Abstract
Mitochondrial respiratory chain disease is caused by a wide range of individually rare genetic disorders that impair cellular energy metabolism. While fluorescence microscopy analysis of nematodes fed MitoTracker Green (MTG) and tetramethylrhodamine ethyl ester (TMRE) can reliably quantify relative mitochondrial density and membrane potential, respectively, in C. elegans, it is a tedious process with limitations in the number and age of animals that can be studied. A novel, large particle, flow cytometry-based method reported here accelerates and automates the relative quantitation of mitochondrial physiology in nematode populations. Relative fluorescence profiles of nematode populations co-labeled with MTG and TMRE were obtained and analyzed by BioSorter (Union Biometrica). Variables tested included genetic mutation (wild-type N2 Bristol versus nuclear-encoded respiratory chain complex I mutant gas-1(fc21) worms), animal age (day 1 versus day 4 adults), classical respiratory chain inhibitor effects (oligomycin, FCCP), and pharmacologic therapy duration (24 hour versus 96 hour treatments with glucose or nicotinic acid). A custom MATLAB script, which can be run on any computer with MATLAB runtime, was written to automatically quantify and analyze results in large animal populations. BioSorter analysis independently validated relative MTG and TMRE changes that we had previously performed by fluorescence microscopy in a variety of experimental conditions, with notably greater animal population sizes and substantially reduced experimental time. Older, fragile animal populations that are difficult to study by microscopy approaches were readily amenable to analysis with the BioSorter method. Overall, this high-throughput method enables efficient relative quantitation of in vivo mitochondrial physiology over time in a living animal in response to gene mutations and candidate therapies, which can be used to accelerate the translation of basic research into optimization of clinical therapies for mitochondrial disease.
Keywords: Mitochondrial disease, flow cytometry, C. elegans, screening
1. Introduction
Primary mitochondrial respiratory chain diseases, individually rare gene disorders that together represent a collectively common group of inherited disorders, are characterized by impaired energy metabolism due to heterogeneous, genetically-based oxidative phosphorylation dysfunction.1 Affected patients are encountered across nearly all pediatric and adult medical disciplines, with a predominance of neurologic, muscular, cardiac, gastrointestinal, and ophthalmologic manifestations.2 Recent advancements in genomic sequencing technologies, such as whole exome sequencing, have led to more systematic identification of genetic causes of mitochondrial disease.3 The minimum prevalence rate for disease-causing mitochondrial DNA (mtDNA) mutations is 1 in 5000 individuals, which is comparable to prior estimates of the minimal prevalence of mitochondrial disease (also 1 in 5000) across all ages.4 Mutations in more than 250 nuclear DNA (nDNA) genes and all 37 mtDNA genes have been linked to a human mitochondrial disease.5 While important to identify disease etiologies, DNA sequencing does not quantify the degree of mitochondrial dysfunction or the resulting changes in flux that occur in intermediary metabolic pathways in a given disorder.6 In vitro biochemical assays can assess the function of key metabolic enzymes in patient cells or tissues, although these are often limited by large minimum sample requirements, lack of spatial and/or temporal information, and lengthy protocols for sample preparation and/or data collection.1 Consequently, a strong need exists to develop fast and reliable analytic methods in animal models that can be used to better characterize different genetic causes of mitochondrial diseases and evaluate effects of candidate drug therapies intended to improve mitochondrial function.
Developing techniques to efficiently quantify mitochondrial physiology in a living model animal will provide distinct investigative advantages in the study of mitochondrial disease. Study of a whole organism as compared to tissue culture can accelerate the translation of basic research into clinical therapies. In particular, Caenorhabditis elegans, a microscopic nematode, is a powerful invertebrate model animal in which to investigate the physiologic mechanisms of genetic diseases. Its genome has been completely sequenced, and publicly accessible collections of genetic mutants exist.7 The reproductive cycle of C. elegans is 3 days per generation, with production of 300 genetically identical offspring per young adult hermaphrodite worm. Thus, C. elegans can generate a large amount of stable genetic mutants in a short period of time. Mitochondrial composition and function is highly conserved between humans and C. elegans, where 84% of human mitochondrial respiratory chain complex I proteins are conserved in C. elegans.8 C. elegans worms have extensive mitochondrial activity, such as in its 8-cell terminal pharyngeal bulb that contracts up to 300 times per minute to break down bacterial food sources.9 Upon reaching adulthood, nematodes are only 1 millimeter in length and comprised of 959 cells, requiring microscopic study. Their transparency allows for ready quantification of the relative fluorescence intensity of different ingested dyes that target distinct organelles to inform their specific physiologic activities.
In the context of mitochondrial biology and disease, lipophilic and cationic fluorescent dyes can be used to interrogate key aspects of in vivo mitochondrial physiology in living animals. Establishment of an electrochemical gradient across the inner mitochondrial membrane by the respiratory chain (RC) creates the mitochondrial membrane potential that drives the generation of chemical energy in the form of adenosine triphosphate (ATP), through the highly conserved process of oxidative phosphorylation (OXPHOS). The same process necessary to generate the membrane potential also makes mitochondria a primary site of free radical species generation and scavenging.10 MitoTracker Green (MTG) is a fluorescent dye used to evaluate overall mitochondrial mass, as MTG selectively accumulates in the negatively-charged mitochondrial matrix due to its high pH and ability to react with mitochondrial proteins at the free thiol groups of cysteine residues.11 Tetramethylrhodamine ethyl ester (TMRE) is a fluorescent dye with a positively charged triphenylphosphonium ion that is used to evaluate relative mitochondrial membrane potential; its accumulation in the negatively charged mitochondrial matrix occurs proportionate to the charge differential established across the inner mitochondrial membrane.12 These two dyes have been extensively used in fluorescence microscopy studies in C. elegans.10 Thus, their concentrations and worm incubation times (typically 24 hours in young adults, as we have shown previously to represent a steady-state) effectively indicate relative mitochondrial amount and integrated function in a synchronized population of C. elegans young adult animals.10,13
Mitochondrial RC complex I (CI) deficiency is the most commonly implicated biochemical site impaired in human RC disease.1 Sequelae of RC CI dysfunction is broadly attributed to the central role of the RC in determining cellular NADH/NAD+ redox balance, integrating energy-generating chemiosmotic processes that include electron transport, proton flux, and ATP production, as well as modulating the balance of reactive oxygen species generation and scavenging.1 The well-characterized C. elegans gas-1(fc21) mutant strain has genetic-based CI deficiency that is caused by a homozygous p.R290K missense mutation in the highly conserved CI subunit homolog gene, NDUFS2.14 gas-1(fc21) worms have significantly decreased lifespan, decreased respiratory chain OXPHOS capacity,8 as well as decreased mitochondrial mass and membrane potential.10,13 Thus, gas-1(fc21) nematodes are a useful animal model in which to study alterations of in vivo mitochondrial physiology that occur relative to wild-type (N2 Bristol) control worms. In addition, our group has previously shown that treatment with nicotinic acid (vitamin B3) rescues the short lifespan of gas-1(fc21) nematodes,13 while a host of other nutritional therapies or supplements have shown benefit in diverse mitochondrial disease models.15 Knowledge of these effects on mitochondrial physiology of previously validated animal models and candidate therapies that were determined by single animal fluorescence microscopy studies can now be used to validate new high-throughput methods.
BioSorter (Union Biometrica) is a large particle flow cytometer and sorter that uses continuous flow to record fluorescence profiles of individual worms within a large population of thousands of animals. Fluorescence profiles can be used to quantify the animal location and relative intensity of distinct mitochondria-targeted fluorescent dyes. While similar information is attainable by manual fluorescence microscopy,10 focusing on and imaging individual worms is highly laborious and time-consuming, thereby limiting the number of animals that can be studied per condition. Indeed, the off-target, non-specific binding of lipophilic dyes (MTG, TMRE) to the fat granules of the anterior intestine10 has required manual microscopic analysis to localize analysis to the mitochondria-dense terminal pharyngeal bulb of C. elegans. Ideally, fluorescence profile analysis of individual worms can be automated in a high-throughput fashion through use of large particle flow cytometry. However, current software from Union Biometrica, Flow Pilot, lacks the needed functionality to reliably select and analyze fluorescence signal intensity in the mitochondria-dense head region of C. elegans. Although FlowPilot can analyze a region of interest as a function of the length of the animal profile, it cannot be used to reliably select a region of interest based on the shape of the fluorescence profile. Therefore, we sought to develop validated experimental and analytic methods to enable use of the BioSorter for relative quantitation of mitochondrial physiology in large nematode populations. This high-throughput screening approach is necessary to both validate and characterize the relative effects on mitochondrial amount and integrated function of different genetic mutant strains, as well as to screen individual gene models to evaluate the efficacy of candidate therapies.13
2. Methods
2.1 C. elegans strain selection and maintenance
A C. elegans stable mutant strain, gas-1(fc21), was studied that harbors a homozygous missense mutation in a nuclear-encoded RC CI subunit, NDUFS2.14 N2 Bristol was the wild-type control strain studied. pKEK strain, a strain with an EGFP-labeled wild-type GAS-1 protein under the control of the gas-1 promoter, was used to validate the staining patterns for MTG and TMRE by fluorescence microscopy. gas-1(fc21) and N2 Bristol worm strains were obtained from the Caenorhabditis Genetics Center (CGC, funded by NIH Office of Research Infrastructure Programs (P40 OD010440)). pKEK strain was obtained from Bernhard Kayser, P.hD., Philip Morgan, M.D., and Margaret Sedensky, M.D., at the University of Washington, Seattle, WA. Nematodes were maintained at 20°C incubators, as previously described.10,13
2.2 Fluorescence microscopy manual quantification of relative mitochondrial membrane potential and mitochondria mass in C. elegans adults
Mitochondrial membrane potential (tetramethylrhodamine ethyl ester, TMRE) and mitochondrial mass (MitoTracker Green FM, MTG) analyses were manually performed at 20°C using in vivo terminal pharyngeal bulb relative fluorescence quantitation, as previously described,10,13 with slight modifications. Briefly, synchronous populations of young adults were moved to 60 mm nematode growth media (NGM) plates spread with OP50 E. coli that contained a desired drug treatment and both 100 nM TMRE (mitochondrial membrane potential) and 2 μM MitoTracker Green FM (mitochondria content) for 24 hours (i.e., nematodes were co-labeled with MTG and TMRE). The next day, nematodes were transferred in S. basal media (5.85 g NaCl, 1 g K2 HPO4, 6 g KH2PO4, and 5 mg cholesterol, in 1 L H2O) onto 60 mm NGM agar plates spread with OP50 E. coli without fluorescent dye for 1 hour, to allow intestinal clearing of residual dye. Live nematodes were then paralyzed in situ with 5 mg/ml levamisole. Photographs were taken in a darkened room at 160× magnification with a Cool Snap cf2 camera (Nikon, Melville, NY) or at 40x magnification with EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific, Waltham, MA). A CY3 fluorescence cube set (MZFLIII, Leica, Bannockburn, IL) was used for TMRE. A GFP2 filter set (Leica) was used for MitoTracker Green FM. Exposure time was 300 ms. The resulting images were background subtracted, and the nematode terminal pharyngeal bulb was manually circled to obtain mean intensity of the region using Fiji Is Just ImageJ.16 Fluorescence data for each strain were normalized to its same day wild-type control to account for day-to-day experimental variation. A minimum of three independent experiments of approximately 50 animals per replicate were performed per strain for each dye.
2.3 Confocal microscopy confirmation of subcellular fluorescence dye localization
Confocal microscopy (TCS SP2, Leica, Bannockburn, IL) was used to confirm subcellular fluorescence localization in mitochondria within the head region of wild-type N2 worms, with the assistance of Andrea Stout, PhD, in the Cell Biology Confocal Core Facility, University of Pennsylvania, Philadelphia, PA. Worms were exposed for 24 hours individually or in combination to MitoTracker Green or TMRE. Following 1 hour intestinal clearing of fluorescent dyes on NGM agar plates spread with unlabeled E. coli, living nematodes were reversibly paralyzed on glass slides in 10 μl of 5 mg/ml levamisole.
2.4 BioSorter based flow cytometry fluorescence profile acquisition in young adult worms co-labeled with MTG and TMRE
Synchronous adult worm populations were simultaneously co-fed MTG and TMRE on NGM agar plates in preparation for flow cytometry analysis on the BioSorter, similarly as described above for fluorescence microscopy analyses. Worm samples were collected in 50 ml conical tubes in a final volume of 7 ml S. basal. 1 ml of 4 mg/ml levamisole was then added to the 50 ml conical tube to reach a final concentration of 25 mM, as was needed to paralyze the worms and minimize the effect of nematode movement on flow cytometry signal detection. For 96 hour treatments, fluorodeoxyuridine (FUDR) was added to plates to a final concentration of 100 μg/ml, as was needed to prevent the development of larvae to unlabeled adult worms from eggs laid by the fluorescent adults being studied. Excitation wavelength for the red channel was 561 nm, and the emission filter was a bandpass filter with center frequency 615 nm and bandwidth of 24 nm. Excitation wavelength for the green channel was 488 nm, and the emission filter was a bandpass filter with center frequency of 510 nm and bandwidth of 23 nm. Photomultiplier tube voltages used for red and green channels were 420 and 560 volts, respectively. The spillover from TMRE to green channel was negligible, whereas spillover from MTG to red channel was readily corrected with compensation of 35% (Supplemental File 1). Initial experiments were conducted with approximately 2,000 worms per condition. Subsequent biological replicate experiments, with a minimum of three independent experiments each with approximately 150 animals per condition, yielded sufficient data to detect statistically significant differences between groups.
2.5 Fluorecence profile data analysis to evaluate relative quantification of mitochondrial membrane potential and mass
FlowPilot is the available analysis software for BioSorter, but currently has limited capability to localize signal and calculate area under the curve for fluorescence intensity in a specific region of interest (e.g., head region) of C. elegans worms. Thus, following nematode population sorting in the BioSorter, fluorescence data profiles of individual worms in each population were analyzed using a custom MATLAB script (Natick, MA, USA; Supplemental File 3). Briefly, compensated profiles were oriented to place the head region directed toward the right. Using the change in derivatives, the head region of each animal was identified as anterior to the large signal peak (which represented non-specific MTG binding in the anterior portion of the intestines) using the green channel (Figures 1, 2, 3). From the green channel peak in the anterior intestinal region toward the head region, the MATLAB script found the point of change of the first derivative that was greater than 50% of the original first derivative after the peak. The sharp change in the first derivative indicated the change from non-specific anterior gut fluorescence to the mitochondria-specific fluorescence signal that occurred in the rest of the head region. The mean values of the red and green signal intensities for each worm were computed with the mean value theorem, using the cumulative trapezoidal numerical integration (i.e., area under the curve of the region of interest divided by the length of the region of interest). The mean and standard deviation of average red and green channel signal intensity per population of each experimental condition were then computed.
Figure 1. Confocal imaging confirmation of fluorescence localization.
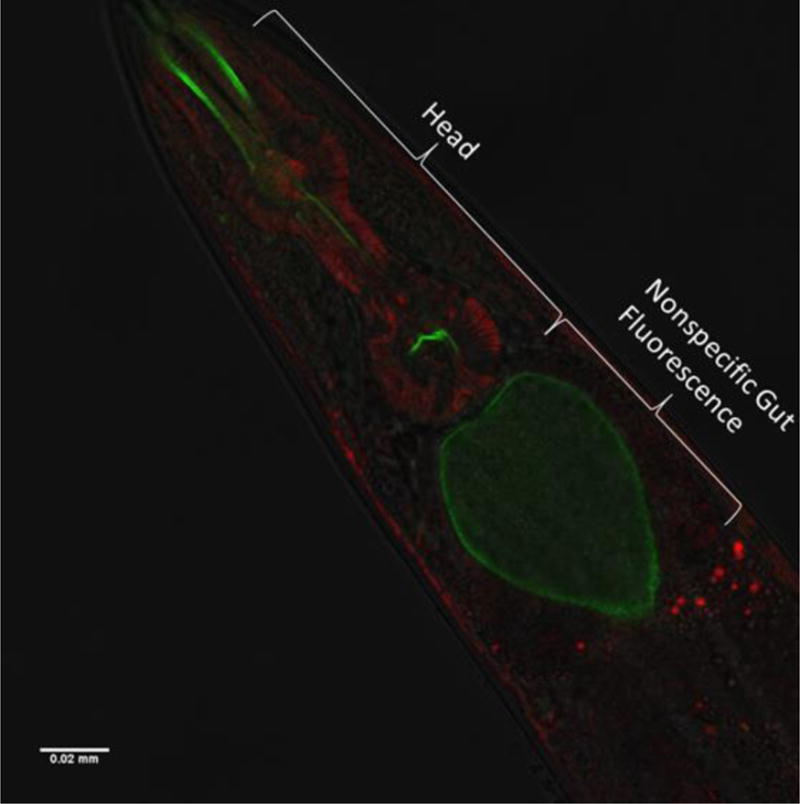
Confocal microscopy of N2 strain C. elegans nematode co-labeled with MitoTracker Green (MTG, green) and Tetramethylrhodamine, ethyl ester (TMRE, red) revealed that MTG accumulated in the anterior gut region in a non-specific manner. However, MTG and TMRE fluorescence accumulated in the head region with a staining pattern more consistent with that of molecular probes binding to mitochondria. Fluorescent stained particles measured at approximately one micrometer in length.
Figure 2. Fluorescence imaging of pKEK strain labeled with TMRE.
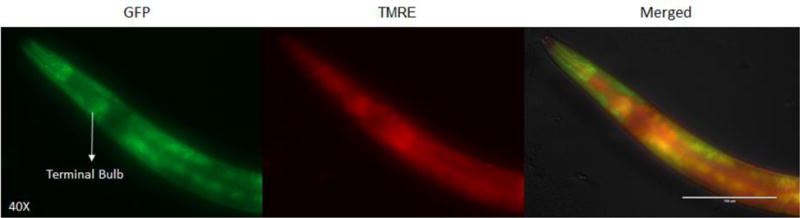
Fluorescence microscopy of pKEK strain labeled with TMRE showed similar staining patterns and co-localization of GFP and TMRE signals. Scale bar indicates 100 μm.
Figure 3. Single worm comparative analysis of fluorescence microscopy and Biosorter profiling of mitochondrial dyes.
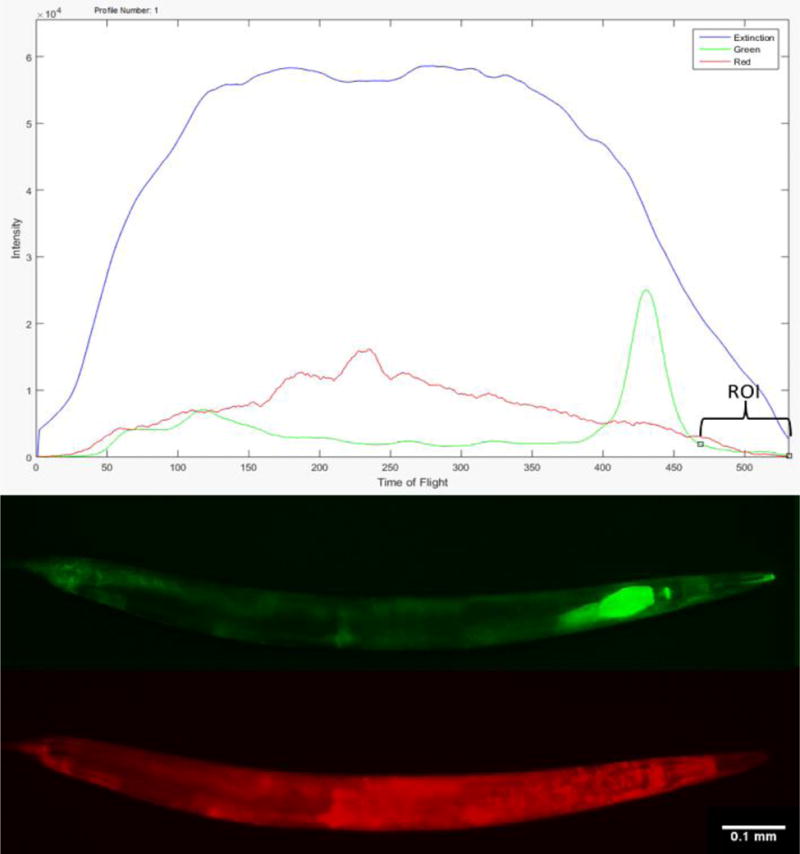
Extinction and fluorescence profiles from MTG and TMRE co-labeled worms on the large particle flow cytometer (top) and in fluorescence microscope images (bottom). In both panels, the worm is oriented with its head toward the right side of the frame. The flow cytometry signal intensity profile accurately captured the high intensity signal from the anterior gut region from MTG (green channel), serving as an effective means by which the analysis software could be programmed to consistently orient each worm and analyze their head region (as noted by the two black open squares on the green signal of the fluorescence profile).
2.6 BioSorter analysis of drug treatment effects on MTG and TMRE fluorescence in synchronized young adult C. elegans worm populations
Nematodes were grown and maintained in 20°C incubators. Synchronized nematode cultures were initiated by standard bleaching protocols to obtain eggs from gravid young adults.17 Collected eggs were allowed to hatch overnight on 10 cm, unspread (i.e., without a bacterial lawn) NGM plates, then L1-arrested larvae were transferred to 10 cm NGM plates spread with OP50 E. coli. Pharmacologic drug treatments were applied from the day 0 of young adulthood stage as follows: nicotinic acid (NA) or glucose (glu) dissolved in water added to a final concentration of 2.5 mM and 10 mM, respectively, on NGM plates. RC toxins including a complex V inhibitor oligomycin (oligo), and an uncoupler, Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), were dissolved in DMSO added to desired variable final concentrations on NGM plates. Upon reaching the first day of egg laying, synchronous young adults were moved to fresh 3.5 cm NGM plates seeded with OP50 E. coli bacteria and treated with the same drug. The same volume of water was added to untreated NGM spread plates with each worm strain as buffer-only control.
2.7 Statistical analysis of BioSorter fluorescence intensity differences between groups
Statistical analyses of the calculated difference in the mean fluorescence intensity between strains or under different experimental conditions were assessed by mixed-effect ANOVA using built-in code in MATLAB. This analysis was used to account for any potential batch effect due to samples being experimentally prepared, processed, and analyzed on different days, by including a batch random effect in the model. A statistical significance threshold was set at p < 0.05.
3. Results
3.1 Confocal and Fluorescence Microscopy of C. elegans Worms
Confocal microscopy of C. elegans worm populations co-labeled with MTG and TMRE demonstrated that MTG accumulated in a non-specific fashion in the anterior gut region (Figure 1). In the head region, however, MTG and TMRE each accumulated with a staining pattern more consistent with their being specifically bound to mitochondria,10 with fluorescent particles speckles measuring about a micrometer in length (Figure 1). However, no co-labeling was seen of individual mitochondria with both MTG and TMRE; that is, no single mitochondrion appeared to simultaneously have both MTG and TMRE staining, similar to results of prior study.10 Fluorescence microscopy of pKEK strain labeled with TMRE demonstrated co-localization of both GFP and TMRE fluorescence signals (Figure 2). Fluorescence microscopy of N2 strain labeled with MTG and pKEK strain showed dissimilar patterns involving the terminal pharyngeal bulb; although the signal was fainter with MTG and had increased labeling of the grinder structure relative to pKEK, there was a significant overlap in location of the signal between N2 + MTG and pKEK, as well as pKEK stained with MitoTracker Deep Red (Supplemental File 2).
3.2 Comparative analysis of Fluorescence Microscopy Imaging and BioSorter Flow Cytometry Analyses of Mitochondrial-Localized Fluorescent Dyes in C. elegans
C. elegans nematodes that were co-labeled with MTG and TMRE demonstrated distinct staining patterns in the head region by fluorescence microscopy (Figure 3). Analysis by the large particle flow cytometer was able to accurately capture the high intensity signal in the anterior gut region from MTG, which could be used to orient the cephalo-caudal position of each worm regardless of its orientation at the time of sorting. The large signal peak (corresponding to non-specific binding of MTG to non-mitochondrial cell fractions) also demarcated an area in the head region located anterior to the peak where green and red channel signals could be readily analyzed, since this was where confocal microscopy showed fluorescent probe staining patterns that were consistent with their exclusively labeling mitochondria.
3.3 Comparison of Relative Fluorescence Quantitative Analyses by Manual Fluorescence Microscopy versus Automated BioSorter Flow Cytometry Techniques
Flow cytometry analyses accurately confirmed the known direction and approximate magnitude of mitochondrial physiology changes in both mitochondrial mass (MTG) and mitochondrial membrane potential (TMRE) that occur in 1 day old gas-1(fc1) young adult worms relative to wild-type (N2) controls (Figure 4).10 Whereas BioSorter flow cytometry results that quantified signal intensity in the entire head region did not precisely match the average percent changes in MTG and TMRE intensity manually measured in the terminal pharyngeal bulb by fluorescence microscopy, it did consistently predict the direction and approximate magnitude of signal change between groups, with similar degrees of statistical significance (Table 1). Although flow cytometry technique yielded higher standard deviation than the fluorescence microscopy technique, standard error of the mean (SEM) for both techniques were within 15% across all conditions and replicates due to the larger population size that could be easily tested with the BioSorter flow cytometry technique. Indeed, the flow cytometry technique enables ready analysis of a substantially larger population size than is possible with manual microscopy and can improve precision and effectively lower SEM values (Figure 5). Furthermore, flow cytometry analysis of 4 day old gas-1(fc1) young adult worms treated with glucose or nicotinic acid for 96 hours showed that nicotinic acid led to an increase in TMRE, a different biological response from what was observed previously at the 24 hour nicotinic acid treatment time when a decrease in TMRE occurred (Figure 5).13 This highlights a distinct advantage of the BioSorter flow cytometry based method to enable time course analyses of mitochondrial physiology in C. elegans, since prolonged 96 hour worm analyses by fluorescence microscopy was not possible due to worm fragility with the experimental methods required.
Figure 4. Population level data comparison of fluorescence microscopy and Biosorter analyses at 24 hours.
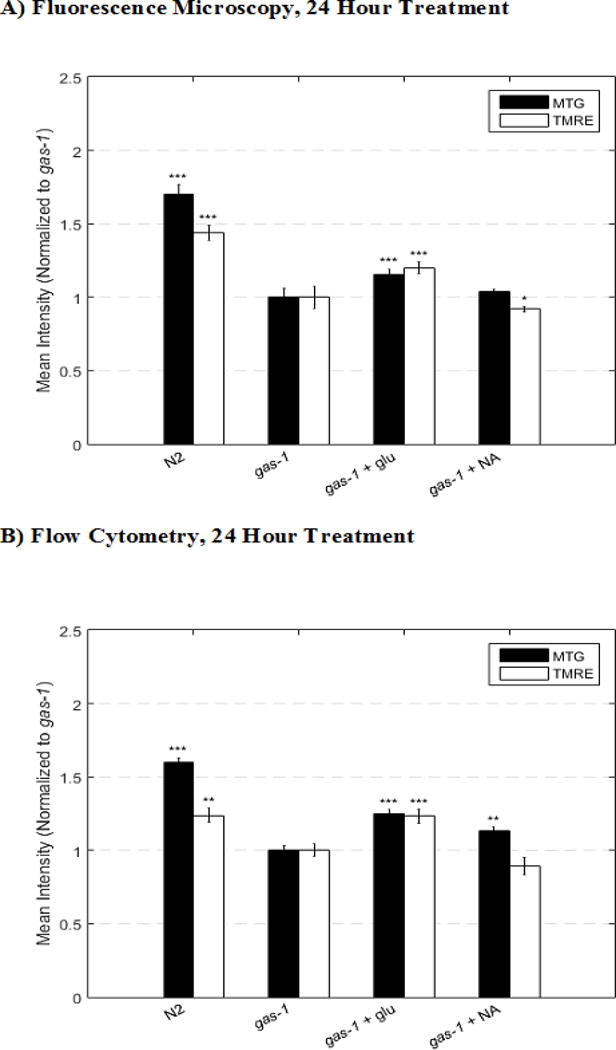
In vivo fluorescence analysis of relative mitochondrial membrane potential and mitochondrial content in gas-1(fc21) mutants treated with glucose and nicotinic acid (NA) for 24 hours and co-labeled with MitoTracker Green FM (MTG) or TMRE for 24 hours. Each drug was compared to the appropriately treated control (i.e., gas-1(fc21) with water). Comparison of (A) manual fluorescence microscopy image analysis and (B) automated flow cytometry analysis validated that flow cytometry confirms direction and magnitude of relative changes in both mitochondrial content (MTG) and mitochondrial membrane potential (TMRE) in 1 day old young adults. P-value conveys the significance of the difference between untreated N2 and untreated gas-1(fc21) (strain effect) or the difference between gas-1(fc21) plus drug and untreated gas-1(fc21) (treatment effect). For each parameter, each drug treatment assay was repeated in 3 independent trials, with 60 worms per trial for fluorescence microscopy and with 1000 to 2000 worms per trial for flow cytometry. Bars and error bars convey mean +/− SEM. *, p < 0.05; **, p < 0.01; ***; p < 0.001 versus concurrent gas-1(fc21) untreated control.
Table 1.
Flow cytometry vs traditional fluorescence microscopy quantitation of anterior worm (entire head) MTG and TMRE mean fluorescence intensity of intergroup comparisons.
| MTG changes (relative to gas-1) |
TMRE changes (relative to gas-1) |
|||
|---|---|---|---|---|
| Condition | Microscopy | Flow Cytometry | Microscopy | Flow Cytometry |
| N2 | p < 0.0001 | p < 0.0001 | p < 0.0001 | p = 0.003 |
| gas-1 + glu | p = 0.0006 | p < 0.0001 | p < 0.0001 | p = 0.0002 |
| gas-1 + NA | p = 0.30 | p = 0.003 | p = 0.02 | p = 0.15 |
We consistently observed consistent direction and similar statistical significance of mean intensity change by total head Biosorter profile quantitation to results of relative quantitation by manual fluorescence microscopy of terminal pharyngeal bulb (posterior head only) fluorescence intensity. was obtained by both methods. Glu, glucose. NA, nicotinic acid.
Figure 5. Time course analysis by BioSorter profiling of mitochondrial dyes.
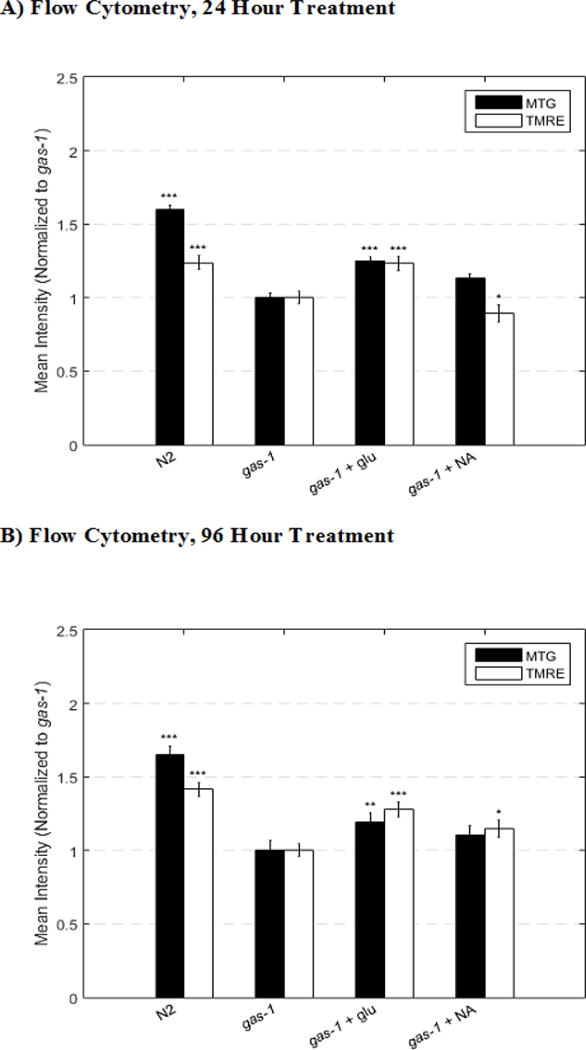
In vivo fluorescence analysis with the BioSorter was performed to quantify relative mitochondrial membrane potential and mitochondrial content in synchronized populations of gas-1(fc21) mutants treated with glucose and nicotinic acid (NA) and co-labeled with MitoTracker Green FM (MTG) or TMRE. Each drug was compared to the appropriately treated buffer control (i.e., gas-1(fc21) with water). Comparison of (A) flow cytometry analysis of 24 hour treatment and (B) flow cytometry analysis of 96 hour treatment confirmed the consistency of direction and magnitude of relative changes in both mitochondrial content (MTG) and mitochondrial membrane potential (TMRE) in both dyes and both drug treatments with the exception of TMRE analysis of NA treatment. Interestingly, whereas 24 hour NA treatment lead to a significant reduction of TMRE signal suggestive of reduced membrane potential (consistent with our prior analysis13), 96 hours of NA treatment led to a significant (although modest) increase in TMRE fluorescence intensity compared to untreated gas-1(fc21) buffer-only control. P-value conveys the significance of the difference between untreated N2 and untreated gas-1(fc21) (strain effect) or the difference between gas-1(fc21) plus drug and untreated gas-1(fc21) (treatment effect). For each parameter, each drug treatment assay was repeated in 3 independent trials, with 1000 to 2000 worms per trial for 24 hour treatment and 200 worms per trial for 96 hour treatment. Of note, 96 hour worm analyses by fluorescence microscopy was not possible due to worm fragility with the experimental methods required. Bars and error bars convey mean +/− SEM. *, p < 0.05; **, p < 0.01; ***; p < 0.001 versus concurrent gas-1(fc21) untreated control.
3.4 MTG and TMRE Validation with Known OXPHOS Membrane Potential Modulators
Flow cytometry analyses accurately confirmed the known direction and approximate magnitude of mitochondrial physiology changes in both mitochondrial mass (MTG) and mitochondrial membrane potential (TMRE) in 1 day old adult wild-type (N2) nematodes exposed for 24 hours to either the RC complex V inhibitor oligomycin, or to the OXPHOS uncoupler FCCP (Figure 6). As expected, oligomycin (1 nM) increased TMRE signals, whereas increasing concentrations of FCCP (10 μM and 20 μM) incrementally decreased TMRE signals.
Figure 6. MTG and TMRE validation using known OXPHOS modulators of mitochondrial membrane potential.
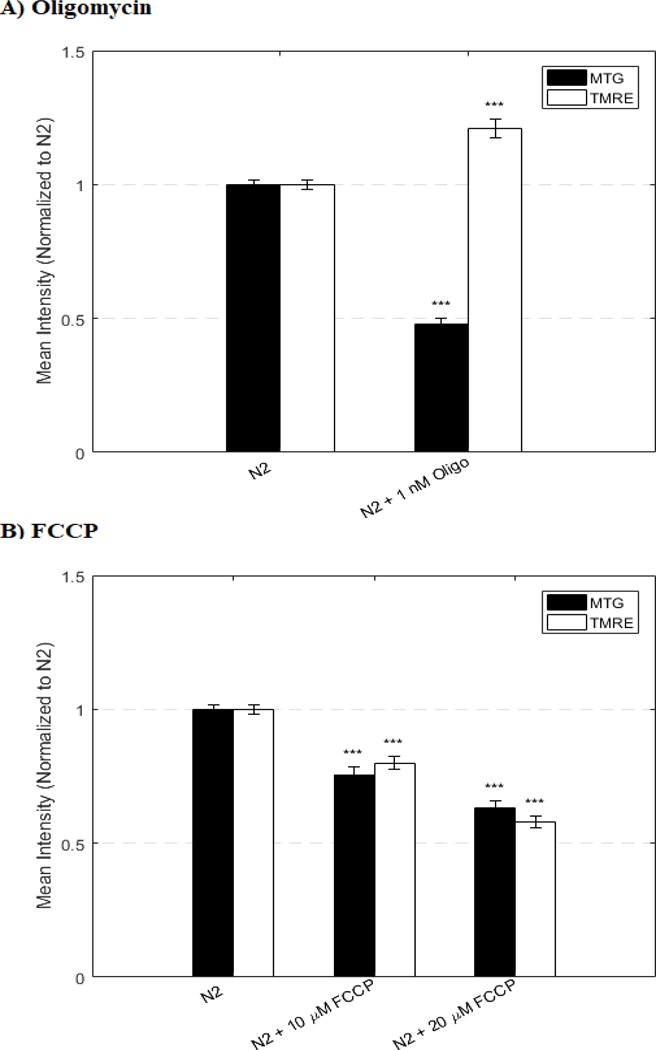
In vivo fluorescence analysis with the BioSorter was performed to quantify relative mitochondrial membrane potential and mitochondrial content in synchronized populations of wild-type N2 strains treated for 24 hours at 20°C with (A) a complex V (ATP synthase) inhibitor, oligomycin, or (B) an OXPHOS uncoupler, FCCP, that were co-labeled with MitoTracker Green FM (MTG) or TMRE. Each inhibitor condition was compared to the appropriately treated buffer control (i.e., N2 treated with DMSO). TMRE signal changed in the predicted direction (increase with oligomycin, decrease with FCCP) in the concentrations tested. For each condition and parameter, approximately 300 worms were studied in multiple biological replicate experiments (n=2 for oligomycin, n=3 for FCCP). Bars and error bars convey mean +/− SEM. *, p < 0.05; **, p < 0.01; ***; p < 0.001 versus concurrent N2 untreated control.
4. Discussion
C. elegans animal models of mitochondrial disease can show variable alterations in mitochondrial physiology over time and in response to pharmacologic therapies. Here, we have developed automated BioSorter-based flow cytometry techniques to efficiently and reliably quantify two key parameters of mitochondrial physiology, mitochondrial mass and mitochondrial membrane potential that is indicative of integrated respiratory chain function, within large nematode populations. Specifically, we have developed and validated this new automated method by replicating results of previous developed manual fluorescence microscopy studies that showed the gas-1(fc21) mutant strain have decreased mitochondrial mass (MTG fluorescence) and decreased mitochondrial membrane potential (TMRE fluorescence) at 24 hours of adult life relative to wild-type N2 Bristol wild type worms. Optimization of the flow cytometry technique to quantify mitochondrial physiology in worms required changing the specific region of interest measured within the animals. Fluorescence microscopy imaging was optimized to focus only on the mitochondria-dense 8-cell region at the posterior portion of the head that is known as the terminal posterior pharyngeal bulb.10 However, profiles acquired on BioSorter are not as precise as those generated by static fluorescence images, most likely due to high speed of nematode samples passing through the laser-based optical assembly. Further, large non-specific binding of the highly lipophilic MTG fluorescent dye within the lipid granules of the anterior gut prevented reliable localization of the terminal pharyngeal bulb on fluorescence raw data profiles generated by flow cytometry (Figure 3). Nonetheless, confocal microscopy demonstrated that the staining patterns of these fluorescent dyes in the worm’s head region were consistent with mitochondria-specific binding (Figures 1, 2, 3). Further, exploiting the non-specific peak of MTG fluorescence proved useful to reliably orient the fluorescence profile of the worm in a consistent cephalad-caudal direction. The MATLAB based software algorithm we developed (Supplemental File 3) was shown to reliably localize the entire head region anterior to the nonspecific area of MTG binding and compute average fluorescence intensity signal in the region of interest. Due to the low noise of each fluorescence profile, the change in the derivative of the green channel fluorescence was used to localize the head region without any additional filtering. However, the broad emission spectrum of MTG had led to fluorescence spillover in the red channel. Since MTG fluorescence spillover contributed linearly to the signal of the red channel, proportional subtraction of MTG-based signal intensity measured in the green channel from the red channel effectively eliminated the effects of fluorescence spillover. Thus, both MTG and TMRE could be used to co-label animals and simultaneously quantify two key parameters of mitochondrial physiology in the same treatment conditions.
BioSorter-based flow cytometry techniques were validated first with confocal microscopy of C. elegans worm populations co-labeled with MTG and TMRE, which demonstrated that MTG and TMRE each accumulated with a staining pattern consistent with binding within mitochondria,10 where fluorescent particles speckles each measured approximately one micrometer in length (Figure 1). While no co-labeling was evident by confocal imaging of individual mitochondria with both MTG and TMRE potentially due to differences in signal intensity or potentially competition within a given mitochondrion, the pKEK strain showed clear co-localization of GAS-1::EGFP signal and TMRE under fluorescence microscopy (Figure 2). Thus, signal from TMRE appears to be indicative of relative mitochondrial membrane potential within the head region. Further, the pKEK strain showed signal overlap by fluorescence microscopy, despite differences in signal intensity, when labeled with MitoTracker Deep Red FM (which was used for validation purposes only given overlap of MTG with pKEK green signal (Supplemental File 3, 4, 5, 6). For high throughput quantification analyses, detecting the direction of change in mitochondrial density alone may help identify candidate therapies to identify for downstream investigations (e.g. previously established fluorescence microscopy, enzymatic assays, etc.). However, any change in mitochondrial content as quantified by the MTG intensity identified in screening studies should be verified with an independent method (e.g. mtDNA content or citrate synthase activity assays) given the nonspecific staining of MTG and the lack of overlap between MTG and TMRE (and between MTR and GAS-1::EGFP). Nonetheless, we further demonstrated here that this new flow cytometry technique accurately detected the predicted change in mitochondrial membrane potential upon exposure to OXPHOS modulators that were seen to increase (oligomycin)or decrease (FCCP) TMRE fluorescence (Figure 6).
Validation of the BioSorter-based flow cytometry technique identified it has several distinct advantages over manual fluorescence microscopy to evaluate the relative mitochondrial physiologic changes in mass and membrane potential of living worms. First, use of the flow cytometry method enabled for chronic treatment effects to be studied in living animal populations over time. Previously, 4 day old young adults (following 96 hour treatment from early young adulthood) were too fragile for manual transfer by platinum pick to permit effective quantitation in animal populations by fluorescence microscopy. Now, using flow cytometry analysis with the BioSorter allowed us to observe that glucose treatment similarly increased both mitochondrial mass and mitochondrial membrane potential in gas-1(fc21) mitochondrial mutant strains toward that seen in wild-type N2 worms at both 24 and 96 hour treatment times (Figure 5). By contrast, BioSorter-based flow cytometry analyses validated that an initial decrease in mitochondrial membrane potential does indeed occur in gas-1(fc21) mitochondrial mutant strains when treated with nicotinic acid for 24 hours, similarly as we prior observed with fluorescence microscopy,13 but for the first time enabled analysis of longer duration treatment effects to reveal that chronic (96 hour) nicotinic acid treatment actually increases mitochondrial membrane potential. Second, flow cytometry provides a technically simpler and likely less technician-dependent method to prepare worm populations for rapid data collection from multiple animal populations at different time points. With the custom MATLAB algorithm (Supplemental File 3), data compilation, visualization, and statistical analysis is automatically performed in minutes, which significantly reduces overall experiment time given the prolonged time (hours) to manually analyze individual worm terminal pharyngeal bulb fluorescence intensity with traditional microscopy. Third, flow cytometry has significantly increased the number of individual worms in which data can be acquired for a given experiment (from ~60 worms to over 1,000 worms per replicate). The ability to readily analyze 10-fold more animals per condition increases the confidence and precision of results obtained.
As suggested above, our custom data analysis software written in MATLAB enables automated data analysis of fluorescence-dye based mitochondrial physiology analyses in C. elegans. Custom data analysis software was necessary, because the existing Union Biometrica software does not have the capability to measure the area under the curve of a pre-defined region of interest with background subtraction. This software code can be readily compiled into a standalone executable (Supplemental File 3) that can be run on any computer with the MATLAB runtime, as well as a stand-alone set of shared libraries that enables the execution of compiled MATLAB applications or components on computers that do not have MATLAB installed. Thus, performance of large particle flow cytometry analyses using the BioSorter requires less training and is likely to provide more robust, consistent results when quantifying mitochondrial physiology in living nematodes, as compared to the more tedious and user-dependent manual fluorescence microscopy.
5. Conclusions
We have developed a novel experimental protocol and analytic method to effectively utilize the BioSorter for large particle flow cytometry to interrogate mitochondrial physiology in living nematode populations. This approach reliably quantified the magnitude and degree of relative changes in mitochondrial density and mitochondrial membrane potential that occur in the gas-1(fc21) C. elegans mutant worm model of mitochondrial disease. We successfully validated this methodology by demonstrating it provides a similar degree of sensitivity and statistical significance in quantifying MTG and TMRE fluorescence in the mutant worms,10 and in response to known OXPHOS modulators as well as pharmacologic treatments13 that were previously achieved with more tedious, manual fluorescence microscopy techniques. This high-throughput BioSorter flow cytometry-based analysis technique, coupled with our custom MATLAB software to rapidly quantify, statistically analyze, and graphically display results of raw signal data, has significantly shortened the time required to obtain and interpret quantitative signal intensity data that are indicative of unique, well-defined aspects of mitochondrial physiology in living nematodes. This technique can be readily adapted for use with Danio rerio (zebrafish). Thus, flow cytometry has brought the potential to rapidly screen additional nematodes per condition, multiple conditions per experiment, and multiple time points that will inform disease mechanisms and treatment effects in diverse models of mitochondrial disease.
Supplementary Material
Highlights.
BioSorter analysis facilitates rapid evaluation of mitochondrial physiology in living nematodes
A custom MATLAB script automates quantitation and statistical analysis in large animal populations
Mitochondrial mass & membrane potential are quantified in response to mutations and therapies
This approach can support accelerated therapy optimization for mitochondrial disease
Acknowledgments
We would like to thank Rui Xiao, PhD, for her guidance in selecting the appropriate statistical analysis for comparison of wild-type to mutant strain, and control group to drug treatment groups. We would like to thank Andrea Stout, PhD, for her guidance with confocal microscopy. CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), provided the N2 Bristol and gas-1(fc21) strains. Bernhard Kayser, Ph.D., Philip G. Morgan, M.D. and Margaret M. Sedensky, M.D. provided the pKEK EGFP mutant strain.
Funding
This work was funded in part by the National Institutes of Health (R01-HD065858 and R01-GM120762-06A1 to MJF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Consent for publication
All authors approved the final manuscript as submitted.
Availability of data and materials
The datasets and microscopy photographs analyzed in the current study are attached in the supplemental files. The software used to analyze the data is attached in the supplemental files.
Declaration of interest
The authors have no financial disclosure or competing interests for this article.
Authors’ contributions
MJF and YJK conceived the experimental design. YJK performed the studies, wrote the MATLAB script, and analyzed the data. YJK, MJF, FT contributed to experimental methodology and wrote the manuscript.
References
- 1.Mitochondrial Medicine Society’s Committee on Diagnosis. Haas RH, Parikh S, et al. The in-depth evaluation of suspected mitochondrial disease. Mol Genet Metab. 2008;94(1):16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haas RH, Parikh S, Falk MJ, et al. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120(6):1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- 3.McCormick E, Place E, Falk MJ. Molecular genetic testing for mitochondrial disease: from one generation to the next. Neurotherapeutics. 2013;10(2):251–261. doi: 10.1007/s13311-012-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorman GS, Schaefer AM, Ng Y, et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol. 2015;77(5):753–759. doi: 10.1002/ana.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopman WJH, Willems PHGM, Smeitink JAM. Monogenic mitochondrial disorders. N Engl J Med. 2012;366(12):1132–1141. doi: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- 6.Schrier Vergano S, Rao M, McCormack S, et al. In vivo metabolic flux profiling with stable isotopes discriminates sites and quantifies effects of mitochondrial dysfunction in C. elegans. Mol Genet Metab. 2014;111(3):331–341. doi: 10.1016/j.ymgme.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corsi AK, Wightman B, Chalfie M. A Transparent Window into Biology: A Primer on Caenorhabditis elegans. Genetics. 2015;200(2):387–407. doi: 10.1534/genetics.115.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falk MJ, Kayser E-B, Morgan PG, Sedensky MM. Mitochondrial complex I function modulates volatile anesthetic sensitivity in C. elegans. Curr Biol. 2006;16(16):1641–1645. doi: 10.1016/j.cub.2006.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKay JP, Raizen DM, Gottschalk A, Schafer WR, Avery L. eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics. 2004;166(1):161–169. doi: 10.1534/genetics.166.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingley S, Polyak E, Lightfoot R, et al. Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion. 2010;10(2):125–136. doi: 10.1016/j.mito.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Presley AD, Fuller KM, Arriaga EA. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr B. 2003;793(1):141–150. doi: 10.1016/S1570-0232(03)00371-4. [DOI] [PubMed] [Google Scholar]
- 12.Mitra K, Lippincott-Schwartz J. Analysis of mitochondrial dynamics and functions using imaging approaches. Curr Protoc Cell Biol. 2010 doi: 10.1002/0471143030.cb0425s46. Chapter 4: Unit 4.25.1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormack S, Polyak E, Ostrovsky J, et al. Pharmacologic targeting of sirtuin and PPAR signaling improves longevity and mitochondrial physiology in respiratory chain complex I mutant Caenorhabditis elegans. Mitochondrion. 2015;22:45–59. doi: 10.1016/j.mito.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayser EB, Morgan PG, Hoppel CL, Sedensky MM. Mitochondrial expression and function of GAS-1 in Caenorhabditis elegans. J Biol Chem. 2001;276(23):20551–20558. doi: 10.1074/jbc.M011066200. [DOI] [PubMed] [Google Scholar]
- 15.Peng M, Ostrovsky J, Kwon YJ, et al. Inhibiting cytosolic translation and autophagy improves health in mitochondrial disease. Hum Mol Genet. 2015;24(17):4829–4847. doi: 10.1093/hmg/ddv207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stiernagle T. Maintenance of C. elegans. WormBook : The Online Review of C Elegans Biology. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


