJ. Prdm14 initiates lymphoblastic leukemia after expanding a population of cells resembling common lymphoid progenitors. Oncogene. 2011;30:2859–2873. doi: 10.1038/onc.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Perez SV, Kim R, Li Z, Marquez VE, Patel S, Plath K, Clark AT. Derivation of new human embryonic stem cell lines reveals rapid epigenetic progression in vitro that can be prevented by chemical modification of chromatin. Hum Mol Genet. 2012;21:751–764. doi: 10.1093/hmg/ddr506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farini D, Scaldaferri ML, Iona S, La Sala G, De Felici M. Growth factors sustain primordial germ cell survival, proliferation and entering into meiosis in the absence of somatic cells. Dev Biol. 2005;285:49–56. doi: 10.1016/j.ydbio.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Fukushima S, Yamashita S, Kobayashi H, Takami H, Fukuoka K, Nakamura T, Yamasaki K, Matsushita Y, Nakamura H, Totoki Y, Kato M, Suzuki T, Mishima K, Yanagisawa T, Mukasa A, Saito N, Kanamori M, Kumabe T, Tominaga T, Nagane M, Iuchi T, Yoshimoto K, Mizoguchi M, Tamura K, Sakai K, Sugiyama K, Nakada M, Yokogami K, Takeshima H, Kanemura Y, Matsuda M, Matsumura A, Kurozumi K, Ueki K, Nonaka M, Asai A, Kawahara N, Hirose Y, Takayama T, Nakazato Y, Narita Y, Shibata T, Matsutani M, Ushijima T, Nishikawa R, Ichimura K. Genome-wide methylation profiles in primary intracranial germ cell tumors indicate a primordial germ cell origin for germinomas. Acta Neuropathol. 2017;133:445–462. doi: 10.1007/s00401-017-1673-2. [DOI] [PubMed] [Google Scholar]
- Gillis AJ, Stoop H, Biermann K, van Gurp RJ, Swartzman E, Cribbes S, Ferlinz A, Shannon M, Oosterhuis JW, Looijenga LH. Expression and interdependencies of pluripotency factors LIN28, OCT3/4, NANOG and SOX2 in human testicular germ cells and tumours of the testis. Int J Androl. 2011;34:e160–174. doi: 10.1111/j.1365-2605.2011.01148.x. [DOI] [PubMed] [Google Scholar]
- Gkountela S, Li Z, Vincent JJ, Zhang KX, Chen A, Pellegrini M, Clark AT. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol. 2013;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, Clark AT. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, Wang W, Li R, Yan J, Zhi X, Zhang Y, Jin H, Zhang W, Hou Y, Zhu P, Li J, Zhang L, Liu S, Ren Y, Zhu X, Wen L, Gao YQ, Tang F, Qiao J. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Heaney JD, Anderson EL, Michelson MV, Zechel JL, Conrad PA, Page DC, Nadeau JH. Germ cell pluripotency, premature differentiation and susceptibility to testicular teratomas in mice. Development. 2012;139:1577–1586. doi: 10.1242/dev.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N, Tang WW, Azim Surani M. Germ cell specification and pluripotency in mammals: a perspective from early embryogenesis. Reprod Med Biol. 2014;13:203–215. doi: 10.1007/s12522-014-0184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N, Weinberger L, Tang WW, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julaton VT, Reijo Pera RA. NANOS3 function in human germ cell development. Hum Mol Genet. 2011;20:2238–2250. doi: 10.1093/hmg/ddr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yamaji M, Seki Y, Saitou M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle. 2008;7:3514–3518. doi: 10.4161/cc.7.22.6979. [DOI] [PubMed] [Google Scholar]
- Leroy X, Augusto D, Leteurtre E, Gosselin B. CD30 and CD117 (c-kit) used in combination are useful for distinguishing embryonal carcinoma from seminoma. J Histochem Cytochem. 2002;50:283–285. doi: 10.1177/002215540205000216. [DOI] [PubMed] [Google Scholar]
- Nakaki F, Saitou M. PRDM14: a unique regulator for pluripotency and epigenetic reprogramming. Trends Biochem Sci. 2014;39:289–298. doi: 10.1016/j.tibs.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Nishikawa N, Toyota M, Suzuki H, Honma T, Fujikane T, Ohmura T, Nishidate T, Ohe-Toyota M, Maruyama R, Sonoda T, Sasaki Y, Urano T, Imai K, Hirata K, Tokino T. Gene amplification and overexpression of PRDM14 in breast cancers. Cancer Res. 2007;67:9649–9657. doi: 10.1158/0008-5472.CAN-06-4111. [DOI] [PubMed] [Google Scholar]
- Oliveros-Etter M, Li Z, Nee K, Hosohama L, Hargan-Calvopina J, Lee SA, Joti P, Yu J, Clark AT. PGC reversion to pluripotency involves erasure of DNA methylation from imprinting control centers followed by locus-specific re-methylation. Stem Cell Rep. 2015;5:337–349. doi: 10.1016/j.stemcr.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashai N, Hao H, All A, Gupta S, Chaerkady R, De Los Angeles A, Gearhart JD, Kerr CL. Genome-wide profiling of pluripotent cells reveals a unique molecular signature of human embryonic germ cells. PLoS One. 2012;7:e39088. doi: 10.1371/journal.pone.0039088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Chen D, Liu W, Kim R, Sahakyan A, Lukianchikov A, Plath K, Jacobsen SE, Clark AT. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell. 2016;18:323–329. doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF, Blasco Lafita MJ, Mills J. Cultured stem-cells from human testicular teratomas: the nature of human embryonal carcinoma, and its comparison with two types of yolk-sac carcinoma. Int J Cancer. 1987;40:334–343. doi: 10.1002/ijc.2910400309. [DOI] [PubMed] [Google Scholar]
- Perrett RM, Turnpenny L, Eckert JJ, O’Shea M, Sonne SB, Cameron IT, Wilson DI, Rajpert-De Meyts E, Hanley NA. The early human germ cell lineage does not express SOX2 during in vivo development or upon in vitro culture. Biol Reprod. 2008;78:852–858. doi: 10.1095/biolreprod.107.066175. [DOI] [PubMed] [Google Scholar]
- Rajpert-De Meyts E, Nielsen JE, Skakkebaek NE, Almstrup K. Diagnostic markers for germ cell neoplasms: from placental-like alkaline phosphatase to micro-RNAs. Folia Histochem Cytobiol. 2015;53:177–188. doi: 10.5603/FHC.a2015.0020. [DOI] [PubMed] [Google Scholar]
- Ruark E, Seal S, McDonald H, Zhang F, Elliot A, Lau K, Perdeaux E, Rapley E, Eeles R, Peto J, Kote-Jarai Z, Muir K, Nsengimana J, Shipley J, Bishop DT, Stratton MR, Easton DF, Huddart RA, Rahman N, Turnbull C U.K.T.C. Collaboration. Identification of nine new susceptibility loci for testicular cancer, including variants near DAZL and PRDM14. Nat Genet. 2013;45:686–689. doi: 10.1038/ng.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, Nakamura S, Sekiguchi K, Sakuma T, Yamamoto T, Mori T, Woltjen K, Nakagawa M, Yamamoto T, Takahashi K, Yamanaka S, Saitou M. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, Yamamoto T, Saitou M. The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev Cell. 2016;39:169–185. doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Schmoll HJ. Extragonadal germ cell tumors. Ann Oncol. 2002;13(Suppl 4):265–272. doi: 10.1093/annonc/mdf669. [DOI] [PubMed] [Google Scholar]
- Sugawa F, Arauzo-Bravo MJ, Yoon J, Kim KP, Aramaki S, Wu G, Stehling M, Psathaki OE, Hubner K, Scholer HR. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015;34:1009–1024. doi: 10.15252/embj.201488049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat Rev Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- Terashima K, Yu A, Chow WY, Hsu WC, Chen P, Wong S, Hung YS, Suzuki T, Nishikawa R, Matsutani M, Nakamura H, Ng HK, Allen JC, Aldape KD, Su JM, Adesina AM, Leung HC, Man TK, Lau CC. Genome-wide analysis of DNA copy number alterations and loss of heterozygosity in intracranial germ cell tumors. Pediatr Blood Cancer. 2014;61:593–600. doi: 10.1002/pbc.24833. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, Lungjangwa T, Imsoonthornruksa S, Stelzer Y, Rangarajan S, D’Alessio A, Zhang J, Gao Q, Dawlaty MM, Young RA, Gray NS, Jaenisch R. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneyoshi N, Sumi T, Onda H, Nojima H, Nakatsuji N, Suemori H. PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem Biophys Res Commun. 2008;367:899–905. doi: 10.1016/j.bbrc.2007.12.189. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- Zhang T, Meng L, Dong W, Shen H, Zhang S, Liu Q, Du J. High expression of PRDM14 correlates with cell differentiation and is a novel prognostic marker in resected non-small cell lung cancer. Med Oncol. 2013;30:605. doi: 10.1007/s12032-013-0605-9. [DOI] [PubMed] [Google Scholar]