Abstract
Caveolin-1 is a principal component of caveolae membranes in vivo. Caveolin-1 mRNA and protein expression are lost or reduced during cell transformation by activated oncogenes. Interestingly, the human caveolin-1 gene is localized to a suspected tumor suppressor locus (7q31.1). However, it remains unknown whether caveolin-1 plays any role in regulating cell cycle progression. Here, we directly demonstrate that caveolin-1 expression arrests cells in the G0/G1 phase of the cell cycle. We show that serum starvation induces up-regulation of endogenous caveolin-1 and arrests cells in the G0/G1 phase of the cell cycle. Moreover, targeted down-regulation of caveolin-1 induces cells to exit the G0/G1 phase. Next, we constructed a green fluorescent protein-tagged caveolin-1 (Cav-1-GFP) to examine the effect of caveolin-1 expression on cell cycle regulation. We directly demonstrate that recombinant expression of Cav-1-GFP induces arrest in the G0/G1 phase of the cell cycle. To examine whether caveolin-1 expression is important for modulating cell cycle progression in vivo, we expressed wild-type caveolin-1 as a transgene in mice. Analysis of primary cultures of mouse embryonic fibroblasts from caveolin-1 transgenic mice reveals that caveolin-1 induces 1) cells to exit the S phase of the cell cycle with a concomitant increase in the G0/G1 population, 2) a reduction in cellular proliferation, and 3) a reduction in the DNA replication rate. Finally, we demonstrate that caveolin-1-mediated cell cycle arrest occurs through a p53/p21-dependent pathway. Taken together, our results provide the first evidence that caveolin-1 expression plays a critical role in the modulation of cell cycle progression in vivo.
INTRODUCTION
Caveolae are 50–100-nm vesicular invaginations of the plasma membrane (Razani et al., 2000b). It has been proposed that caveolae participate in vesicular trafficking events and signal transduction processes (Lisanti et al., 1994; Couet et al., 1997b; Okamoto et al., 1998; Smart et al., 1999). Caveolin-1, a 21–24-kDa integral membrane protein, is a principal component of caveolae membranes in vivo (Glenney, 1989, 1992; Glenney and Soppet, 1992; Kurzchalia et al., 1992; Rothberg et al., 1992). The mammalian caveolin gene family consists of caveolin-1, -2, and -3 (Parton, 1996; Scherer et al., 1996; Tang et al., 1996; Okamoto et al., 1998). Caveolin-1 and -2 are coexpressed and form a hetero-oligomeric complex (Scherer et al., 1997) in many cell types, with particularly high levels in adipocytes, whereas expression of caveolin-3 is muscle-specific and found in both cardiac and skeletal muscle, as well as smooth muscle cells (Song et al., 1996b).
It has been proposed that caveolin family members function as scaffolding proteins (Sargiacomo et al., 1995) to organize and concentrate specific lipids (cholesterol and glycosphingolipids [Fra et al., 1995; Murata et al., 1995; Li et al., 1996b]) and lipid-modified signaling molecules (Src-like kinases, H-Ras, endothelial nitric-oxide synthase, and G proteins [Li et al., 1995; Garcia-Cardena et al., 1996; Li et al., 1996a,b; Shaul et al., 1996; Song et al., 1996a]) within caveolae membranes. The direct interaction of caveolin with signaling molecules leads to their inactivation (Lisanti et al., 1994; Couet et al., 1997a,b,c; Okamoto et al., 1998). In support of this idea, caveolin-1 was found to suppress the kinase activity of Src family tyrosine kinases (c-Src/Fyn), epidermal growth factor receptor, Neu, and protein kinase C through the caveolin-scaffolding domain, a modular protein domain that recognizes a specific caveolin binding motif in various signaling molecules (Couet et al., 1997a,b,c; Okamoto et al., 1998). In addition, the scaffolding domain of caveolin-1 was shown to inhibit endothelial nitric-oxide synthase activity and the GTPase activity of heterotrimeric G proteins (Li et al., 1995; Li et al., 1996a,b; Song et al., 1996a; Garcia-Cardena et al., 1997; Razani et al., 1999).
Because many of these signaling molecules can cause cellular transformation when constitutively activated, it is reasonable to speculate that caveolin itself may possess transformation suppressor activity. In fact, caveolin-1 mRNA and protein expression are lost or reduced during cell transformation by activated oncogenes such as v-Abl and H-Ras (G12V); caveolae are absent from these cell lines (Koleske et al., 1995). In addition, induction of caveolin-1 expression in v-Abl- and H-ras (G12V)-transformed NIH 3T3 cells abrogated the anchorage-independent growth of these cells in soft agar and resulted in the de novo formation of caveolae (Engelman et al., 1997).
Several other lines of evidence are consistent with the idea that caveolin-1 functions as transformation suppressor protein. Recently, we have shown that antisense-mediated reductions in caveolin-1 protein expression in NIH 3T3 cells are sufficient to drive oncogenic transformation and constitutively activate the p42/44 mitogen-activated protein kinase cascade (Galbiati et al., 1998). Mutations in the adenomatous polyposis coli gene or the β-catenin gene lead to the accumulation of β-catenin in human colon carcinoma (Powell et al., 1992; Polakis, 1997) and melanoma (Korinek et al., 1997; Morin et al., 1997; Peifer, 1997; Rubinfeld et al., 1997) and other types of cancer (Ben-Ze'ev and Geiger, 1998). Elevation of β-catenin expression in such tumors is believed to induce uncontrolled activation of gene transcription by the β-catenin–Lef-1 complex, thereby contributing to tumor progression (Gumbiner, 1997; Peifer, 1997; Ben-Ze'ev and Geiger, 1998). We have recently demonstrated that activation of β-catenin–Lef-1 signaling by Wnt-1 or by overexpression of β-catenin itself is inhibited by caveolin-1 expression (Galbiati et al., 2000). Finally, the human caveolin-1 gene is localized to a suspected tumor suppressor locus (D7S522; 7q31.1), a known fragile site (FRA7G) that is deleted in many types of cancer (Engelman et al., 1998c,d,e, 1999). Thus, down-regulation of caveolin-1 expression and caveolae organelles may be critical for maintaining the transformed phenotype.
We have previously shown that in nontransformed NIH 3T3 cells, caveolin-1 levels are down-regulated in rapidly dividing cells and dramatically elevated in confluent cells (Galbiati et al., 1998; Volonte' et al., 1999), where caveolin-1 is concentrated at the areas of cell-cell contact. These observations may be related to the ability of caveolin-1 to regulate contact inhibition and growth arrest in nontransformed cells (Galbiati et al., 1998; Volonte' et al., 1999). However, this hypothesis has not been formally tested.
The p53 tumor suppressor protein plays a critical role in regulating cell growth arrest. Mutations in p53 or inactivation of p53 through interactions with viral or cellular proteins are the most frequent alterations observed in cancer cells (Levine, 1997). p53 prevents the accumulation of genetic alterations through the induction of growth arrest or senescence to block the replication of damaged DNA. p53 is also involved in mediating programmed cell death (apoptosis), which is important for eliminating defective cells (Amundson et al., 1998; Sionov and Haupt, 1998; Bates and Vousden, 1999). Whether the cell enters growth arrest or undergoes apoptosis depends on a combination of different incoming signals.
p53 is directly involved in the arrest in the G1 phase of the cell cycle through the induction of p21WAF1/Cip1 (reviewed in el-Deiry, 1998)). p21 mediates p53-dependent G1 arrest by inhibiting the activity of cyclin-dependent kinases (CDKs), which phosphorylate the retinoblastoma (pRb) gene product, as well as other substrates. p21 also induces growth arrest by preventing PCNA from activating DNA polymerase δ, which is essential for DNA replication (Waga et al., 1994). p21-deficient fibroblasts show impairment of G1 arrest (Deng et al., 1995), confirming a key role for p21 in cell cycle arrest.
In addition to G1 arrest, p53 is also capable of inducing G2 arrest. p53-induced G2 arrest is mediated by the product of the 14-3-3ς gene, which sequesters the phosphorylated form of cdc25, a phosphatase of the cyclin B/cdc2 complex that is essential for the G2/M transition (Peng et al., 1997). Also, p53 inhibits the cyclinB/cdc2 complex through the induction of GADD45, which disrupts the complex, probably through direct interaction with cdc2 (Wang et al., 1999).
Here, we have addressed the possible role of caveolin-1 in mediating cell cycle arrest. We show that either transient overexpression of caveolin-1 in NIH 3T3 cells, or stable transgenic expression of caveolin-1 in mouse embryonic fibroblasts, induces arrest in the G0/G1 phase of the cell cycle. Also, we demonstrate that caveolin-1-mediated cell cycle arrest occurs through the classical p53/p21-dependent pathway.
EXPERIMENTAL PROCEDURES
Materials
Antibodies and their sources were as follows: anti-caveolin-1 IgG (mAb 2297; gift of Dr. Roberto Campos-Gonzalez, BD Transduction Laboratories); anti-caveolin-2 IgG (mAb 65; gift of Dr. Roberto Campos-Gonzalez, BD Transduction Laboratories); anti-caveolin-3 IgG (mAb 26; gift of Dr. Roberto Campos-Gonzalez, BD Transduction Laboratories); anti-p21WAF1/Cip1 IgG (polyclonal antobody C-19; Santa Cruz Biotechnologies, Santa Cruz, CA). A variety of other reagents were purchased commercially: DMEM (Cellgro); donor bovine calf serum (JRH Biosciences). The pTA-p53 luciferase reporter was from CLONTECH (Palo Alto, CA). p53-WT and p53-MUT expression vectors were from CLONTECH. All other biochemicals used were of the highest purity available and were obtained from regular commercial sources. Lipoprotein-deficient fetal bovine serum (LPDS) was obtained from Intracel, Rockville, MD.
Cell Culture
NIH 3T3 cells were grown in DMEM supplemented with glutamine, antibiotics (penicillin and streptomycin) and 10% donor bovine calf serum, as previously described (Koleske et al., 1995; Engelman et al., 1997). C2C12 cells were cultured in high mitogen medium (DMEM containing 15% fetal bovine serum and 1% chicken embryo extract) and induced to differentiate at confluence in low mitogen medium (DMEM containing 3% horse serum).
Immunoblotting
Cells were collected in boiling sample buffer and homogenized with the use of a 26-gauge needle. Cellular proteins were resolved by SDS-PAGE (12.5% acrylamide) and transferred to BA83 nitrocellulose membranes (0.2 μm; Schleicher & Schuell, Riehen, Switzerland). Blots were incubated for 2 h in Tris-buffered saline/Tween 20 (TBST) (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.2% Tween 20) containing 2% powdered skim milk and 1% bovine serum albumin. After three washes with TBST, membranes were incubated for 2 h with the primary antibody (∼1000-fold diluted in TBST) and for 1 h with horseradish peroxidase-conjugated goat anti-rabbit/mouse IgG (∼5000-fold diluted). Bound antibodies were detected with the use of an enhanced chemiluminescence detection kit (Amersham Pharmacia Biotech).
Cell Cycle Analysis by Fluorescence-activated Cell Sorter (FACS)
Cells were collected by trypsinization, pelleted, and resuspended in complete growth medium at a final concentration of 1 × 106 cells/ml. Cells were incubated for 30 min at 37°C with 3.0 μg/ml Hoechst 33342. Cells were then subjected to FACS analysis with the use of a fluorescence-activated cell sorter (FACStar plus; Becton Dickinson). For each analysis 20,000–50,000 gated events were collected to permit cell cycle analysis of both green fluorescent protein (GFP)-positive and GFP-negative cell subpopulations. Data analysis was performed with the use of CellQuest software. For each condition, at least three independent experiments were performed that yielded virtually identical results. A representative cell cycle analysis experiment is shown.
Induction of G0 and G1 Synchronization
NIH 3T3 cells were grown for 24 h in the absence of serum (0%) to induce G0 arrest, or treated for 16 h with staurosporine (10 nM) in complete medium to induce G1 arrest (Nishi et al., 1998). Cells were then subjected to Western blot analysis and cell cycle analysis.
p53 Luciferase Reporter Assays
Cells were seeded in six-well plates at 300,000 cells/well. The following day, cells were transiently transfected, with the use of a modified calcium-phosphate precipitation method, with 1 μg of the luciferase reporter (pTA-p53), 0.5 μg of pSV-β-galactosidase, and 1 μg of the indicated cDNA. Twelve hours posttransfection, cells were rinsed twice with PBS and incubated in normal media containing 10% serum or 0.5% serum for another 24–36 h. Normal media containing 10% serum was routinely used, unless specifically indicated otherwise. Cells were then lysed in 500 μl of extraction buffer; 200 μl was used to measure luciferase activity and 100 μl of which was used to measure β-galactosidase activity. The p53 luciferase reporter activity was controlled for transfection efficiency and potential toxicity with the use of β-galactosidase activity. For each condition, at least three independent experiments were performed.
Generation of Caveolin-1 Transgenic Mice
The full-length untagged c-DNA encoding caveolin-1 was subcloned into the EcoRI site of the multiple cloning site (MCS) region of the transgenic expression vector pCAGGS (gift of Armin Rehm, Ploegh Laboratory, Harvard Medical School, Boston, MA [Niwa et al., 1991]). In the pCAGGS vector, the cytomegalovirus (CMV) enhancer and the chicken β-actin promoter sequence are located upstream of the MCS region. In addition, a rabbit β-globin poly(A) sequence is located downstream from the MCS region. The resulting plasmid, pCAGGS-Cav-1, was digested with SalI and HindIII to isolate the transgenic cassette consisting of the CMV enhancer, the chicken β-actin promoter, the caveolin-1 cDNA, and the rabbit β-globin poly(A) sequence. The isolated region was purified for pronuclear injection into mouse embryos from FVB mice (Taconic Farms, Germantown, NY). Mouse embryos (fertilized one-cell zygotes) were injected and then implanted in female CD-1 mice (Charles River Breeding Laboratories) at the Transgenic Facility here at The Albert Einstein College of Medicine. Caveolin-1 transgenic mice were identified by slot blot analysis with the use of genomic DNA prepared from mouse tail biopsies. A fragment containing the rabbit β-globin poly(A) sequence, obtained by digesting the pCAGGS vector with EcoRI and HindIII, was radiolabeled and used as the probe for slot blot analysis. Caveolin-1-positive founder transgenic mice then were back-crossed at least three times with C57BL/6 mice (Jackson Immunoresearch, West Grove, PA).
Interestingly, caveolin-1 transgenic mice did not show any overt “clinical” phenotype. To identify a phenotype associated with caveolin-1 overexpression in caveolin-1 transgenic mice, tissue sections from these mice were hematoxylin/eosin-stained and examined by light microscopy. Interestingly, no pathological changes were observed (our unpublished results). Importantly, several pathologists carefully assessed the tissues where caveolin-1 is transgenically overexpressed (fat, kidney, liver, brain, lung, spleen, skeletal muscle, and heart, among others) and we did not observe any noticeable pathological changes.
Primary Culture of Mouse Embryonic Fibroblasts (MEFs)
Pregnant mice were sacrificed and E13.5 embryos were isolated; the head and liver were separated and discarded. The remaining embryonic tissue was minced with a razor blade and trypsinized in a 10-cm dish at 37°C for 5 min. Next, 20 ml of DMEM supplemented with 10% fetal bovine serum, glutamine, and antibiotics (penicillin and streptomycin) was added to the dish and the cells were incubated at 37°C. After 24 h, the medium was replaced and cells were cultured for an additional 48 h. When confluent, mouse embryonic fibroblasts were split 1:4. After 48 h, the MEFs (representing passage 1) were frozen. Experiments were performed with the use of MEFs at passage 1 or 2.
Analysis of Cellular Proliferation
Growth Curves.
Primary cultures of MEFs were seeded in 10-cm dishes at a density of 40,000 cells/dish at day 0. Cell number was then counted with the use of a cytometer after 1, 2, 3, 4, and 5 d of incubation at 37°C. Each point represents the average of four independent determinations.
BrdU Incorporation.
Primary cultures of MEFs were seeded in 96-well plates at a density of 10,000 cells/well at day 0. Each MEF clone was seeded in quadruplicate. After 2 d, BrdU incorporation was assessed with the use of the Cell Proliferation ELISA/BrdU (colorimetric) kit from Roche, according to the manufacture's recommendations.
Caspase-3-like Activity Assay
Caspase-3-like activity was assessed with the use of a slightly modified ApoAlert CPP32/caspase-3 fluorescent assay. Briefly, after treatment with staurosporine (0.3 μM) for 6 h, incubation medium was removed and the cells were lysed for 10 min in ice-cold lysis buffer containing 20 mM Tris-HCl (pH 7.4), 137 mM NaCl, 1% NP-40, 10% glycerol, 1 μg/ml aprotinin, 1 μM leupeptin, and 1 mM phenylmethylsulfonyl fluoride. Total cell lysates (10 μg each) were incubated at 37°C for 60 min in enzyme assay buffer containing 20 mM HEPES (pH 7.5), 10% glycerol, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 100 μM caspase 3 substrate DEVD-AMC. The reaction was terminated by adding 1 ml of distilled H2O. Fluorescence was measured with the use of an LS50B luminescence spectrometer (PerkinElmer Cetus, Norwalk, CT) equipped with a 380-nm excitation filter and a 460-nm emission filter.
RESULTS
Endogenous Caveolin-1 Protein Expression Is Up-Regulated during the G0/G1 Phase of the Cell Cycle
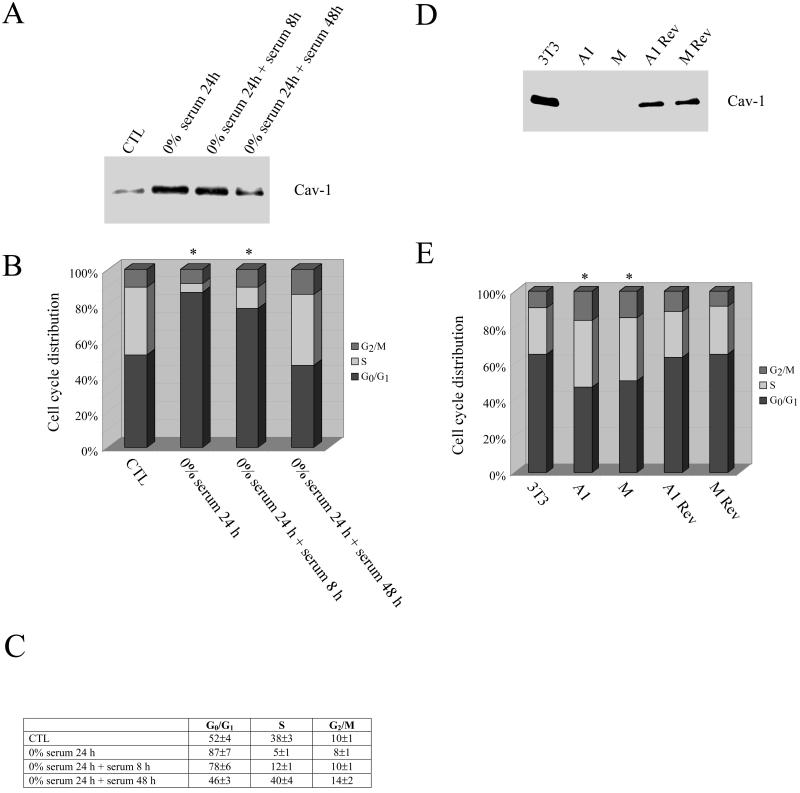
Figure 1A shows that addition of serum to serum-deprived NIH 3T3 cells induces a time-dependent down-regulation of caveolin-1 expression. FACS analysis of the cell cycle demonstrated that NIH 3T3 cells were arrested in the G0/G1 phase when endogenous caveolin-1 expression was up-regulated by serum starvation (from 52 to 87%; Figure 1B). Conversely, addition of serum induced cells to exit the G0/G1 phase of the cell cycle. Interestingly, there is a direct correlation between the time-dependent reduction of caveolin-1 expression and exit from G0/G1 phase (Figure 1, A and B). Figure 1C shows a table representing the averages and SDs of three independent experiments of cell cycle analysis.
Figure 1.
Endogenous caveolin-1 protein expression is up-regulated during the G0/G1 phase of the cell cycle. (A–C) Nontransfected NIH 3T3 cells were serum-starved for 24 h before the addition of serum for 8, 24, and 48 h. Each point was performed in triplicate. (A) Western blotting. Cell lysates were subjected to SDS-PAGE and Western blotting analysis with the use of a caveolin-1 specific mAb probe. A representative immunoblot is shown. Note that serum starvation up-regulates caveolin-1 protein expression, whereas the addition of serum to serum-starved NIH 3T3 cells causes a time-dependent reduction in caveolin-1 expression. Each lane contains equal amount of total proteins. (B) Cell cycle analysis. Cells were subjected to FACS analysis (in EXPERIMENTAL PROCEDURES). The cell cycle analysis shown represents the average from three independent experiments. Note that up-regulation of caveolin-1 protein expression is associated with a G0/G1 arrest. (C) Quantitation. Table representing the averages and SDs from the three independent experiments of the cell cycle analysis shown in B. (D and E) Parental NIH 3T3 cells, NIH 3T3 clones harboring caveolin-1 antisense (A1 and M), and revertant NIH 3T3 clones (A1 Rev, M Rev) were analyzed. (D) Western blot analysis. Cell lysates were subjected to SDS-PAGE and Western blotting analysis with the use of a caveolin-1-specific mAb probe (a representative immunoblot is shown; each lane contains equal amount of total protein). (E) Cell cycle analysis. Each point was performed in triplicate. A representative cell cycle analysis is shown. Note that the G0/G1 population is reduced when caveolin-1 expression is down-regulated. In B and E, the asterisk (∗) indicates significant differences in the cell cycle distribution.
We have previously used an antisense approach to derive stable NIH 3T3 cell lines that expressed dramatically reduced levels of caveolin-1 (Galbiati et al., 1998). NIH 3T3 cells harboring antisense caveolin-1 exhibit a transformed phenotype (Galbiati et al., 1998). However, loss of the caveolin-1 antisense vector results in a complete reversion of the transformed phenotype (Galbiati et al., 1998). Here, we demonstrate that two independent clones (A1 and M), which express reduced levels of caveolin-1 (Figure 1D), are characterized by a significantly reduced G0/G1 population (from 65% [WT 3T3] to 47% [A1] and 50% [M)]; Figure 1E). In addition, the two antisense clones showed a significant increase in the S phase component of the cell cycle (from 25% [WT 3T3] to 37% [A1] and 35% [M]). Importantly, reductions in G0/G1 phase and increases in S phase, induced by down-regulation of caveolin-1, are reversed when caveolin-1 protein levels are restored to normal by loss of the caveolin-1 antisense vector (Figure 1, D and E). Thus, loss of endogenous caveolin-1 expression significantly decreases the number of cells in the G0/G1 phase of the cell cycle.
Recombinant Expression of Caveolin-1 Induces G0/G1 Arrest in NIH 3T3 Cells
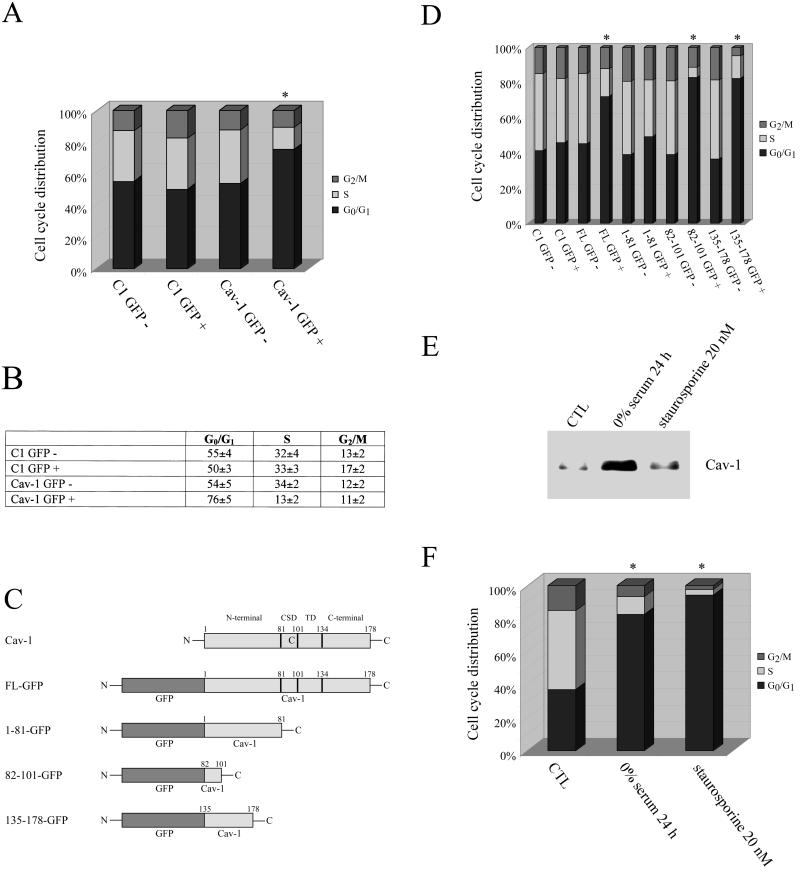
To better evaluate the effects of caveolin-1 expression on cell cycle progression, we next generated a green fluorescent protein-tagged caveolin-1 (Cav-1-GFP). In this construct, wild-type caveolin-1 is placed at the C terminus of GFP. We have previously demonstrated that the fusion protein Cav-1-GFP behaves identically to endogenous caveolin-1 (Volonte' et al., 1999). We transiently transfected NIH 3T3 cells with cDNAs encoding Cav-1-GFP or GFP alone. After 48 h, living cells were subjected to DNA staining with Hoechst 33342 followed by FACS analysis. We analyzed the cell cycle characteristics of GFP-positive as well as GFP-negative cells from the same plate by gating with the use of the medium intensity green fluorescence signal.
Cav-1-GFP expression in NIH 3T3 cells was associated with a significant increase in the G0/G1 population (from 54 to 76%) and a decrease in the S phase population (from 34 to 13%) compared with the nonfluorescent cell population (Figure 2A). Importantly, overexpression of GFP alone did not alter the cell cycle distribution (Figure 2A). Figure 2B shows a tabulation representing the averages and SDs of three independent experiments of cell cycle analysis.
Figure 2.
Recombinant expression of caveolin-1 induces G0/G1 arrest in NIH 3T3 cells. (A and B) NIH 3T3 cells were transiently transfected with GFP alone (C1-GFP) or caveolin-1-GFP fusion protein (Cav-1 WT-GFP). (A) Cell cycle analysis. After 48 h, cells were subjected FACS analysis of the cell cycle (in EXPERIMENTAL PROCEDURES). The cell cycle analysis shown represents the average from three independent experiments. Note that only overexpression of Cav-1-GFP fusion protein causes G0/G1 arrest. (B) Quantitation. Table representing the averages and SDs of three independent experiments of the cell cycle analysis shown in A. (C and D) NIH 3T3 cells were transiently transfected with GFP alone (C1-GFP), the caveolin-1-full-length-GFP fusion protein (Cav-1 FL-GFP), and a variety of GFP-tagged caveolin-1 deletion mutants: caveolin-1 residues 1–81 [Cav-1 (1–81)-GFP], caveolin-1 residues 82–101 [Cav-1 (82–101)-GFP], and caveolin-1 residues 135–178 [Cav-1 (135–178)-GFP]. (C) Cav-1-GFP deletion mutants. The caveolin-1 deletion mutants are shown schematically. CSD, caveolin scaffolding domain; TD, potential membrane spanning/transmembrane domain. (D) Cell cycle analysis. After 48 h, cells were subjected to FACS analysis of the cell cycle (in EXPERIMENTAL PROCEDURES). Each point was performed in triplicate. A representative cell cycle analysis is shown. Interestingly, only the caveolin scaffolding domain (residues 82–101) and the C-terminal domain (residues 135–178), but not the N-terminal domain (residues 1–81) of caveolin-1, induce a G0/G1 block. (E and F) NIH 3T3 cells were left untreated (CTL) or treated for 16 h with 10 nM staurosporine to induce a G1 block. Also, cells were grown for 24 h with 0% serum to induce a G0 block. (E) Western blot analysis. Cells were then subjected to Western blotting analysis with the use of a caveolin-1-specific mAb probe (a representative immunoblot is shown; each lane contains equal amount of total proteins). (F) Cell cycle analysis. Each point was performed in triplicate. A representative cell cycle analysis is shown. Note that caveolin-1 protein expression is up-regulated only in G0 arrested-cells, but not in G1-arrested cells. In A, D, and F, an asterisk (∗) indicates significant differences in the cell cycle distribution.
We next performed cell cycle analysis with the use of a variety of caveolin-1 deletion mutants to identify a given region of the caveolin-1 molecule responsible for mediating arrest in the G0/G1 phase of the cell cycle. These caveolin-1 deletion mutants are shown schematically in Figure 2C. Note that full-length caveolin-1-GFP contains residues 1–178 of caveolin-1, Cav-1 (1–81)-GFP contains the first 81 residues, Cav-1 (82–101)-GFP contains the caveolin scaffolding domain, and Cav-1 (135–178)-GFP contains the C-terminal domain of caveolin-1. Importantly, these constructs are expressed at equivalent levels in transfected cells (Schlegel and Lisanti, 2000).
Figure 2D shows the results of this mutational analysis. We analyzed the cell cycle characteristics of GFP-positive as well as GFP-negative cells from the same plate by gating of the medium intensity green fluorescent signal. Virtually identical G0/G1 arrests were obtained with full-length caveolin-1-GFP, Cav-1 (82–101)-GFP, and Cav-1 (135–178)-GFP. Cav-1 (1–81)-GFP and GFP alone (C1 GFP) did not alter the cell cycle distribution. From this mutational analysis, we can conclude that both the caveolin scaffolding domain and the C terminus of caveolin-1 are important in mediating G0/G1 arrest. Both of these domains have been previously implicated as negative regulators of a variety of different signal transduction events (Ju et al., 1997; Engelman et al., 1998a,b; Razani et al., 1999).
To assess whether the G0/G1 arrest we observed represents a G0 arrest rather than a G1 arrest, we treated NIH 3T3 cells with 0% serum for 24 h to induce G0 arrest, and with 10 nM staurosporine for 16 h to induce G1 arrest. Serum starvation and staurosporine at low concentrations are known to arrest cells in G0 and G1, respectively (Crissman et al., 1991; Nishi et al., 1998).
Figure 2E shows that we successfully blocked NIH 3T3 cells in the G0/G1 phase of the cell cycle. The same samples were subjected to immunoblot analysis with the use of a caveolin-1-specific mAb probe. Figure 2F shows that only when cells are arrested in the G0 phase of the cell cycle, endogenous caveolin-1 protein expression is up-regulated. This result suggests that caveolin-1 is responsible for a selective G0 arrest. Importantly, we did not detect apoptotic cells after treatment with 10 nM staurosporine (our unpublished results), indicating that the low dose of staurosporine we used was not sufficient to induce programmed cell death.
Caveolin-1 Induces G0/G1 Arrest through a p53-dependent Mechanism
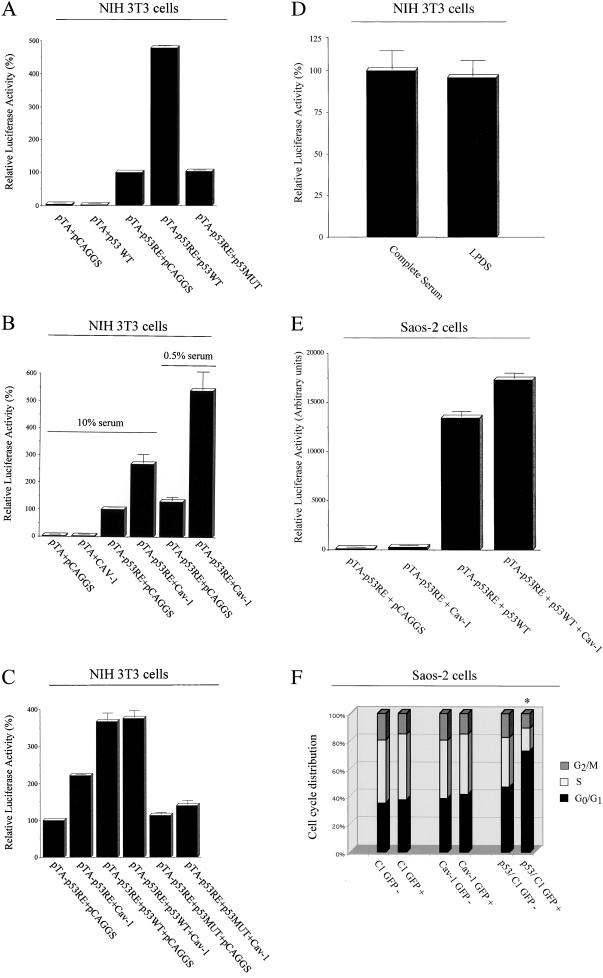
The p53 tumor suppressor protein plays a pivotal role in the regulation of cell growth arrest. To explore the possibility that caveolin-1 may induce cell cycle arrest by activating a p53-dependent pathway, we took advantage of a luciferase reporter plasmid containing a promoter specifically responsive to p53 (pTA-p53RE). First, we performed control experiments to show that the reporter specifically responds to the p53 tumor suppressor protein. We transiently transfected NIH 3T3 cells with the reporter vector pTA-p53RE in combination with wild-type p53 or a dominant negative mutant p53 (p53MUT). The p53 dominant negative mutant contains a G-to-A conversion at nucleotide 1017 and, if coexpressed with wild-type p53, it is responsible for the formation of a mixed tetramer that cannot interact with p53-binding sites, leading to a block in the downstream effects of p53. p53MUT is an important internal negative control that can be used to verify the specific activation of the p53 responsive element. Figure 3A shows that wild-type p53 activates its own responsive element by four- to fivefold. Conversely, the dominant negative form of p53 was unable to induce activation of the p53 responsive element. Importantly, wild-type p53 did not activate the luciferase reporter vector pTA, which lacks of the p53 responsive element (Figure 3A).
Figure 3.
Caveolin-1 induces G0/G1 arrest through a p53-dependent mechanism. (A–D) NIH 3T3 cells. (A) p53 WT and a dominant negative mutant of p53 (p53 MUT) were transiently cotransfected with the luciferase reporter plasmid (pTA-p53RE) in NIH 3T3 cells and luciferase activity was determined. Note that only expression of p53 WT activates the p53 responsive element by approximately four- to fivefold; in contrast, p53 MUT and the empty vector pCAGGS have no effect. Also, note that p53 WT has no effect on the luciferase reporter plasmid pTA, which lacks of the p53 responsive element. (B) NIH 3T3 cells were transiently transfected with caveolin-1 and the luciferase reporter plasmid (pTA-p53RE) and luciferase activity was determined. Note that only the expression of caveolin-1, but not the empty vector used to express caveolin-1 (pCAGGS), activates the p53 responsive element by approximately two- to threefold. This effect was augmented by the use of low serum treatment (0.5%). Importantly, caveolin-1 has no effect on the luciferase reporter plasmid pTA, which lacks of the p53 responsive element. (C) NIH 3T3 cells were transiently transfected with p53 (WT or MUT), caveolin-1 or the empty vector pCAGGS, in the presence of the luciferase reporter plasmid pTA-p53RE. Also, NIH 3T3 cells were transiently transfected with caveolin-1 and the empty vector pCAGGS, in the presence of the luciferase reporter plasmid pTA-p53RE. The luciferase activity was then determined. Note that the activation of the p53 responsive element by caveolin-1 is blunted when caveolin-1 is coexpressed with p53 MUT. (D) NIH 3T3 cells were transiently transfected with the luciferase reporter plasmid (pTA-p53RE) and the vector containing caveolin-1 cDNA. After 24 h, the media was removed and cells were washed with PBS. Fresh media containing either 10% normal fetal bovine serum (“complete serum”) or 10% LPDS was then added to the cells. Luciferase activity was determined the after day. Note that similar results were obtained with complete serum and LPDS, indicating that the effect of caveolin-1 on the p53 reporter is independent of the cholesterol content of the serum. (E and F) Saos-2 cells, a p53 negative cell line. (E) Saos-2 cells were transiently transfected with the luciferase reporter plasmid (pTA-p53RE) in combination with different vectors and luciferase activity was determined. Note that only transfection with the p53 cDNA or p53 plus the caveolin-1 cDNA dramatically activated the p53 responsive element. In contrast, the caveolin-1 cDNA alone or the empty vector used to express caveolin-1 (pCAGGS) had no effect. In addition, the combination of caveolin-1 plus p53 resulted in a synergistic effect on the activation of the p53 responsive element (i.e., caveolin-1 plus p53 activated pTA-p53RE more than the sum resulting from caveolin-1 alone or p53 alone). (F) Saos-2 cells were transiently transfected with GFP alone (C1-GFP) or caveolin-1-GFP fusion protein (Cav-1 WT-GFP). After 48 h, cells were subjected to FACS analysis and separated into GFP-negative and GFP-positive populations (in EXPERIMENTAL PROCEDURES). The cell cycle analysis shown represents the average from several independent experiments. Note that overexpression of the Cav-1-GFP fusion protein fails to causes G0/G1 arrest in these p53 negative cells. In contrast, transfection with p53 induces cell cycle arrest, as expected. Transfection with p53 served as an important positive control for these studies.
We next transiently transfected the luciferase vector pTA-p53RE with caveolin-1 or pCAGGS, the vector used to drive the expression of caveolin-1. Figure 3B shows that caveolin-1 expression activates the p53 responsive element by approximately two- to threefold. Interestingly, when cells are grown in 0.5% serum, the ability of caveolin-1 to activate the p53 responsive element was increased, resulting in approximately fivefold activation. These results indicate that caveolin-1 expression mediates activation of the p53 responsive element.
To verify that the ability of caveolin-1 to activate the p53 responsible element is mediated by p53, we transiently transfected caveolin-1 with wild-type p53 or the p53 dominant negative mutant and assessed the activation of the p53 responsive element by using the luciferase reporter vector pTA-p53RE. Figure 3C shows that when caveolin-1 is coexpressed with the p53 dominant negative mutant, its ability to activate the p53 responsive element is blocked, suggesting that expression of a functional wild-type p53 is required for caveolin-1 to achieve activation of the p53 responsive element. Also, wild-type p53 alone or in combination with caveolin-1 induced a very similar activation of its own responsive element. These results provide strong evidence that activation of the p53 responsive element by caveolin-1 is mediated by wild-type p53.
To examine a possible role for serum cholesterol in the caveolin-1-mediated activation of p53, we next compared the activity of caveolin-1 in the presence of normal control serum and lipoprotein-deficient serum. NIH 3T3 cells were transiently transfected with the luciferase reporter plasmid pTA-p53RE and the vector containing caveolin-1 cDNA. After 24 h, media were removed and cells were washed with PBS. Fresh media containing either 10% normal fetal bovine serum (“complete serum”) or 10% LPDS was then added to the cells. Luciferase activity was determined the following day. Figure 3D shows that similar results were obtained with complete and LPDS serum, indicating that the effect of caveolin-1 on the p53 reporter is independent of the cholesterol content of the serum.
To examine whether p53 is required to mediate the effects of caveolin-1, we next used a well-established p53 negative cell line, termed Saos-2 cells (Chen et al., 1990; Diller et al., 1990; Chandar et al., 1992). Briefly, Saos-2 cells were transiently transfected with the luciferase reporter plasmid pTA-p53RE in combination with different vectors and luciferase activity was determined. Figure 3E shows that only transfection with the p53 cDNA or p53 plus the caveolin-1 cDNA dramatically activated the p53 responsive element. Importantly, the caveolin-1 cDNA alone and the empty vector used to express caveolin-1 (pCAGGS) had no effect. In addition, the combination of caveolin-1 plus p53 resulted in a synergistic effect on the activation of the p53 responsive element (i.e., caveolin-1 plus p53 activated pTA-p53RE more than the sum resulting from caveolin-1 alone or p53 alone).
p53 also appears to be required for caveolin-1-mediated cell cycle arrest. Saos-2 cells were transiently transfected with GFP alone (C1-GFP) or the caveolin-1-GFP fusion protein (Cav-1 WT-GFP). After 48 h, cells were subjected to FACS analysis and separated into GFP-negative and GFP-positive populations (in EXPERIMENTAL PROCEDURES). Figure 3F shows that overexpression of the Cav-1-GFP fusion protein fails to causes G0/G1 arrest in these p53 negative cells. In contrast, transfection with p53 induces cell cycle arrest, as expected (Diller et al., 1990). Transfection with p53 is an important positive control for these studies.
Expression of Caveolin-1 in Mice as a Transgene
Our results with NIH 3T3 cells indicate that caveolin-1 expression induces cell cycle arrest. However, NIH 3T3 cells are immortalized. Thus, we next decided to examine the potential role of caveolin-1 as a cell cycle regulator with the use of primary cultures of mouse embryo fibroblasts. For this purpose, we generated transgenic mice that recombinantly express caveolin-1.
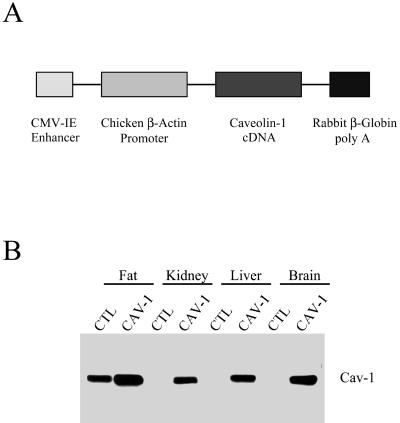
To achieve broad expression of caveolin-1 in mice as a transgene, the cDNA for caveolin-1 was inserted into a vector (pCAGGS) driven by the CMV enhancer and the chicken β-actin promoter, followed by the rabbit β-globin polyadenylation signal (Figure 4A; Niwa et al., 1991). Positive mice harboring the caveolin-1 transgene were identified by slot blot analysis of their genomic DNA, with the use of the rabbit β-globin sequence as a probe. To confirm expression of the caveolin-1 protein, their organs were harvested and subjected to Western blot analysis with the use of a caveolin-1-specific mAb probe.
Figure 4.
Expression of caveolin-1 in mice as a transgene. (A) The caveolin-1 transgenic expression cassette. The full-length untagged cDNA encoding murine caveolin-1 was subcloned into the expression vector pCAGGS. Note that in the pCAGGS-Cav-1 construct, the CMV enhancer, and the β-actin promoter sequence are located upstream of the caveolin-1 cDNA, whereas the rabbit β-globin poly(A) sequence is located downstream. The CMV enhancer/β-actin promoter sequences are expected to drive constitutive expression of caveolin-1 in a wide variety of tissue types. (B) Tissue distribution of caveolin-1 transgene expression. The expression of caveolin-1 in normal control mice (CTL) and caveolin-1 transgenic mice (Cav-1) is shown. Protein lysates were prepared from a variety of mouse tissues. Immunoblotting was performed with a monospecific antibody probe that recognizes only caveolin-1 (mAb 2297). Note that the expression of caveolin-1, in caveolin-1 transgenic mice, is significantly higher compared with the endogenous expression in normal control mice in all tissues examined.
Figure 4B shows that high levels of caveolin-1 transgene expression were observed in fat, kidney, liver, and brain. Overexpression of caveolin-1 in caveolin-1 transgenic mice was also observed in lung, spleen, skeletal muscle, and heart (our unpublished results). In contrast, normal control mice lacking the caveolin-1 transgene showed high levels of caveolin-1 in fat only, consistent with the known tissue distribution of caveolin-1 mRNA and protein.
Generating Mouse Embryonic Fibroblasts That Transgenically Express Caveolin-1
To derive mouse embryonic fibroblasts overexpressing caveolin-1, a caveolin-1 transgenic male mouse was crossed with a C57Bl/6 female mouse. After 13.5 d, the pregnant female was sacrificed and MEFs were derived (in EXPERIMENTAL PROCEDURES).
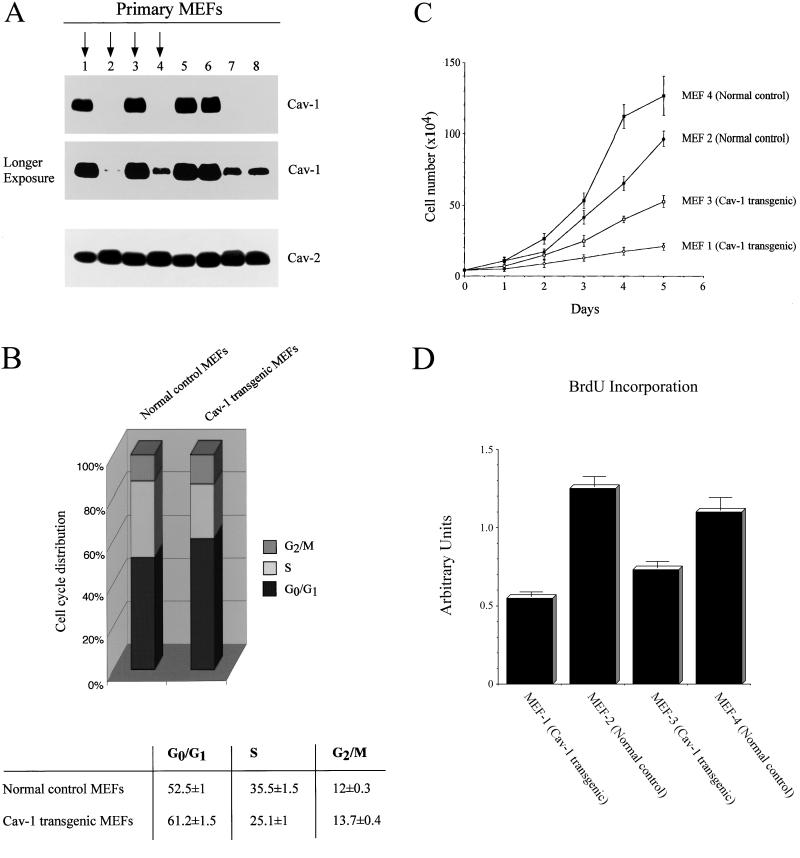
Figure 5A shows that we successfully derived four independent mouse embryonic fibroblast primary cell lines overexpressing caveolin-1 (1, 3, 5, and 6), and four independent MEF primary cell lines expressing normal levels of caveolin-1 (2, 4, 7, and 8). A longer exposure is included to show that MEFs lacking the caveolin-1 transgene express endogenous caveolin-1. Note also that expression levels of endogenous caveolin-2 are not affected by the overexpression of caveolin-1 (Figure 5A).
Figure 5.
Primary cultures of MEFs that transgenically express caveolin-1 show phenotypic changes in cell cycle regulation and cellular proliferation. (A) Immunoblot analysis. Eight independent primary cultures of MEFs were derived from littermate embryos and analyzed by Western blotting with the use of a monospecific antibody probe that recognizes only caveolin-1 (mAb 2297), and a monospecific antibody probe that recognizes only caveolin-2 (mAb 65). Note that four independent MEFs transgenically express caveolin-1; a longer exposure is also shown to illustrate the presence of endogenous caveolin-1. Arrows point at two Cav-1 transgenic MEF cultures (1 and 3) and normal control MEF cultures (2 and 4) that were subjected to further characterization. (B) Cell cycle analysis. The cell cycle distribution of Cav-1 transgenic MEFs is shown, compared with normal control MEFs. Note that transgenic expression of caveolin-1 in primary MEFs leads to arrest in the G0/G1 phase of the cell cycle. (C) Cellular proliferation. Cells were initially plated in 10-cm dishes at a density of 40,000 cells/dish at day 0 and then cell numbers were counted after 1, 2, 3, 4, and 5 d of incubation at 37°C. Each point represents the average of four independent measurements. Note that MEFs that transgenically express caveolin-1 show a significantly reduction in cellular proliferation, compared with normal control MEFs. (D) BrdU incorporation. Cells were plated in 96-well plates at a density of 10,000 cells/well at day 0. Each MEF primary cell line was seeded in quadruplicate. After 2 d, BrdU incorporation was carried out. Note transgenic expression of caveolin-1 results in reduced DNA replication.
MEFs That Transgenically Express Caveolin-1 Show Phenotypic Changes in Cell Cycle Regulation and Cellular Proliferation
Our results indicate that transient overexpression of caveolin-1 in NIH 3T3 cells is responsible for arrest in the G0 phase of the cell cycle. However, MEFs represent a better in vivo model to study the role of caveolin-1 in cell cycle regulation. Based on our studies with NIH 3T3 cells, we would predict that MEFs derived from caveolin-1 transgenic embryos would show an increased G0/G1 population. Figure 5B shows that MEFs derived from caveolin-1 transgenic embryos have a significant reduction in the S phase population and a corresponding increase in the G0/G1 population, compared with MEFs derived from normal control littermate embryos. Interestingly, we also observed a minor increase in the G2/M population that we did not observe in our transient transfection experiments with NIH 3T3 cells. This difference may be explained by the different nature of the two models we used, primary cultures (MEF) versus an immortalized cell line (NIH 3T3).
Cells that are arrested in the G0/G1 and G2/M phases of the cell cycle should be characterized by a reduction in cellular proliferation. To test this hypothesis, primary MEF cultures were plated in 10-cm dishes at a density of 40,000 cells/dish and the cell numbers were counted after 1, 2, 3, 4, and 5 d of incubation at 37°C. MEFs derived from caveolin-1 transgenic mouse embryos (MEFs 1 and 3) showed significant growth inhibition compared with MEFs derived from normal control littermate embryos (MEFs 2 and 4). Figure 5C shows that after 5 d in complete medium, Cav-1 transgenic MEFs demonstrate an approximate three- to fourfold reduction in total cell number, consistent with the cell cycle analysis data (Figure 5B).
To independently assess whether caveolin-1 expression induces a reduction in the S phase population in vivo, we used BrdU incorporation to determine the population of cells in S phase (Figure 5D). Note that Cav-1 transgenic MEFs (1 and 3) show a significant degree of S phase suppression compared with MEFs (2 and 4) derived from normal control littermate embryos. Taken together, these results clearly indicate that transgenic expression of caveolin-1 in mouse embryonic fibroblasts is responsible for initiating growth arrest.
Caveolin-1 Transgenic MEFs Undergo Cell Cycle Arrest through Activation of a p53-dependent Pathway
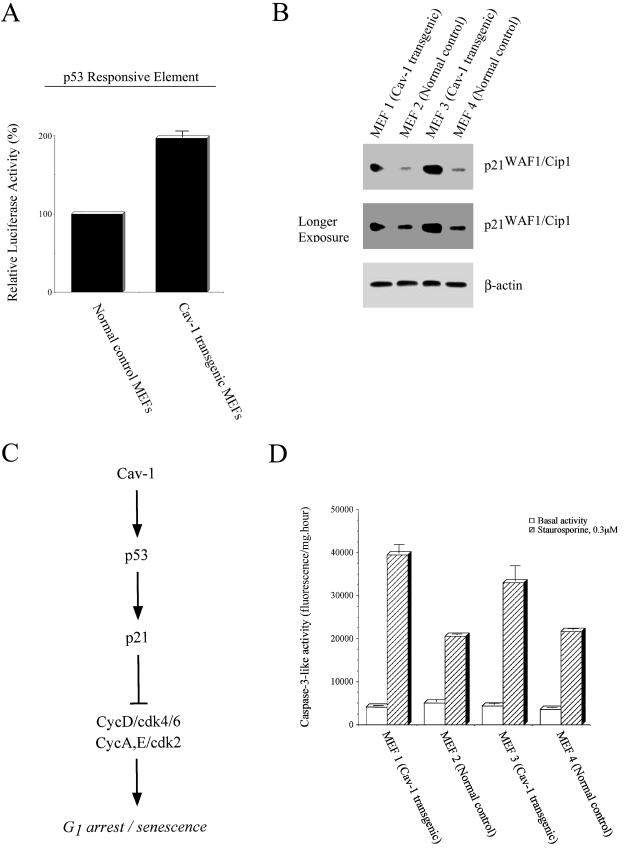
Our previous results (this report) indicate that transient expression of caveolin-1 in NIH 3T3 cells induces activation of a p53 responsive element (Figure 3B). Thus, we decided to independently assess whether the cell cycle arrest induced by caveolin-1 in mouse embryonic fibroblasts is due to p53 activation. We transiently transfected mouse embryonic fibroblasts with the p53 responsive element reporter, pTA-p53RE. Figure 6A shows that the p53 responsive element is activated by approximately twofold in Cav-1 transgenic MEFs (1 and 3) compared with normal control MEFs (2 and 4).
Figure 6.
Caveolin-1 transgenic MEFs undergo cell cycle arrest through activation of a p53-dependent pathway. (A) p53 activity. MEFs derived from caveolin-1 transgenic mice and from normal control mice were transiently transfected with the luciferase reporter plasmid (pTA-p53RE), and the luciferase activity was determined. Note that the p53 responsive element is activated in Cav-1 transgenic MEFs. (B) p21WAF1/Cip1 expression. MEFs derived from caveolin-1 transgenic mice (MEF 1 and MEF 3) and from normal control mice (MEF 2 and MEF 4) were subjected to Western blot analysis with the use of a monospecific antibody probe that recognizes p21. Note that p21 protein expression is up-regulated in Cav-1 transgenic MEFs. Two exposures are shown to better illustrate the degree of p21 up-regulation and the presence of p21 expression in normal control MEFs. Western blotting with the use of a monospecific antibody probe that recognizes β-actin was used to verify equal protein loading. (C) Schematic representation of the activation of the p53/p21-pathway by caveolin-1. Caveolin-1 induces G1 arrest/senescence through activation of the p53 tumor suppressor protein. Activation of p53 results in up-regulation of p21 protein expression leading to inactivation of the kinase activity of CycD/cdk4/6 and CycA, E/cdk2 and cell cycle arrest in the G1 phase. (D) Caspase-3 activation. Caspase-3-like activity was measured in MEFs derived from caveolin-1 transgenic mice (MEF 1 and MEF 3) and MEFs derived from normal control mice (MEF 2 and MEF 4). Note that caveolin-1 expression in MEFs does not affect basal caspase-3 activity, but significantly sensitizes cells toward staurosporine-induced caspase-3 activation.
p53 activation is known to result in the induction of p21WAF1/Cip1. Thus, we next assessed p21 protein expression in these primary MEF cultures. Figure 6B indicates that p21 protein expression is significantly increased in Cav-1 transgenic MEFs (1 and 3). Thus, transgenic expression of caveolin-1 in MEFs induces activation of p53 and overexpression of p21WAF1/Cip1. Taken together, these results clearly indicate that transgenic expression of caveolin-1 in mouse embryonic fibroblasts induces cell cycle arrest through activation of the p53/p21 pathway (Figure 6C).
Transgenic Expression of Caveolin-1 Does Not Induce Apoptosis, but Caveolin-1 Sensitizes MEFs to Apoptotic Stimulation
Our current results indicate that caveolin-1 expression is directly involved in mediating cell cycle arrest. To rule out the possibility that caveolin-1 induces apoptosis, rather than cell cycle arrest, we assessed caspase-3 activity in primary cultures of Cav-1 transgenic MEFs and MEFs derived from their normal control littermates.
Caspase-3 is a key element in the signal cascade leading to programmed cell death. Activation of caspase-3 is positively associated with apoptosis. Figure 6D shows that transgenic expression of caveolin-1 did not increase basal caspase-3 activity. In contrast, when apoptosis was induced by treatment with 0.3 μM staurosporine for 6 h, caspase-3 activity was significantly increased in Cav-1 transgenic MEFs compared with MEFs derived from their normal control littermates (Figure 6D). These results indicate that expression of caveolin-1 by itself is not sufficient to induce apoptosis, but rather caveolin-1 expression sensitizes MEFs to staurosporine-induced apoptosis. This is consistent with the idea that caveolin-1 expression mediates cell cycle arrest.
The inability of caveolin-1 expression by itself to induce apoptosis was also noted during FACS analysis of the cell cycle. In all the FACS analysis experiments we performed, we did not identify any increases in an apoptotic subpopulation (our unpublished results).
Caveolin-3 Expression also Induces Cell Cycle Arrest in the G0/G1 Phase
To explore whether other members of the caveolin gene family can mediate cell cycle arrest in the G0/G1 phase, we decided to study the effect of caveolin-3 expression via cell cycle analysis. Of the three known caveolin gene family members, caveolins 1 and 3 show the closest homology. However, the expression of caveolin-3 is muscle-specific.
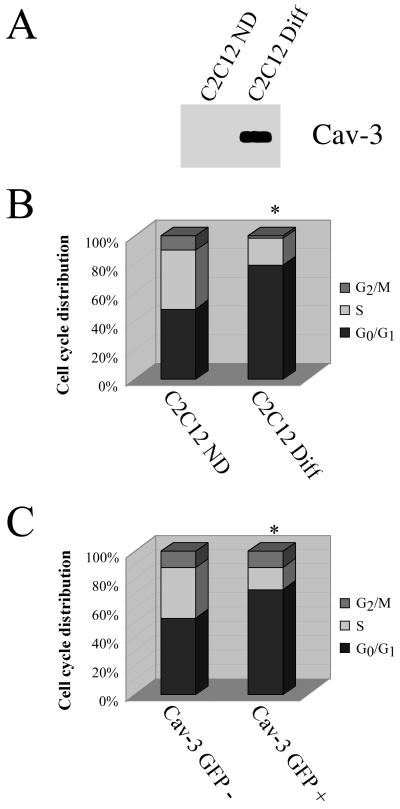
Cultured C2C12 cells can be induced to differentiate from myoblasts into myotubes bearing an embryonic phenotype in low mitogen medium over a period of 2 d. We have previously shown that both mRNA and protein levels of caveolin-3 are dramatically induced during the course of differentiation of C2C12 cells from myoblasts to myotubes (Song et al., 1996; Tang et al., 1996). Figure 7 shows that caveolin-3 expression is induced in differentiated C2C12 cells (Figure 7A) and that C2C12 cells undergo arrest in the G0/G1 phase of the cell cycle after differentiation (Figure 7B). These results indicate that caveolin-3 expression is associated with a block in the G0/G1 phase of the cell cycle in C2C12 cells.
Figure 7.
Caveolin-3 expression also induces arrest in the G0/G1 phase of the cell cycle. (A and B) Endogenous expression in C2C12 cells. Undifferentiated C2C12 cells (C2C12 ND) or C2C12 cells differentiated for 48 h with 3% horse serum (C2C12 Diff) were subjected to SDS-PAGE and Western blot analysis with the use of a caveolin-3-specific mAb probe (a representative immunoblot is shown; each lane contains an equal amount of total protein) (A) and cell cycle analysis (in EXPERIMENTAL PROCEDURES) (B) The cell cycle analysis shown represents the average from three independent experiments (C2C12 ND: G0/G1 = 49 ± 5; S = 41 ± 4; G2/M = 10 ± 1.5; C2C12 Diff: G0/G1 = 79 ± 5; S = 19 ± 2; G2/M = 2 ± 1). Note that the expression of endogenous caveolin-3 in differentiated C2C12 cells is associated with arrest in the G0/G1 phase of the cell cycle. (C) Transient expression in NIH 3T3 cells. NIH 3T3 cells were transiently transfected with a GFP-tagged caveolin-3 expression vector and subjected to cell cycle analysis (in EXPERIMENTAL PROCEDURES). The cell cycle analysis shown represents the averages of three independent experiments (Cav-3 GFP-: G0/G1= 53 ± 5; S = 36 ± 4; G2/M = 11 ± 1; Cav-3 GFP+: G0/G1= 72 ± 6; S = 16 ± 2; G2/M = 12 ± 2). Note that transient expression of caveolin-3 leads to arrest in the G0/G1 phase of the cell cycle. In B and C, an asterisk (∗) indicates significant differences in the cell cycle distribution.
Next, we evaluated the effects of transient caveolin-3 expression on cell cycle progression. NIH 3T3 cells were transiently transfected with a GFP-tagged form of caveolin-3 (Galbiati et al., 1999) and subjected to cell cycle analysis. In these experiments, we analyzed the cell cycle characteristics of GFP-positive as well as GFP-negative cells from the same plate by gating the medium intensity green fluorescence signal. Figure 7C indicates that recombinant expression of caveolin-3 in NIH 3T3 cells induces a G0/G1 block. These results suggest that both caveolins 1 and 3 can induce cell cycle arrest in the G0/G1 phase of the cell cycle.
DISCUSSION
In recent years, several independent lines of evidence have emerged that suggest that caveolin-1 functions as a tumor suppressor protein in mammalian cells. In fact, modification and/or inactivation of caveolin-1 expression appears to be a common feature of the transformed phenotype. Caveolin-1 expression, for example, is absent in several transformed cell lines derived from human mammary carcinomas, including MT-1, MCF-7, ZR-75–1, T47D, MDA-MB-361, and MDA-MB-474 (Sager et al., 1994). Recently, we have shown that antisense-mediated down-regulation of caveolin-1 is sufficient to induce cell transformation in NIH 3T3 cells (Galbiati et al., 1998). NIH 3T3 cells harboring antisense caveolin-1 spontaneously formed foci, exhibited anchorage-independent growth in soft agar, formed tumors in immunodeficient mice, and appeared morphologically transformed as seen by scanning electron microscopy (Galbiati et al., 1998). Cell transformation is usually characterized by loss of contact inhibition and cell cycle control. Interestingly, we have recently shown that caveolin-1 protein expression is up-regulated when NIH 3T3 cells reach confluence (Galbiati et al., 1998), suggesting a possible role for caveolin-1 in mediating contact inhibition. However, the role of caveolin-1 in the regulation of cell cycle progression remains unknown.
Here, we test this hypothesis directly by using two different, but complementary, approaches: 1) transient expression of caveolin-1 in NIH 3T3 cells, and 2) recombinant expression of caveolin-1 as a transgene in mice and characterization of the phenotype of MEFs derived from these mice.
Transient expression of caveolin-1 in NIH 3T3 cells resulted in the cell cycle arrest in the G0/G1 phase. Caveolin-1-expressing cells showed a significant reduction in the S phase population as well. We also showed that antisense-mediated down-regulation of caveolin-1 in NIH 3T3 cells is associated with a reduced G0/G1 population and increased S phase population. These results indicate that caveolin-1 expression may have a direct role in the regulation of cell cycle progression.
In support of this hypothesis, we demonstrate that when cells are grown without serum, to induce cell cycle arrest, caveolin-1 expression is up-regulated. Conversely, when serum-deprived cells are forced to enter the cell cycle by addition of serum, caveolin-1 expression is significantly down-regulated. These data are consistent with the previous observations that both the number of caveolae organelles and caveolin-1 protein expression are highest expressed in terminally differentiated cells (adipocytes, endothelial cells, and muscle cells [Simionescu and Simionsecu, 1983; Scherer et al., 1994; Scherer et al., 1996]), which are in the G0 phase of the cell cycle. Interestingly, when we selectively induce G1 arrest by treating NIH 3T3 cells with low doses (10 nM) of staurosporine, we did not observed variation in caveolin-1 expression. In contrast, G0 arrest induced by serum deprivation resulted in up-regulation of caveolin-1. Taken together, these results indicate that caveolin-1 protein expression is responsible for a selective G0 arrest of the cell cycle.
Caveolin-1 is most closely related to caveolin-3 based on protein sequence homology: caveolin-1 and caveolin-3 are ∼65% identical and ∼85% similar. Thus, we would predict that caveolin-3 may have similar properties in inhibiting cell cycle progression. In fact, we demonstrate that caveolin-3 overexpression in NIH 3T3 cells induces a G0/G1 block of the cell cycle. Also, we show that caveolin-3 expression is induced during the differentiation of C2C12 cells from myoblasts to myotubes, and that differentiated C2C12 cells are arrested in the G0/G1 phase of the cell cycle. As myoblasts have to enter the G0 phase of the cell cycle to differentiate, these data support the idea that caveolin-3 expression is responsible for a selective arrest in the G0 phase of the cell cycle.
Recently, we examined the functional role of caveolin-1 in regulating signaling along the mitogen-activated protein kinase cascade, a major pathway involved in cell proliferation (Engelman et al., 1998a,b). We have demonstrated that the caveolin scaffolding domain of caveolin-1 (residues 82–101) inhibited the in vitro kinase activity of purified MEK-1 and ERK-2. In this report, we also demonstrated that the caveolin scaffolding domain, but not the N-terminal domain of caveolin-1 (residues 1–81), is responsible for cell cycle arrest. Interestingly, we observed that the C-terminal domain of caveolin-1 (residues 135–178) and the caveolin scaffolding domain were similarly effective in promoting cell cycle arrest. We conclude that both the caveolin scaffolding domain and the C-terminal domain are critical regions of caveolin-1 involved in the arrest in the G0/G1 phase of the cell cycle. This result was not surprising because we and others have previously shown that the caveolin scaffolding domain and the C-terminal domain of caveolin-1 were similarly effective in binding and inhibiting catalytic activity of a number of known signaling molecules (Ju et al., 1997; Engelman et al., 1998a,b; Razani et al., 1999).
Primary cultures of MEFs are an important tool for studying the function of a given protein in a nonimmortalized setting. To further study the role of caveolin-1 in cell cycle regulation, we next created transgenic mice that recombinantly express caveolin-1 and derived primary MEF cultures from these mice. We showed that transgenic expression of caveolin-1 in these MEFs induced arrest in the G0/G1 phase of the cell cycle and a corresponding reduction in the S phase population. Similarly, the cellular proliferation of Cav-1 transgenic MEFs was dramatically reduced. Taken together, these results indicate that transgenic expression of caveolin-1 in vivo is sufficient to inhibit cell proliferation via cell cycle arrest in the G0/G1 phase of the cell cycle. We speculate that the down-regulation of caveolin-1 protein expression observed during cell transformation is, at least in part, responsible for the abnormal growth characteristics of oncogenically transformed cells.
How does caveolin-1 expression induce cell cycle arrest? The p53 tumor suppressor protein is a major intracellular regulator involved in modulating cell cycle progression. Our results indicate that p53 activity is increased approximately two- to threefold when caveolin-1 is transiently expressed in NIH 3T3 cells. A G-to-A conversion at nucleotide 1017 in the p53 protein generates a dominant negative mutant, that if coexpressed with wild-type p53, forms a mixed tetramer that is unable to interact with p53 binding sites, leading to inhibition of the downstream effects of p53.
Interestingly, when caveolin-1 is transiently coexpressed with this dominant negative mutant of p53 (p53MUT), caveolin-1's ability to stimulate p53 activity is blocked. These results indicate that caveolin-1 activation of the p53 responsive element is selectively mediated by wild-type p53. Importantly, Cav-1 transgenic MEFs showed a similar twofold increase in p53 activity compared with MEFs derived from normal littermate control mice.
p53-mediated arrest in the G0/G1 phase of the cell cycle is mediated by the induction of p21WAF1/Cip1 expression. As we have shown here that Cav-1 transgenic MEFs have increased p53 activity, we also examined the level of p21 expression in these cells. As predicted, we demonstrated that p21 protein expression is up-regulated in Cav-1 transgenic MEFs. Taken together, these results clearly indicate that transgenic expression of caveolin-1 in MEFs results in cell cycle arrest via the p53/p21-pathway (see Figure 6C for a schematic representation of this pathway).
The tumor suppressor protein p53 has also been implicated in mediating programmed cell death (apoptosis) through either sequence-specific transactivation-dependent or -independent pathways (Amundson et al., 1998; Sionov and Haupt, 1998; Bates and Vousden, 1999). To determine the specific role of p53 in mediating cell cycle arrest rather than apoptosis, it is essential to demonstrate that apoptotic pathways are not activated and that cells do not undergo apoptosis. Indeed, we demonstrated that caveolin-1-mediated activation of the p53/p21-pathway does not result in apoptosis. In fact, we showed that transgenic expression of caveolin-1 in MEFs does not increase the basal activity of caspase-3, a critical step in mediating programmed cell death. Also, when we subjected mouse embryonic fibroblasts to FACS analysis of the cell cycle, we did not observe any apoptotic subpopulations. We conclude that caveolin-1 expression is sufficient to activate the p53 pathway and induce cell cycle arrest, but that caveolin-1 expression by itself is not sufficient to induce apoptosis.
Interestingly, when we treated primary MEF cultures with an apoptotic stimulus (staurosporine), we found that transgenic expression of caveolin-1 sensitized these cells to apoptosis, indicating that an apoptotic stimulus is still necessary to mediate cell death. These results are consistent with previous data showing that expression of caveolin-1 is sufficient to sensitize Rat1a fibroblasts to ceramide-induced cell death (Zundel et al., 2000).
Because transgenic expression of caveolin-1 in MEFs induced cell cycle arrest, we speculate that caveolin-1 transgenic mice may be protected against tumor development. However, additional experiments, i.e., tumor induction, are necessary to directly test this hypothesis.
Recently, we have demonstrated that p53 is a positive regulator of caveolin-1 gene transcription and protein expression (Razani et al., 2000a). p53-mediated induction of caveolin-1 gene expression is intriguing, because this would indicate that caveolin-1 is a previously unrecognized p53-regulated gene in cancers. Because the observed induction was on the order of 6–12 h, caveolin-1 is likely to be an immediate target. Interestingly, MEF cultures derived from p53-deficient mice also display a distinct lack of caveolin-1 expression (Lee et al., 1998). Furthermore, p53 has also been directly implicated in the up-regulation of caveolin-1 during free cholesterol-mediated cellular growth (Bist et al., 2000). However, it is also possible that p53 could additionally act on its many bona fide target genes (e.g., p21, mdm-2, GADD45, cyclin G, etc.) (Ko and Prives, 1996), activating signaling cascades, thereby indirectly leading to caveolin-1 induction. We conclude that p53 positively regulates caveolin-1 gene expression (Razani et al., 2000a) and that caveolin-1 expression can increase the activity of p53 (this report). Thus, caveolin-1 and p53 may act synergistically.
Hayashi and colleagues have recently identified a common sporadic mutation in caveolin-1 in human breast cancers (Hayashi et al., 2001). Interestingly, this mutation is analogous to one of the inherited mutations previously identified in human caveolin-3 (P104L) in limb-girdle muscular dystrophy (LGMD-1C) patients (Galbiati et al., 2001). More specifically, the same invariant proline residue in caveolin-1 is mutated to leucine (P132L). Recombinant expression of the caveolin-1 cDNA containing this mutation is sufficient to induce oncogenic transformation in NIH 3T3 cells and to hyperactivate the p42/44 MAP kinase cascade (Hayashi et al., 2001). Similarly, ablation of caveolin-1 expression in NIH 3T3 cells using an anti-sense approach induced oncogenic transformation, activates the p42/44 MAP kinase pathways, and confers tumorigenicity in nude mice (Galbiati et al., 1998). As the caveolin-1 (P132L) mutant mimics the caveolin-1 anti-sense phenotype, these results suggest that caveolin-1 (P-132L) behaves in a dominant-negative fashion, as has been previously documented for caveolin-3 (P104L) (Galbiati et al., 1999). Thus, it is likely that dominant negative mutations in the 12 invariant caveolin-1 residues will be identified in other forms of human cancer.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health, the Muscular Dystrophy Association, the American Heart Association, and the Susan B. Komen Breast Cancer Foundation (to M.P.L.). M.P.L. is the recipient of a Hirschl/Weil-Caulier Career Scientist Award. P.G.F. was supported by postdoctoral fellowships from the Heart and Stroke Foundation of Canada and the Canadian Institutes of Health Research. R.G.P. was supported in part by grants from the National Institutes of Health (R01-CA-70897, R01-CA-75503, and P50-HL-56399); R.G.P. is a recipient of the Irma T. Hirschl award and an award from the Susan G. Komen Breast Cancer Foundation. F.G. was the recipient of a fellowship from Telethon-Italia (no. 470/bi) and a Scientist Development Grant from the American Heart Association. J.L. was the recipient of a Scientist Development Grant from the American Heart Association. L.Z. was supported by grants from the National Institutes of Health, American Cancer Society, U.S. Army, and a scholarship from the Leukemia and Lymphoma Society of America.
Note added in proof.
We have now shown that MEFs derived from Cav-1 knock-out mice (CAV-1 −/−) proliferate ∼2- to 3-fold faster than their wild-type counterparts (Razani, B., Engelman, J.A., Wang, X.B., Schubert, W., Zhang, X.L., Marks, C.B., Macaluso, F., Russell, R.G., Li, M., Pestell, R.G., Di Vizio, D., Hou, H., Knietz, B., Lagaud, G., Christ, G.J., Edelmann, W., and Lisanti, M.P. (2001). Caveolin-1 null mice are viable, but show evidence of hyper-proliferative and vascular abnormalities. J. Biol. Chem., In press.). Thus, loss of caveolin-1 expression clearly augments cell proliferation. These data directly support our current observations and are consistent with the idea that caveolin-1 may function as a tumor suppressor gene.
REFERENCES
- Amundson SA, Myers TG, Fornace AJ., Jr Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene. 1998;17:3287–3299. doi: 10.1038/sj.onc.1202576. [DOI] [PubMed] [Google Scholar]
- Bates S, Vousden KH. Mechanisms of p53-mediated apoptosis. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ze'ev A, Geiger B. Differential molecular interactions of beta-catenin and plakoglobin in adhesion, signaling and cancer. Curr Opin Cell Biol. 1998;10:629–639. doi: 10.1016/s0955-0674(98)80039-2. [DOI] [PubMed] [Google Scholar]
- Bist A, Fielding CJ, Fielding PE. p53 regulates caveolin gene transcription, cell cholesterol, and growth by a novel mechanism. Biochemistry. 2000;39:1966–1972. doi: 10.1021/bi991721h. [DOI] [PubMed] [Google Scholar]
- Chandar N, Billig B, McMaster J, Novak J. Inactivation of p53 gene in human and murine osteosarcoma cells. Br J Cancer. 1992;65:208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Chen YM, Bookstein R, Lee WH. Genetic mechanisms of tumor suppression by the human p53 gene. Science. 1990;250:1576–1580. doi: 10.1126/science.2274789. [DOI] [PubMed] [Google Scholar]
- Couet J, Li S, Okamoto T, Ikezu T, Lisanti MP. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J Biol Chem. 1997a;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- Couet J, Li S, Okamoto T, Scherer PS, Lisanti MP. Molecular and cellular biology of caveolae: paradoxes and plasticities. Trends Cardiovasc Med. 1997b;7:103–110. doi: 10.1016/S1050-1738(97)00001-7. [DOI] [PubMed] [Google Scholar]
- Couet J, Sargiacomo M, Lisanti MP. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J Biol Chem. 1997c;272:30429–30438. doi: 10.1074/jbc.272.48.30429. [DOI] [PubMed] [Google Scholar]
- Crissman HA, Gadbois DM, Tobey RA, Bradbury EM. Transformed mammalian cells are deficient in kinase-mediated control of progression through the G1 phase of the cell cycle. Proc Natl Acad Sci USA. 1991;88:7580–7584. doi: 10.1073/pnas.88.17.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Diller L, Kassel J, Nelson CE, Gryka MA, Litwak G, Gebhardt M, Bressac B, Ozturk M, Baker SJ, Vogelstein BEA. p53 functions as a cell cycle control protein in osteosarcomas. Mol Cell Biol. 1990;10:5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS. p21/p53, cellular growth control and genomic integrity. Curr Top Microbiol Immunol. 1998;227:121–137. doi: 10.1007/978-3-642-71941-7_6. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chu C, Lin A, Jo H, Ikezu T, Okamoto T, Kohtz DS, Lisanti MP. Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo. A role for the caveolin-scaffolding domain. FEBS Lett. 1998a;428:205–211. doi: 10.1016/s0014-5793(98)00470-0. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Lee RJ, Karnezis A, Bearss DJ, Webster M, Siegel P, Muller WJ, Windle JJ, Pestell RG, Lisanti MP. Reciprocal regulation of Neu tyrosine kinase activity and caveolin-1 protein expression in vitro and in vivo. Implications for mammary tumorigenesis. J Biol Chem. 1998b;273:20448–20455. doi: 10.1074/jbc.273.32.20448. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Wycoff CC, Yasuhara S, Song KS, Okamoto T, Lisanti MP. Recombinant expression of caveolin-1 in oncogenically transformed cells abrogates anchorage-independent growth. J Biol Chem. 1997;272:16374–16381. doi: 10.1074/jbc.272.26.16374. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zhang XL, Galbiati F, Lisanti MP. Chromosomal localization, genomic organization, and developmental expression of the murine caveolin gene family (Cav-1, -2, and -3). Cav-1 and Cav-2 genes map to a known tumor suppressor locus (6-A2/7q31) FEBS Lett. 1998c;429:330–336. doi: 10.1016/s0014-5793(98)00619-x. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zhang XL, Galbiati F, Volonte D, Sotgia F, Pestell RG, Minetti C, Scherer PE, Okamoto T, Lisanti MP. Molecular genetics of the caveolin gene family: implications for human cancers, diabetes, Alzheimer's disease, and muscular dystrophy. Am J Hum Genet. 1998d;63:1578–1587. doi: 10.1086/302172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zhang XL, Lisanti MP. Genes encoding human caveolin-1 and -2 are co-localized to the D7S522 locus (7q31.1), a known fragile site (FRA7G) that is frequently deleted in human cancers. FEBS Lett. 1998e;436:403–410. doi: 10.1016/s0014-5793(98)01134-x. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zhang XL, Lisanti MP. Sequence and detailed organization of the human caveolin-1 and -2 genes located near the D7S522 locus (7q31.1). Methylation of a CpG island in the 5′ promoter region of the caveolin-1 gene in human breast cancer cell lines. FEBS Lett. 1999;448:221–230. doi: 10.1016/s0014-5793(99)00365-8. [DOI] [PubMed] [Google Scholar]
- Fra AM, Masserini M, Palestini P, Sonnino S, Simons K. A photo-reactive derivative of ganglioside GM1 specifically cross-links VIP21-caveolin on the cell surface. FEBS Lett. 1995;375:11–14. doi: 10.1016/0014-5793(95)95228-o. [DOI] [PubMed] [Google Scholar]
- Galbiati, F., Razani, B., and Lisanti, M.P. (2001). Role of caveolae and caveolin-3 in muscular dystrophy. Trends Molec Med. In press. [DOI] [PubMed]
- Galbiati F, Volonte D, Brown AM, Weinstein DE, Ben-Ze'ev A, Pestell RG, Lisanti MP. Caveolin-1 expression inhibits Wnt/beta-catenin/Lef-1 signaling by recruiting beta-catenin to caveolae membrane domains. J Biol Chem. 2000;275:23368–23377. doi: 10.1074/jbc.M002020200. [DOI] [PubMed] [Google Scholar]
- Galbiati F, Volonte' D, Engelman JA, Watanabe G, Burk R, Pestell RG, Lisanti MP. Targeted down-regulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J. 1998;17:6633–6648. doi: 10.1093/emboj/17.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Volonte D, Minetti C, Chu JB, Lisanti MP. Phenotypic behavior of caveolin-3 mutations that cause autosomal dominant limb girdle muscular dystrophy (LGMD-1C). Retention of LGMD-1C caveolin-3 mutants within the Golgi complex. J Biol Chem. 1999;274:25632–25641. doi: 10.1074/jbc.274.36.25632. [DOI] [PubMed] [Google Scholar]
- Garcia-Cardena G, Martasek P, Masters BS, Skidd PM, Couet J, Li S, Lisanti MP, Sessa WC. Dissecting the interaction between nitric oxide synthase (NOS) and caveolin. Functional significance of the NOS caveolin binding domain in vivo. J Biol Chem. 1997;272:25437–25440. doi: 10.1074/jbc.272.41.25437. [DOI] [PubMed] [Google Scholar]
- Garcia-Cardena G, Oh P, Liu J, Schnitzer JE, Sessa WC. Targeting of nitric oxide synthase to endothelilal cell caveolae via palmitoylation: implications for caveolae localization. Proc Natl Acad Sci USA. 1996;93:6448–6453. doi: 10.1073/pnas.93.13.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenney JR. Tyrosine phosphorylation of a 22 kD protein is correlated with transformation with Rous sarcoma virus. J Biol Chem. 1989;264:20163–20166. [PubMed] [Google Scholar]
- Glenney JR. The sequence of human caveolin reveals identity with VIP 21, a component of transport vesicles. FEBS Lett. 1992;314:45–48. doi: 10.1016/0014-5793(92)81458-x. [DOI] [PubMed] [Google Scholar]
- Glenney JR, Soppet D. Sequence and expression of caveolin, a protein component of caveolae plasma membrane domains phosphorylated on tyrosine in RSV-transformed fibroblasts. Proc Natl Acad Sci USA. 1992;89:10517–10521. doi: 10.1073/pnas.89.21.10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Carcinogenesis: a balance between beta-catenin and APC. Curr Biol. 1997;7:R443–R446. doi: 10.1016/s0960-9822(06)00214-4. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Matsuda S, Machida K, Yamamoto T, Fukuda Y, Nimura Y, Hayakawa T, Hamaguchi M. Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancr Res. 2001;61:2361–2364. [PubMed] [Google Scholar]
- Ju H, Zou R, Venema VJ, Venema RC. Direct interaction of endothelial nitric-oxide synthase and caveolin-1 inhibits synthase activity. J Biol Chem. 1997;272:18522–18525. doi: 10.1074/jbc.272.30.18522. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Baltimore D, Lisanti MP. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA. 1995;92:1381–1385. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Kurzchalia T, Dupree P, Parton RG, Kellner R, Virta H, Lehnert M, Simons K. VIP 21, A 21-kD membrane protein is an integral component of trans-Golgi-network-derived transport vesicles. J Cell Biol. 1992;118:1003–1014. doi: 10.1083/jcb.118.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Reimer CL, Oh P, Campbel l DB, Schnitzer JE. Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene. 1998;16:1391–1397. doi: 10.1038/sj.onc.1201661. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Li S, Couet J, Lisanti MP. Src tyrosine kinases, G alpha subunits and H-Ras share a common membrane-anchored scaffolding protein, caveolin. Caveolin binding negatively regulates the auto-activation of Src tyrosine kinases. J Biol Chem. 1996a;271:29182–29190. doi: 10.1074/jbc.271.46.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Okamoto T, Chun M, Sargiacomo M, Casanova JE, Hansen SH, Nishimoto I, Lisanti MP. Evidence for a regulated interaction of hetero-trimeric G proteins with caveolin. J Biol Chem. 1995;270:15693–15701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- Li S, Song KS, Lisanti MP. Expression and characterization of recombinant caveolin: purification by poly-histidine tagging and cholesterol-dependent incorporation into defined lipid membranes. J Biol Chem. 1996b;271:568–573. [PubMed] [Google Scholar]
- Lisanti MP, Scherer P, Tang Z-L, Sargiacomo M. Caveolae, caveolin and caveolin-rich membrane domains: a signaling hypothesis. Trends Cell Biol. 1994;4:231–235. doi: 10.1016/0962-8924(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC [see comments] Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Murata M, Peranen J, Schreiner R, Weiland F, Kurzchalia T, Simons K. VIP21/caveolin is a cholesterol-binding protein. Proc Natl Acad Sci USA. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Schnier JB, Bradbury EM. The accumulation of cyclin-dependent kinase inhibitor p27kip1 is a primary response to staurosporine and independent of G1 cell cycle arrest. Exp Cell Res. 1998;243:222–231. doi: 10.1006/excr.1998.4166. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, A family of scaffolding proteins for organizing “pre-assembled signaling complexes” at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- Parton RG. Caveolae and caveolins. Curr Opin Cell Biol. 1996;8:542–548. doi: 10.1016/s0955-0674(96)80033-0. [DOI] [PubMed] [Google Scholar]
- Peifer M. Beta-catenin as oncogene: the smoking gun [comment] Science. 1997;275:1752–1753. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216 [see comments] Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta. 1997;1332:F127–F147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- Razani B, Altschuler Y, Zhu L, Pestell RG, Mostov KE, Lisanti MP. Caveolin-1 expression is down-regulated in cells transformed by the human papilloma virus (HPV), in a p53-dependent manner: replacement of caveolin-1 expression suppresses HPV-mediated cell transformation. Biochemistry. 2000a;39:13916–13924. doi: 10.1021/bi001489b. [DOI] [PubMed] [Google Scholar]
- Razani B, Rubin CS, Lisanti MP. Regulation of cAMP-mediated signal transduction via interaction of caveolins with the catalytic subunit of protein kinase A. J Biol Chem. 1999;274:26353–26360. doi: 10.1074/jbc.274.37.26353. [DOI] [PubMed] [Google Scholar]
- Razani B, Schlegel A, Lisanti MP. Caveolin proteins in signaling, oncogenic transformation and muscular dystrophy. J Cell Sci. 2000b;113:2103–2109. doi: 10.1242/jcs.113.12.2103. [DOI] [PubMed] [Google Scholar]
- Rothberg KG, Heuser JE, Donzell WC, Ying Y, Glenney JR, Anderson RGW. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines [see comments] Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- Sager R, Sheng S, Anisowicz A, Sotiropoulou G, Zou Z, Stenman G, Swisshelm K, Chen Z, Hendrix MJC, Pemberton P, Rafidi K, Ryan K. RNA genetics of breast cancer: maspin as a paradigm. Cold Spring Harbor Symp Quant Biol. 1994;LIX:537–546. doi: 10.1101/sqb.1994.059.01.060. [DOI] [PubMed] [Google Scholar]
- Sargiacomo M, Scherer PE, Tang Z-L, Kubler E, Song KS, Sanders MC, Lisanti MP. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc Natl Acad Sci USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer PE, Lewis RY, Volonte D, Engelman JA, Galbiati F, Couet J, Kohtz DS, van Donselaar E, Peters P, Lisanti MP. Cell-type and tissue-specific expression of caveolin-2. Caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. J Biol Chem. 1997;272:29337–29346. doi: 10.1074/jbc.272.46.29337. [DOI] [PubMed] [Google Scholar]
- Scherer PE, Lisanti MP, Baldini G, Sargiacomo M, Corley-Mastick C, Lodish HF. Induction of caveolin during adipogenesis and association of GLUT4 with caveolin-rich vesicles. J Cell Biol. 1994;127:1233–1243. doi: 10.1083/jcb.127.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer PE, Okamoto T, Chun M, Nishimoto I, Lodish HF, Lisanti MP. Identification, sequence and expression of caveolin-2 defines a caveolin gene family. Proc Natl Acad Sci USA. 1996;93:131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A, Lisanti MP. A molecular dissection of caveolin-1 membrane attachment and oligomerization. Two separate regions of the caveolin-1 C-terminal domain mediate membrane binding and oligomer/oligomer interactions in vivo. J Biol Chem. 2000;275:21605–21617. doi: 10.1074/jbc.M002558200. [DOI] [PubMed] [Google Scholar]
- Shaul PW, Smart EJ, Robinson LJ, German Z, Yuhanna IS, Ying Y, Anderson RGW, Michel T. Acylation targets endothelial nitric-oxide synthase to plasmalemmal caveolae. J Biol Chem. 1996;271:6518–6522. doi: 10.1074/jbc.271.11.6518. [DOI] [PubMed] [Google Scholar]
- Simionescu N, Simionsecu M. The cardiovascular system. In: Weiss L, editor. Histology: Cell and Tissue Biology. New York: Elsevier Biomedical; 1983. pp. 371–433. [Google Scholar]
- Sionov RV, Haupt Y. Apoptosis by p53: mechanisms, regulation, and clinical implications. Springer Semin Immunopathol. 1998;19:345–362. doi: 10.1007/BF00787230. [DOI] [PubMed] [Google Scholar]
- Smart EJ, Graf GA, McNiven MA, Sessa WC, Engelman JA, Scherer PE, Okamoto T, Lisanti MP. Caveolins, liquid-ordered domains, and signal transduction. Mol Cell Biol. 1999;19:7289–7304. doi: 10.1128/mcb.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KS, Li S, Okamoto T, Quilliam L, Sargiacomo M, Lisanti MP. Copurification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent free purification of caveolae membranes. J Biol Chem. 1996a;271:9690–9697. doi: 10.1074/jbc.271.16.9690. [DOI] [PubMed] [Google Scholar]
- Song KS, Scherer PE, Tang Z-L, Okamoto T, Li S, Chafel M, Chu C, Kohtz DS, Lisanti MP. Expression of caveolin-3 in skeletal, cardiac, and smooth muscle cells. Caveolin-3 is a component of the sarcolemma and co-fractionates with dystrophin and dystrophin-associated glycoproteins. J Biol Chem. 1996b;271:15160–15165. doi: 10.1074/jbc.271.25.15160. [DOI] [PubMed] [Google Scholar]
- Tang Z-L, Scherer PE, Okamoto T, Song K, Chu C, Kohtz DS, Nishimoto I, Lodish HF, Lisanti MP. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J Biol Chem. 1996;271:2255–2261. doi: 10.1074/jbc.271.4.2255. [DOI] [PubMed] [Google Scholar]
- Volonte' D, Galbiati F, Lisanti MP. Visualization of caveolin-1, a caveolar marker protein, in living cells using green fluorescent protein (GFP) chimeras: the subcellular distribution of caveolin-1 is modulated by cell-cell contact. FEBS Lett. 1999;445:431–439. doi: 10.1016/s0014-5793(99)00164-7. [DOI] [PubMed] [Google Scholar]
- Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA [see comments] Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- Wang XW, Zhan Q, Coursen JD, Khan MA, Kontny HU, Yu L, Hollander MC, O'Connor PM, Fornace AJ, Jr, Harris CC. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zundel W, Swiersz LM, Giaccia A. Caveolin 1-mediated regulation of receptor tyrosine kinase-associated phosphatidylinositol 3-kinase activity by ceramide. Mol Cell Biol. 2000;20:1507–1514. doi: 10.1128/mcb.20.5.1507-1514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]