Abstract
In this short review, we highlight recent findings in the emerging field of epitranscriptomic mechanisms and discuss their potential role in neural plasticity, learning and memory. These include the influence of RNA modifications on activity-induced RNA structure states, RNA editing and RNA localization, and how qualitative state changes in RNA increase the functional diversity and information-carrying capacity of RNA molecules. We predict that RNA modifications may be just as important for synaptic plasticity and memory as quantitative changes in transcript and protein abundance, but with the added advantage of not being required to signal back to the nucleus, and therefore better suited to be coordinated with the temporal dynamics of learning.
Keywords: epitranscriptomics, RNA modification, neuron, memory, plasticity, brain
Introduction
In recent years, rapid advances in next generation sequencing and their convergence with new chemogenetic RNA labeling techniques has led to an unprecedented level of understanding of RNA biology and the emergence of a new field called “epitranscriptomics” (Zhao et al., 2017; Song and Yi, 2017). Epitranscriptomic mechanisms are defined as the ensemble of functionally relevant changes to RNA that serve to influence the complexity of information processing mediated by RNA, and which move beyond the classic boundaries of a linear DNA-RNA-protein continuum. Given the extraordinary diversity of their functional influence at all levels of RNA metabolism, it is perhaps unsurprising that post-transcriptional RNA modifications have now been implicated in neural plasticity, learning, and memory (Hess et al., 2013; Walters et al., 2017; Widagdo et al., 2016). What defines this as a breakthrough moment in contemporary neuroscience, however, is the emerging appreciation that RNA modifications may be essential for neurons to adapt in real-time and in an experience-dependent manner, with the added advantage of exerting their effects without the need to signal back to the nucleus as is the case with activity-dependent Ca2+ signalling that is commonly associated with plasticity, transcription and memory formation (Flavell and Greenberg, 2008), and in some instances, effectively bypassing the requirement for a linear relationship between mRNA and protein levels in the brain.
RNA modifications can influence the ‘what’, ‘where’, ‘when’, ‘why’ and ‘how’ a specific RNA will become functionally relevant in a neuron. For example, RNA modification can directly influence the rate of mRNA translation and decay (Wang et al., 2015; Zhou et al., 2015; Choi et al., 2016; Du et al., 2016; Mauer et al., 2017; Slobodin et al., 2017) although there is some debate as to whether these effects are benefical or detrimental for overall RNA function. RNA modification also directs the dynamic switching of long noncoding RNA (lncRNA) structure states that control their interaction with RNA binding proteins (RBPs), a process which is fundamental for non-coding RNA-directed epigenetic regulation of gene expression (Shafik et al., 2016; Spitale et al., 2015; Liu et al., 2015; Patel et al., 2016). Perhaps most importantly, RNA modifications may also promote the localization of RNAs into specific cellular compartments, including the synapse where they could be selectively modified. Indeed, it has recently been shown that enzymes required for regulating RNA methylation are present at the synapse (Walters et al, 2017). This may be particularly important for mediating rapid effects on plasticity at individual synapses, such as that which occurs during learning and in the formation of memory.
We predict that the cell-type-specific and context-dependent functional state of RNA will soon be considered just as important as traditional measures of RNA abundance or activity-induced RNA expression patterns for understanding the role of RNA in the network of molecular mechanisms underlying learning and memory. The purpose of this short review is to introduce the neuroscience community to the field of epitranscriptomics, and to provide a few suggestions as to where the continued discovery and functional characterization of novel mechanisms of RNA metabolism may lead.
RNA modifications
It has long been known that the covalent modification of DNA has profound effects on its structure and function. Likewise, RNA is also subject to covalent modification as an important epigenetic regulatory mechanism (Liu & Pan, 2016; Roundtree et al., 2017; Zhao et al., 2017). RNA modifications, however, are much more diverse than those that occur on DNA, with more than 100 identified to date (Machnicka et al., 2012). Additionally, specific RNA modifications are targeted to different functional classes of RNA, including ribosomal RNA (rRNA), transfer RNA (tRNA), messenger RNA (mRNA) and long non-coding RNA (lncRNA). This complexity represents a highly-sophisticated layer of control over RNA function and translation that appears to be involved in the fine-tuning of transcriptomic responses to the environment. Although we are only just beginning to understand the importance of this dimension of RNA function (Frye et al., 2016), several of the most prevalent RNA marks have been profiled, alongside basic characterization of the proteins which read, write and erase them (Table 1). Several of these comparatively well understood RNA modifications are described below.
Table 1.
Summary of the most well characterized epitranscriptomic mechanisms to date, each of which may play important roles in conferring functionality to RNA in the brain to mediate experience-dependent plasticity, learning and memory.
| RNA modification | Enzyme | Function |
|---|---|---|
| pseudoU | pseudouridine synthase (PUS 1–10) | Promotes RBP interaction RNA structure stability mRNA translation |
| 5mC | NSUN2 RNA methyltransferase DNMT2 RNA methyltransferase ALYREF 5mC reader |
Potentially localization |
| 5hmC | Tet1 | Enriched in brain, function in RNA unknown |
| m6A | METTL3/METTL14 (RNA methyltransferase) FTO/ALKBH5 (RNA demethylase) YTHDF1-3 (readers) |
RNA stability Splicing Translation Localization Decay Structure states RBP interactions |
| m6Am | ? | RNA stability |
| m1A | ? | Translation |
| A to I editing | ADAR/AID | Alternate protein synthesis |
| C to U editing | APOBEC | Alternate protein synthesis |
Pseudouridine (Ψ)
The isomerization of uridine bases to pseudouridine (also known as pseudoU or Ψ) is the most abundant of all RNA modifications (Ofengand, 2002), and has recently been shown to occur on both mRNAs and ncRNAs in the brain and to respond dynamically to stress (Li et al., 2015). The accumulation of Ψ is thought to be involved in RNA structure stability and mRNA translation, and to promote RBP interactions (Yu & Meier, 2014); however, it remains to be determined whether this RNA modification is functionally relevant in the context of experience-dependent plasticity. An interesting hint that Ψ may be involved in the experience-dependent regulation of RNA states in the brain comes from observations of the enzymes and ncRNAs that catalyze the formation of Ψ. For example, the pseudouridine synthase Trub1 is highly expressed in the brain, and its target RNAs are also enriched relative to other tissues (Li et al., 2015, Safra et al., 2017). In a preliminary series of studies, we have found that Trub1 is dynamically expressed in the prefrontal cortex following fear conditioning, and that knockdown of this enzyme leads to altered memory (unpublished observation). Furthermore, small nucleolar RNAs (snoRNAs) serve as guides for the site-directed action of other pseudouridine synthases (Ge & Yu, 2013). This class of small ncRNAs is also dynamically expressed in the hippocampus in response to contextual fear conditioning (Rogelj et al., 2013; Poplawski et al., 2016). In additional unpublished studies in our lab, we have also observed dynamic changes in snoRNA expression in the prefrontal cortex in response to cued fear conditioning, including several known to serve as guides for Ψ. Together, these data indicate that the mechanism by which Ψ is generated in the brain is dynamic and, although purely speculative this stage, suggest that Ψ-mediated post-transcriptional regulation of RNA could be involved in learning and memory.
5-methylcytosine (m5C) and 5-hydroxymethylcytosine (hm5C)
The methylation of cytosine in RNA has been mapped in the eukaryotic transcriptome and has been shown to occur in both mRNA and ncRNA (Squires et al., 2012), and targets polyadenylated RNA in the mouse brain (Amort et al., 2017). The formation of 5-methylcytosine (m5C) in RNA is catalyzed primarily by the RNA methyltransferase NSUN2, and to some extent by DNMT2 (Khoddami & Cairns, 2013, Tuorto et al., 2012). Mutations in NSUN2 have been implicated in intellectual disability (Abbasi-Moheb et al., 2012), but a direct role for this protein in neural plasticity, learning and memory has yet to be demonstrated. Reader proteins for m5C are also being characterized. One recently identified 5mC reader is Aly/REF export factor (ALYREF), which directs the movement of mRNAs from the nucleus into the cytoplasm (Yang et al., 2017); this finding suggests a potential role for 5mC in marking mRNAs for localization within the cell, which is an important feature of synaptic plasticity.
The oxidized derivative of m5C, 5-hydroxymethylcytosine (hm5C), also accumulates within polyadenylated RNA, and influences translation in the Drosophila brain (Delatte et al., 2016). This epitranscriptomic mark is present globally within RNA in different regions of the mouse brain, with the highest levels being found in the brainstem, hippocampus and cerebellum (Miao et al., 2016). In a recent study, it was discovered that hm5C in RNA can be converted by Tet1 to produce 5-formylcytosine (f5C) and 5-carboxylcyotsine (ca5C) in RNA, similar to that which occurs during active DNA demethylation (Basanta-Sanchez, 2017); however, little is known about the functional relevance of these rare, but potentially important, RNA modifications in the brain.
N6-methyladenosine (m6A) and N1-methyladenosine (m1A)
The methylation of adenosine is perhaps the most well characterized RNA modification, with N6-methyladenosine (m6A) having been shown to participate in a variety of biological processes, including translation, splicing, RNA decay, localization and structure-state induced changes in RBP interaction (Wang et al., 2015, Zhou et al., 2015; Lu and Pan, 2016; Zhou et al., 2016). This RNA modification has a preference for a particular sequence motif (RRm6ACH) and has been shown to accumulate at the stop codon, along the coding sequence and within the 3′ untranslated region (UTR) (Dominissini et al., 2012, Meyer et al., 2012). In the mouse brain, the level of m6A increases across the lifespan (Meyer et al., 2012) with recent studies demonstrating an important role for the accumulation of m6A in learning and memory (Hess et al., 2013, Walters et al., 2017, Widagdo et al., 2016).
A protein complex that comprises the active subunits methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14), and the regulatory subunit Wilms’ tumor 1-associating protein (WTAP) catalyzes the formation of m6A. Conversely, m6A is removed from RNA by fat mass and obesity-associated protein (FTO) and alkylation repair homologue protein 5 (ALKBH5). Moreover, m6A is also detected by specific readers, including the YTH domain family (YTHDF1, YTHDF2, and YTHDF3), which promote the translocation of m6A-modified RNAs to different cellular compartments for translation and decay (reviewed by Song & Yi, 2017). N6, 2′-O-dimethyladenosine (m6Am), which is a close cousin of m6A, also impacts RNA metabolism; this RNA modification promotes RNA stability by protecting RNA from degradation by mRNA-decapping enzyme 2 (DCP2) (Mauer et al., 2017). In contrast, methylation at the N1 position of adenosine (m1A) is also present in mRNA and lncRNA but found predominantly around the start codon, where it preferentially accumulates proximal to canonical and alternative translation initiation sites (Dominissini et al., 2016). These unique features are highly conserved between mouse and human cells, indicating a functional role for m1A in promoting translation.
From this evidence, it is clear that RNA modifications are essential for coordinating many aspects of cellular function. The grand challenge is to elucidate how they regulate experience-dependent gene expression in the adult brain, and to determine to what extent these epitranscriptomic mechanisms are involved in learning and memory. What follows is a brief discussion of what we believe are fruitful areas of investigation into how this might occur.
RNA modifications may affect activity-induced RNA structure states in the brain
The process of learning, and the associated neural plasticity that leads to memory formation, requires the ability to detect and rapidly respond to dynamic changes in the environment. At the level of individual neurons, these responses occur on a timescale that is faster than activity-induced transcription via the coordinated, activity-induced switching of internal molecular states and cellular metabolism. In recent years, our understanding of experience-dependent gene regulation and neuronal adaptation has advanced significantly with the recognition that the structure state of RNA can provide the modifiable context in which this can occur. Structurally labile RNA elements are able to react to changes in ion concentration and metabolite flux, which can lead to altered RBP and RNA-RNA interactions within the cell (Mortimer et al., 2014). For example, a stem-loop structure in the 3′UTR of brain-derived neurotrophic factor (BDNF) mRNA is structurally responsive to calcium influx, thereby stabilizing this transcript in response to neuronal activity (Fukuchi & Tsuda, 2010, Vanevski & Xu, 2015). This structure state also promotes the interaction of BDNF mRNA with the RNA binding protein HuD, which has a direct impact on translation of BDNF (Allen et al., 2013, Vanevski & Xu, 2015). Further, an important role for the G-quadruplex RNA structures, which are non-canonical RNA structures organized in stacks of tetrads or G-quartets, in which four guanines are assembled in a planar arrangement by Hoogsteen hydrogen bonding. G-quadruplex RNA has been shown to be critically involved in mediating the localization of CamKIIα and PSD-95 to neurites, which are essential for synaptic plasticity (Subramian et al, 2011).
Importantly, the dynamic switching of RNA structure states in response to changes in the cellular environment can be influenced by RNA modification. An interesting example of structural lability conferred by RNA modification is the brain-enriched lncRNA MALAT1, which influences synaptogenesis (Bernard et al., 2010) and is found in nuclear paraspeckles within hippocampal neurons, implying a key role in alternative splicing. When MALAT1 accumulates m6A modifications, its interaction with the RBP heterogeneous nuclear ribonucleoprotein C (HNRNPC) is enhanced, which then promotes its accumulation in paraspeckles (Liu et al., 2015, Zhou et al., 2016). We found that a significant number of lncRNAs, including MALAT1, are dynamically expressed in the adult brain in response to fear-related learning (Spadaro et al., 2015) and that the majority of these lncRNAs contain motifs for m6A and Ψ. The purpose of these modifications on neuronal lncRNAs remains to be determined; direct induction of an experience-dependent structure state change could promote the downstream influence of lncRNAs on RNA-directed epigenetic regulation in learning and memory, and this hypothesis warrants further investigation.
RNA modifications may affect activity-induced RNA editing in the brain
RNA editing is an enzymatic process by which the RNA sequence is altered after transcription, often by conversion of canonical bases into other nucleobases, such as adenosine to inosine with thousands of sites having been shown in the human brain (Sakurai et al, 2014). RNA editing is mediated by two major classes of enzymes: adenosine deaminases (ADARs) and the lesser known vertebrate-specific APOBEC family, which are cytidine deaminases (Li & Church, 2013, Prohaska et al., 2014). ADAR-mediated RNA editing involves the deamination of adenosine and its subsequent conversion to inosine, which is functionally similar to guanine and preferentially base-pairs with cytosine (O’Neil et al., 2017). Adenosine-to-inosine editing alters RNA base-pairing to itself (with potential effects on RNA secondary structure) and to other RNAs, including tRNAs; this can lead to the translation of proteins which differ in sequence despite arising from the same genomic locus, and may serve as an important context-specific mechanism of functional diversity involved in the fine-tuning of the genomic response to rapid changes in the environment (Li & Church, 2013, O’Neil et al., 2017). Similarly, cytosine deamination in RNA converts a cytosine to uridine, which also alters the base-pairing capacity of the RNA (Prohaska et al., 2014). Although it is increasingly recognized that RNA editing is a fundamental feature of neural plasticity and is involved in learning and memory (Behm & Öhman, 2016), little is known about how the RNA editing machinery can so efficiently select its target RNAs for downstream processing.
A hint as to how this might occur comes from observations of the role of deamination in DNA editing. For example, activation-induced cytidine deaminase (AID), originally characterized within the context of antibody class diversification, was recently shown to have a preferred affinity for methylated adenosine (Franchini et al., 2014). AID has also been proposed to target hydroxymethylcytosine in DNA as part of the active DNA demethylation pathway in the brain (Guo et al., 2011). In Trypanosoma brucei, C-to-U editing occurs at specific sites in tRNA that harbor a 3-methylcytosine modification (Rubio et al., 2017). Given that deaminases can target both DNA and RNA, it is plausible that RNA modification is a prerequisite for the efficient targeting of select RNAs for editing, which might then confer the specificity required to control RNA editing in a dynamic, experience-dependent manner in the brain.
RNA modifications may affect activity-induced RNA localization in the brain
Local, stimulus-responsive, translation of plasticity-related genes is involved in key aspects of brain function, including synaptic plasticity and memory (Kang & Schuman, 1996, Miller et al., 2002, Wang et al., 2009). This process involves synaptic localization of ribosomes and their respective regulatory RNAs, as well as RNA-binding proteins such as TDP-43, and specific mRNAs which are subject to local synaptic translation. In fact, over 2000 dynamically expressed mRNAs, including CamKIIα and Arc, can be found in the neuropil and particularly within dendrites (Cajigas et al., 2012; and reviewed by Fernandez-Moya et al., 2014, Rangaraju et al., 2017). The localized expression of RNA is not restricted to protein-coding genes; the RNA-induced silencing complex and associated microRNAs (including miR-9 and miR-134) are also found in dendrites and are functionally involved in plasticity and memory (Sambandan et al., 2017, Schratt et al., 2006). Other classes of small ncRNA can also occur in dendrites; the small ncRNA BC1 is involved in local translational control at the dendrite (Muslimov et al., 2006), and some neuronal Piwi-interacting RNAs (piRNAs) occur within the dendritic compartment and affect dendritic spine morphogenesis in mouse neurons (Lee et al., 2011).
A critical issue that remains to be resolved is how nascent RNAs are directed to specific dendritic compartments when and where they are most needed, and rendered functional in a temporally controlled manner (such as the adaptive local translation that occurs at individual synapses following learning). A recent study reported that there is abundant expression of the m6A demethylase FTO at the synapse, and that FTO contributes to contextual fear memory, potentially by influencing the local translation potential of mRNAs expressed at hippocampal synapses (Walters et al., 2017). A role for RNA modification in regulating plasticity states in the brain is in agreement with our findings demonstrating that the experience-dependent transcriptome-wide accumulation of m6A is necessary for the formation of cued fear memory (Widagdo et al., 2016). We also observed that a significant number of m6A-modified neuronal transcripts encode proteins that influence synaptic plasticity and memory, including CamKIIα. We predict that further research will identify more synaptically localized proteins, which act as readers and modifiers, and that are essential regulators of synaptic plasticity through local control over RNA stability and translation mediated by m6A, m5C and other RNA modifications.
Outlook
It is becoming increasingly evident that RNA modifications may have a profound impact on RNA structure, RNA editing, and the localization of both coding and non-coding RNAs to specific subcellular compartments (Figure 1). Together, these effects vastly increase the functional diversity and information-carrying capacity of RNA molecules, and these qualitative changes may be just as important for synaptic plasticity and memory as quantitative changes in transcript and protein abundance. RNA modification typically does not require signaling back to the nucleus in order to become functional, which means these processes can therefore be initiated, and exert their effects, very rapidly. More than any other cell type, neurons must be able to respond to altered environmental inputs with extraordinary speed, therefore, RNA modifications represent a clear candidate for a molecular mechanism capable of aligning with the timing required for experience-dependent plasticity, and we predict that they play an integral role in controlling the organizational architecture of neuronal networks involved in learning and memory.
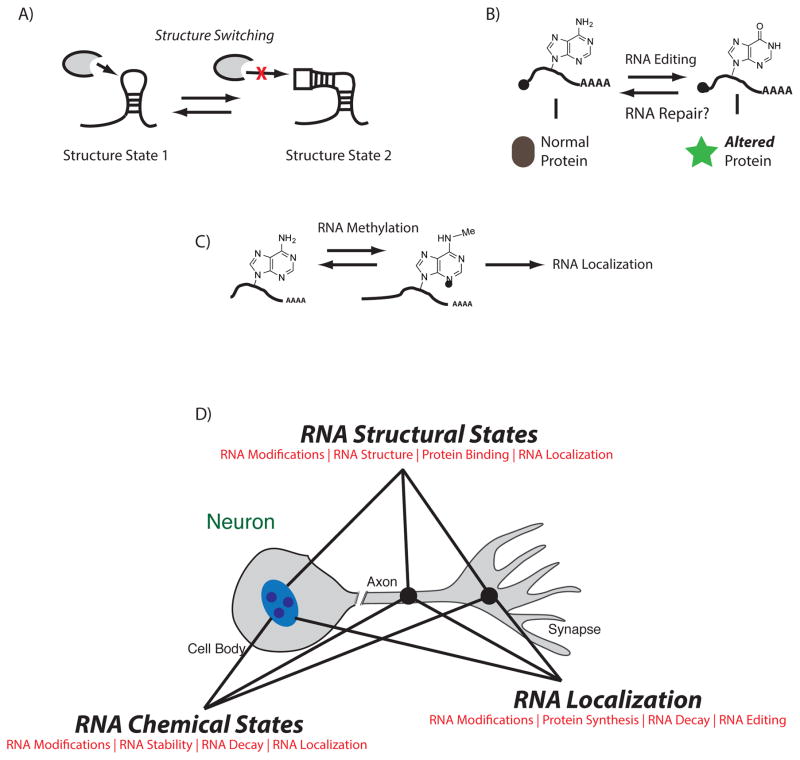
Figure 1.
A) RNA modifications can alter the structure of RNA, which then influences its interaction with RNA binding proteins. This is particularly relevant for long noncoding RNAs which become functionalized depending on their structure state, B) RNA modifications may influence RNA editing as m6A is a known target for deamination (A–I conversion). This would result in altered protein translation driven from the same initial transcript, C) RNA modification on mRNA can alter its ability to be sequestered to the ribosome for translation or to other nuclear subcompartments for further processing (alternative splicing) or degradation as well as transport to the synapse. D) In each compartment of the neuron, RNA modifications may impact the fine-tuning of RNA function, this would be particularly important at activated synapses where specificity is critical plasticity, learning and memory.
Within the past few years, the field of epitranscriptomics has benefited from rapid advances in technology and in our appreciation of RNA modification. However, of more than 100 known covalent RNA modifications, very few have been profiled across the transcriptome and even fewer have been functionally investigated in neurons. A deeper understanding of all RNA modifications in the brain, including their localized patterns of accumulation in different cell types and regions of the brain in response to a variety of learning conditions, is urgently required. In order to achieve this, we must be able to directly quantify the temporal and spatial dynamics of RNA modifications, and elucidate how they regulate protein abundance and localization to drive changes in synaptic plasticity. Furthermore, in order to determine the functional relevance of different RNA species, structural states, and modifications for cognition and memory, innovative new methods for temporally precise and spatially restricted sequence-specific causal manipulations will be required. Fortunately, new methods are rapidly coming online to assist in understanding the functional role of the epitranscriptome (see Chen and Engel in this issue; Nainar et al., 2016, and Li, Xiong and Yi, 2017). Armed with new sequencing technologies, including cell-type specific and state-dependent profiling approaches, we are ready to move beyond a linear understanding of inducible gene expression and plasticity in the brain, and to elucidate the qualitative nature of RNA and its role in cognition and memory.
Acknowledgments
The authors gratefully acknowledge grant support from the NIH (5R01MH105398-TWB and 1R01MH109588-TWB and RCS) and the NIGMS (1DP2GM119164-RCS). ELZ and LJL are supported by the University of Queensland and LJL is a recipient of a Westpac Future Leaders Scholarship. The authors thank Ms. Rowan Tweedale for helpful editing of the manuscript.
References
- Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM, Esmaeeli-Nieh S, Cremer K. Mutations in NSUN2 cause autosomal-recessive intellectual disability. The American Journal of Human Genetics. 2012;90:847–855. doi: 10.1016/j.ajhg.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M, Bird C, Feng W, Liu G, Li W, Perrone-Bizzozero NI, Feng Y. HuD promotes BDNF expression in brain neurons via selective stabilization of the BDNF long 3′ UTR mRNA. PloS one. 2013;8:e55718. doi: 10.1371/journal.pone.0055718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, Jia XY, Micura R, Lusser A. Distinct 5-methylcytosine profiles in poly (A) RNA from mouse embryonic stem cells and brain. Genome biology. 2017;18:1. doi: 10.1186/s13059-016-1139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basanta-Sanchez M, Wang R, Liu Z, Ye X, Li M, Shi X, Agris PF, Zhou Y, Huang Y, Sheng J. TET1-Mediated Oxidation of 5-Formylcytosine (5fC) to 5-Carboxycytosine (5caC) in RNA. ChemBioChem. 2017;18:72–76. doi: 10.1002/cbic.201600328. [DOI] [PubMed] [Google Scholar]
- Behm M, Öhman M. RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends in Genetics. 2016;32:165–175. doi: 10.1016/j.tig.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. The EMBO journal. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74(3):453–66. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Ieong KW, Demirci H, Chen J, Petrov A, Prabhakar A, O’Leary SE, Dominissini D, Rechavi G, Soltis SM, Ehrenberg M, Puglisi JD. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat Struct Mol Biol. 2016;23(2):110–5. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moya SM, Bauer KE, Kiebler MA. Meet the players: local translation at the synapse. Frontiers in molecular neuroscience. 2014:7. doi: 10.3389/fnmol.2014.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–90. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini DM, Chan CF, Morgan H, Incorvaia E, Rangam G, Dean W, Santos F, Reik W, Petersen-Mahrt SK. Processive DNA demethylation via DNA deaminase-induced lesion resolution. PloS one. 2014;9:e97754. doi: 10.1371/journal.pone.0097754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA modifications: what have we learned and where are we headed? Nature Reviews Genetics. 2016;17:365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- Fukuchi M, Tsuda M. Involvement of the 3′-untranslated region of the brain-derived neurotrophic factor gene in activity-dependent mRNA stabilization. Journal of neurochemistry. 2010;115:1222–1233. doi: 10.1111/j.1471-4159.2010.07016.x. [DOI] [PubMed] [Google Scholar]
- Ge J, Yu YT. RNA pseudouridylation: new insights into an old modification. Trends in biochemical sciences. 2013;38:210–218. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming G-l, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Brönneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento O, Belgardt BF, Franz T, Horvath TL, Rüther U, Jaffrey SR, Kloppenburg P, Brüning JC. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16(8):1042–8. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nature biotechnology. 2013;31:458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath B, Kosik KS. Identification of piRNAs in the central nervous system. Rna. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Church GM. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nature neuroscience. 2013;16:1518–1522. doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nature chemical biology. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 2017;14(1):23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nature structural & molecular biology. 2016;23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic acids research. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic acids research. 2012;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Xin N, Wei B, Hua X, Zhang G, Leng C, Zhao C, Wu D, Li J, Ge W. 5-hydroxymethylcytosine is detected in RNA from mouse brain tissues. Brain research. 2016;1642:546–552. doi: 10.1016/j.brainres.2016.04.055. [DOI] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nature reviews Genetics. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- Muslimov IA, Iacoangeli A, Brosius J, Tiedge H. Spatial codes in dendritic BC1 RNA. The Journal of cell biology. 2006;175:427–439. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nainar S, Marshall PR, Tyler CR, Spitale RC, Bredy TW. Evolving insights into RNA modifications and their functional diversity in the brain. Nature neuroscience. 2016;19:1292–1298. doi: 10.1038/nn.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil RT, Wang X, Morabito MV, Emeson RB. Comparative analysis of A-to-I editing in human and non-human primate brains reveals conserved patterns and context-dependent regulation of RNA editing. Molecular brain. 2017;10:11. doi: 10.1186/s13041-017-0291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS letters. 2002;514:17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplawski SG, Peixoto L, Porcari GS, Wimmer ME, McNally AG, Mizuno K, Giese KP, Chatterjee S, Koberstein JN, Risso D. Contextual fear conditioning induces differential alternative splicing. Neurobiology of learning and memory. 2016;134:221–235. doi: 10.1016/j.nlm.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohaska KM, Bennett RP, Salter JD, Smith HC. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdisciplinary Reviews: RNA. 2014;5:493–508. doi: 10.1002/wrna.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V, tom Dieck S, Schuman EM. Local translation in neuronal compartments: how local is local? EMBO reports. 2017;18:693–711. doi: 10.15252/embr.201744045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogelj B, Hartmann CE, Yeo CH, Hunt SP, Giese KP. Contextual fear conditioning regulates the expression of brain-specific small nucleolar RNAs in hippocampus. Eur J Neurosci. 2003;(11):3089–96. doi: 10.1111/j.1460-9568.2003.03026.x. [DOI] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MAT, Gaston KW, McKenney KM, Fleming IM, Paris Z, Limbach PA, Alfonzo JD. Editing and methylation at a single site by functionally interdependent activities. Nature. 2017;542:494–497. doi: 10.1038/nature21396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Nir R, Farouq D, Slutzkin IV, Schwartz S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome research. 2017;27:393–406. doi: 10.1101/gr.207613.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Ueda H, Yano T, Okada S, Terajima H, Mitsuyama T, Toyoda A, Fujiyama A, Kawabata H, Suzuki T. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 2014;24(3):522–34. doi: 10.1101/gr.162537.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan S, Akbalik G, Kochen L, Rinne J, Kahlstatt J, Glock C, Tushev G, Alvarez-Castelao B, Heckel A, Schuman EM. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science. 2017;355:634–637. doi: 10.1126/science.aaf8995. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Shafik A, Schumann U, Evers M, Sibbritt T, Preiss T. The emerging epitranscriptomics of long noncoding RNAs. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2016;1859:59–70. doi: 10.1016/j.bbagrm.2015.10.019. [DOI] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell research. 2017 doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Han R, Calderone V, Vrielink JAO, Loayza-Puch F, Elkon R, Agami R. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell. 2017;169:326–337. e312. doi: 10.1016/j.cell.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Yi C. Chemical modifications to RNA: a new layer of gene expression regulation. ACS chemical biology. 2017;12:316–325. doi: 10.1021/acschembio.6b00960. [DOI] [PubMed] [Google Scholar]
- Spadaro PA, Flavell CR, Widagdo J, Ratnu VS, Troup M, Ragan C, Mattick JS, Bredy TW. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biological psychiatry. 2015;78:848–859. doi: 10.1016/j.biopsych.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic acids research. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. G–quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO reports. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nature structural & molecular biology. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- Vanevski F, Xu B. HuD interacts with BDNF mRNA and is essential for activity-induced BDNF synthesis in dendrites. PloS one. 2015;10:e0117264. doi: 10.1371/journal.pone.0117264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters BJ, Mercaldo V, Gillon CJ, Yip M, Neve RL, Boyce FM, Frankland PW, Josselyn SA. The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology. 2017 doi: 10.1038/npp.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC. Synapse-and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N 6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo J, Zhao QY, Kempen MJ, Tan MC, Ratnu VS, Wei W, Leighton L, Spadaro PA, Edson J, Anggono V. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. Journal of Neuroscience. 2016;36:6771–6777. doi: 10.1523/JNEUROSCI.4053-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Research. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Meier UT. RNA-guided isomerization of uridine to pseudouridine—pseudouridylation. RNA biology. 2014;11:1483–1494. doi: 10.4161/15476286.2014.972855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nature reviews Molecular cell biology. 2017;18:31. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, Pan T. N 6-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. Journal of molecular biology. 2016;428:822–833. doi: 10.1016/j.jmb.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]