Abstract
Salinity is a major constraint for intrinsically salt sensitive grain legume chickpea. Chickpea exhibits large genetic variation amongst cultivars, which show better yields in saline conditions but still need to be improved further for sustainable crop production. Based on previous multi-location physiological screening, JG 11 (salt tolerant) and ICCV 2 (salt sensitive) were subjected to salt stress to evaluate their physiological and transcriptional responses. A total of ~480 million RNA-Seq reads were sequenced from root tissues which resulted in identification of 3,053 differentially expressed genes (DEGs) in response to salt stress. Reproductive stage shows high number of DEGs suggesting major transcriptional reorganization in response to salt to enable tolerance. Importantly, cationic peroxidase, Aspartic ase, NRT1/PTR, phosphatidylinositol phosphate kinase, DREB1E and ERF genes were significantly up-regulated in tolerant genotype. In addition, we identified a suite of important genes involved in cell wall modification and root morphogenesis such as dirigent proteins, expansin and casparian strip membrane proteins that could potentially confer salt tolerance. Further, phytohormonal cross-talk between ERF and PIN-FORMED genes which modulate the root growth was observed. The gene set enrichment analysis and functional annotation of these genes suggests they may be utilised as potential candidates for improving chickpea salt tolerance.
Introduction
Chickpea is the second most important grain legume crop grown in the tropics and sub-tropics for its high nutritional value and natural ability to fix atmospheric nitrogen1. Over the past 30 years, chickpea production has increased from 6.4 to 13.1 million metric tons (FAOSTAT, 2013). Major producers are India, Australia, Turkey and Myanmar who collectively contribute more than 75% of the global production. From 2009 to 2013, the global area under chickpea increased from 11.5 to 13.5 million hectares and production increased by 25% (from 10.4 to 13.1 million metric tons) (FAOSTAT, 2013). Given its high nutritional content, market value, adaptability and nitrogen fixation ability, chickpea is being increasingly recognised as a global staple food crop of the future.
Salinity is a major problem for crop production especially in the arid and semi-arid regions where chickpea is mainly cultivated2,3. Chickpea is challenged by a number of abiotic stresses but salinity is a major constraint that alone accounts for 8–10% of crop yield losses worldwide4,5. Chickpea has a narrow genetic base, however, considerable genetic variability has been observed amongst the germplasm in response to salt stress5–8.
In order to harness this genetic variation for crop improvement, it is crucial to unravel the molecular mechanisms and potential candidate genes that would allow overcoming its phenotypic plasticity.
Salt stress causes accumulation of Na+ and Cl− ions in various plant tissues and impairs the plant growth especially during the reproductive stage9 Khan et al.5,10. Salinity disturbs the cellular ionic and osmotic environment within the plant. This severely affects important processes like photosynthesis and other important metabolic processes leading to retarded plant growth and poor reproductive success11,12. Based on a number of physiological studies in chickpea, researchers have proposed accumulation of ions in various tissues as detrimental to crop productivity13,14 Pushpavalli et al.1). However, there is little knowledge on the effect of salinity on various developmental stages in chickpea. Further, salt tolerance mechanisms such as ion-exclusion, vacuolar compartmentalization and detoxification that are reported in other crops have not been well studied in chickpea15–17.
Several studies have reported that salt stress induces complex regulatory mechanisms through major transcriptional reorganisation18–21. To understand this, many candidate genes have been identified in response to salt stress through evolving high-throughput approaches like microarrays, SuperSAGE, and deep sequencing22,23,21. Considerable efforts were made to map the tolerance trait and identify the genes underlying the tightly linked DNA markers in JG 11 × ICCV 2 population1. However, full gene information could not be retrieved due to lack of chickpea reference genome at the time. In recent studies, salt responsive genes including cell wall biogenesis, heat shock proteins and transcription factors were identified to be differentially regulated between contrasting genotypes at various developmental stages21,18. Despite considerable effort, the cis-acting genes and their gene networks that actually regulate this multigenic trait could not be intuitively elucidated. In addition, important genes involved in morphogenesis and organ modulation resulting in salt stress avoidance or tolerance could not be underpinned in chickpea. Meanwhile, there has been considerable progress on improving salt tolerance in crops like wheat, rice, and soybean where identification and expression of potential candidate genes such as transmembrane ion-transporters led to improved tolerance. However such knowledge is lacking in chickpea. Therefore, the aim of this study was to identify the comprehensive set of genes involved in modulation of salt stress tolerance in chickpea at various developmental stages.
Results and Discussion
Phenotypic Response to Salt Stress
Phenotypic Characteristics at the Vegetative Stage
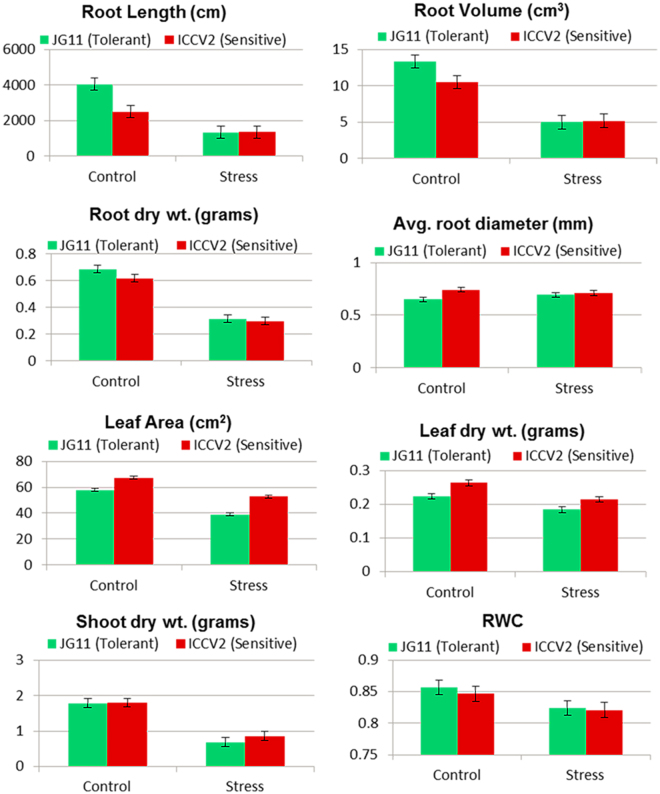
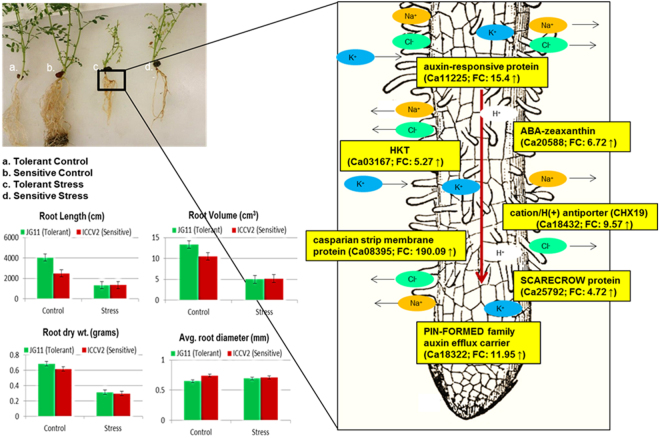
The root length of tolerant genotype was 50% higher as compared to the sensitive genotype in the control condition, however it reduced drastically by 70% in both the genotypes under salt stress (Fig. 1). Similarly, the root volume was 24% higher in the tolerant genotype in the control condition while it reduced by 70% in both genotypes under salt stress. Importantly, the tolerant genotype showed an increase in average root diameter as compared to the sensitive genotype. These results are in agreement to previous studies that reported agravitropic growth of roots in response to salt stress, i.e., growth in diameter rather than length24. The root dry weight, shoot dry weight and surface leaf area were almost same in both the genotypes in control condition while it reduced by 50% in both the genotypes in the stress condition.
Figure 1.
Physiological screening of chickpea genotypes in response to salt stress. Important parameters like root length, root biomass, root diameter, leaf area and shoot biomass were measured at the vegetative stage.
Phenotypic Characteristics at the Reproductive Stage
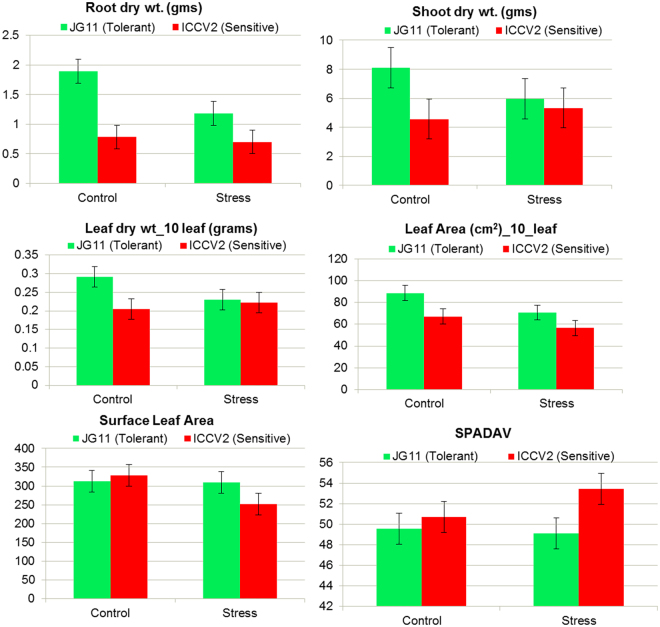
At reproductive stage, the root dry weight decreased by almost 50% in the tolerant genotype during stress while it was constant in the sensitive genotype (Fig. 2). This may suggest that ion accumulation in the sensitive genotype added to the weight in stressed condition while the tolerant genotype show decrease in root biomass through ion-exclusion. Interestingly, the shoot biomass also decreased by 25% in the tolerant genotype while it increased by 20% in the sensitive genotype which again could be due to higher absorption and accumulation of salt ions during the stress. In addition, leaf dry weight also decreased in the tolerant genotype while it increased in the sensitive genotype during the salt stress condition.
Figure 2.
Physiological screening of chickpea genotypes in response to salt stress. Important parameters like root length, root biomass, root diameter, leaf area and shoot biomass were measured at the reproductive stage.
RNA-sequencing, mapping and Gene Set Enrichment Analysis (GSEA)
Reference genome guided assembly of RNA reads
The root transcriptome profiles of JG 11 (salt tolerant genotype) and ICCV 2 (salt sensitive genotype) at the vegetative and reproductive stages were studied by RNA-sequencing. More than 480 million reads were generated from the three biological replicates of the two genotypes (2 genotypes × 2 time-points × 2 conditions × 3 biological replicates) with a library size of ~20 million single-end reads per sample. Approximately 92% reads passed the quality trimming and 72% clean reads mapped to the improved CDC frontier kabuli v2.6.3 reference genome (http://doi.org/10.7946/P2G596) using tophat2 (tophat-gcc/2.0.13). The accepted hits from this mapped assembly were used to identify the number of counts per gene in response to salt stress at the vegetative and reproductive stages of the tolerant and the sensitive genotypes. The reference-guided assembly produced 47,031 transcripts representing 35,855 gene loci which are 7% more than the reference annotation (Edwards et al., 2016). In addition to this, 3,912 potentially novel fragments which have at least one splice junction shared with a reference transcript were found. The fragments mapped against the reference to Exons (99.3%), Introns (0.66%) and Exon-Exon (0%) regions by their types. A gene was considered to be expressed if normalised counts of reads mapping to that gene(s) was greater than one. Using this cut-off, 21,622 gene loci were identified as expressed in at least one condition in response to salt stress. Out of the 33,351 total genes annotated in reference genome, 21,518 (64%) in ICCV 2 C, 21,250 (63%) in JG 11 C, 20,865 (62%) in ICCV 2 LC and 21,622 (64%) in JG 11 LC were considered as expressed for further differential gene expression analysis. Based on the different number of counts in a particular stress condition and developmental stage, these gene(s) were assigned as differentially expressed. To check if the gene expression difference is due to salt treatment, we established a hierarchical clustering amongst the three biological replicates of the control and stress conditions at the reproductive stage. The r-log transformation values of gene counts clustered the three biological replicates of each condition in one group. This also shows that the diversity amongst the samples is due to different gene expressions in response to salt stress and not due to slight differences in the experimental setup.
Identification of Differentially Expressed Genes (DEGs)
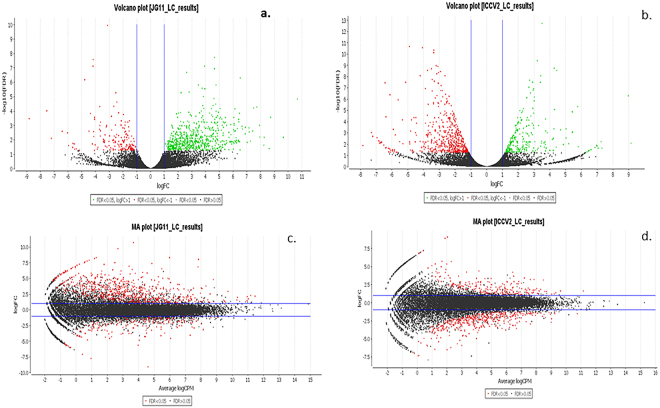
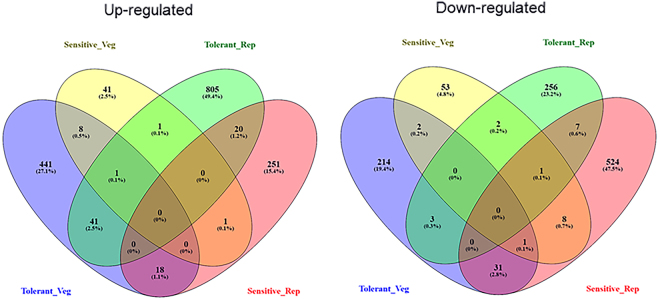
The gene-counts for the RNA-Seq libraries from the three biological replicates of the tolerant and the sensitive genotypes at the vegetative and reproductive stages were extracted using HTSeq counts. The log2 fold-change of gene-counts was calculated for the stress against control for the salt-tolerant (JG 11) and salt-sensitive (ICCV 2) genotypes and compared at the two developmental stages. EdgeR (GLM-likelihood ratio test) was performed using Blast2GO PRO software to identify the potential differentially expressed genes (fold change > 1 and FDR < 0.05) (Fig. 3). A total of 3,053 differentially expressed genes (DEGs) were identified amongst the two contrasting genotypes at the two developmental stages. Interestingly, the number of DEGs varied from 1,309 in the tolerant genotype at reproductive stage to 863 in the sensitive genotype at reproductive stage. At vegetative stage, the number of DEGs varied from 761 in the tolerant genotype to 120 in the sensitive genotype (Fig. 4). At the vegetative stage, the salt-tolerant genotype had more up-regulated genes (510) and less down-regulated genes (251) whereas the salt-sensitive genotype had less up-regulated genes (53) and more down-regulated genes (67). During the reproductive stage, the salt-tolerant genotype had more up-regulated genes (1000) and less down-regulated genes (309) whereas the salt-sensitive genotype had less up-regulated genes (291) and more down-regulated genes (572). In both the cases, the salt tolerant genotype possessed more number of total DEGs as compared to the salt-sensitive genotype. Both the genotypes show a large number of differentially expressed genes at the reproductive stage, suggesting a major transcriptional re-programming in response to salt stress at this important developmental stage. On comparing the commonalities amongst DEGs expressed in the salt-sensitive and salt-tolerant genotypes, 588 genes in JG 11-Veg, 74 genes in ICCV 2-Veg, 872 in genes JG 11-Rep and 612 genes in ICCV 2-Rep were exclusively expressed in the individual genotype/developmental condition (Fig. 5). In comparison, 144 DEGs were commonly expressed amongst ‘ICCV 2-Rep vs. JG 11-Rep’, 11 common DEGs in ‘ICCV 2-Veg vs. ICCV 2-Rep’, 17 common DEGs in ‘ICCV 2-Veg vs. JG 11-Rep’, and 57 common DEGs in ‘ICCV 2-Rep vs. JG 11-Veg’.
Figure 3.
Volcano plot showing the significantly expressed differential genes in response to salt stress.
Figure 4.
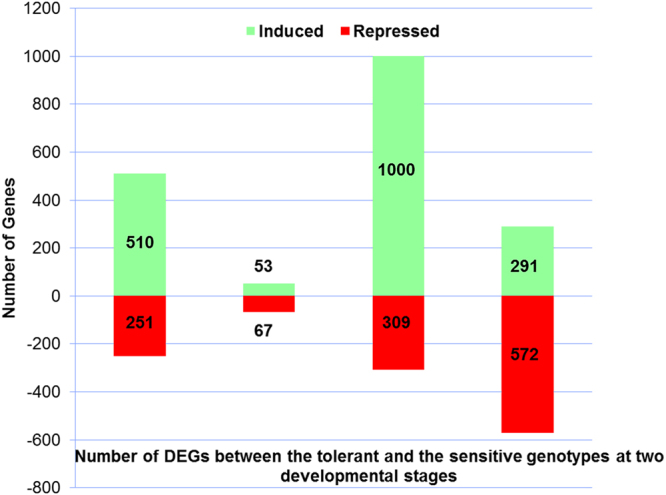
Significant Differentially Expressed Genes (DEGs) between the two genotypes at two developmental stages in response to the salt stress. More number of genes was up-regulated in the tolerant while more number of genes was down-regulated in the sensitive genotype.
Figure 5.
Venn diagram showing the commonly up- and down-regulated differentially expressed genes amongst the genotypes at the two developmental stages in response to the salt stress.
Mapping the functions of genes to GO categories and KEGG database
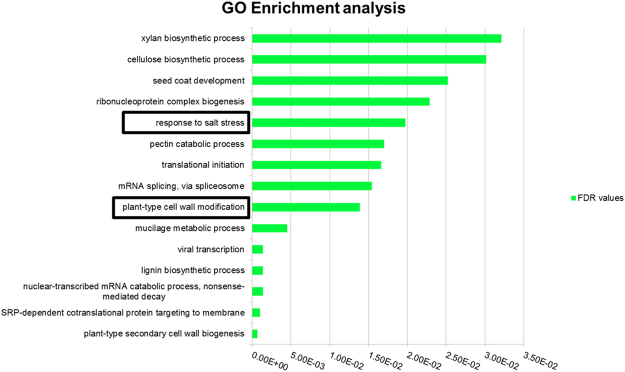
The FASTA sequences of DEGs were BLAST searched against nr-databases to functionally annotate and map sequences to gene ontology GO terms using Bast2GO. From the total DEGs identified, only 76% (1,937) could be significantly annotated and mapped to molecular, cellular and biological GO categories. Subsequently, Gene Set Enrichment Test (GSEA) was performed for the induced/repressed genes using Fisher’s test (FDR < 0.05) to find the over-represented and under-represented categories (Fig. 6). Amongst these, biological processes like response to salt (GO: 0009651), plant-type secondary cell wall biogenesis (GO: 0009834), SRP-dependent co-translational protein targeting to membrane (GO: 0006614), mRNA splicing via spliceosome (GO: 0000398) and cellulose biosynthetic process (GO: 0030244) were significantly represented (Table 1). The KEGG pathway map analysis showed differentially expressed genes representing mainly glycerophosholipid metabolism, phenylpropanoid biosynthesis, inositol metabolism and starch metabolism.
Figure 6.
Gene set enrichment analysis shows enriched GO categories based on differentially expressed genes between tolerant and sensitive genotypes in response to salt stress.
Table 1.
List of significantly enriched Gene Ontology based on Fisher’s Test (FDR < 0.05) amongst tolerant vs. sensitive genotype in response to salt stress.
| GO-ID | Term | Over/Under |
|---|---|---|
| GO:0009651 | Response to salt stress | OVER |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | OVER |
| GO:0000184 | Nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | OVER |
| GO:0009809 | Lignin biosynthetic process | OVER |
| GO:0019083 | Viral transcription | OVER |
| GO:0010191 | Mucilage metabolic process | OVER |
| GO:0009827 | Plant-type cell wall modification | UNDER |
| GO:0000398 | mRNA splicing, via spliceosome | OVER |
| GO:0006413 | Translational initiation | OVER |
| GO:0045490 | Pectin catabolic process | UNDER |
| GO:0022613 | Ribonucleoprotein complex biogenesis | OVER |
| GO:0010214 | Seed coat development | UNDER |
| GO:0030244 | Cellulose biosynthetic process | UNDER |
| GO:0045492 | Xylan biosynthetic process | UNDER |
| GO:0016072 | rRNA metabolic process | OVER |
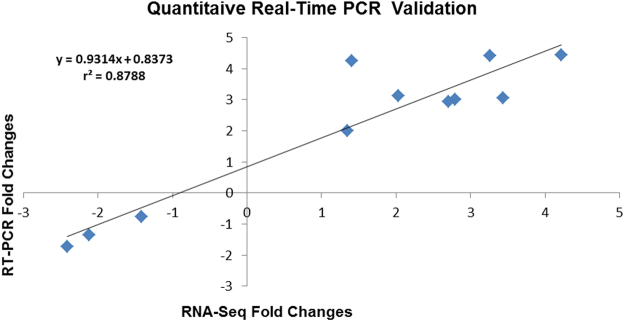
Validation of RNA-Seq results with quantitative Real-Time PCR
The reliability of RNA-Seq gene expression was conformed with 10 salt responsive genes including cation exchanger, blue copper, glutaredoxin, ascorbate oxidase and chloride-channel protein. Fold changes obtained from RT-PCR results correlated well with fold changes of cation exchanger, blue copper and glutaredoxin, calcium-transporting ATPase and ascorbate oxidase which confirms the validity of RNA-Seq data (Fig. 7).
Figure 7.
Quantitative Real-Time validation with 10 salt responsive genes with r2 > 0.87 as the significant threshold.
Transcriptomic responses to salt stress
Biological significance of DEGs in response to salt
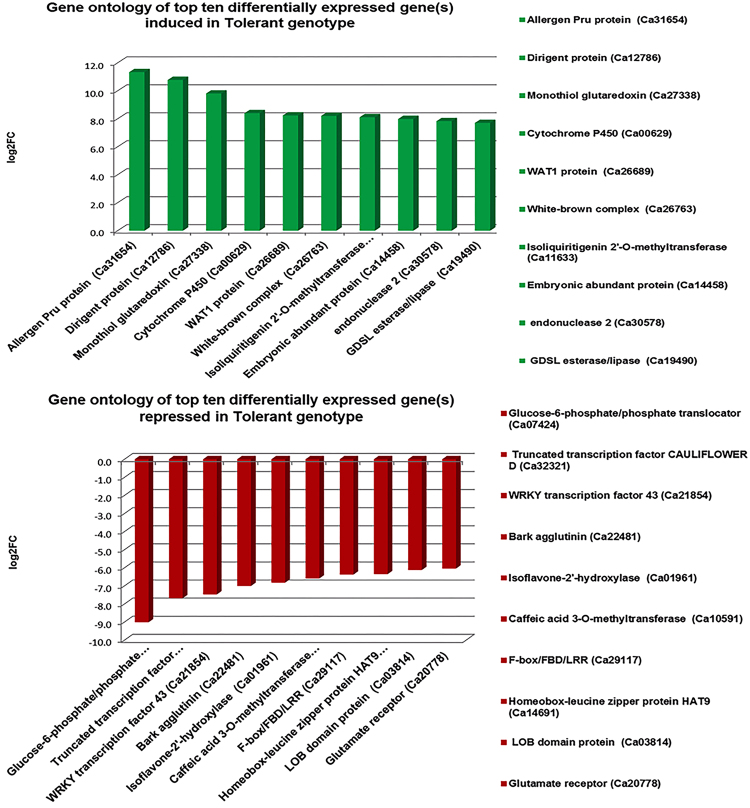
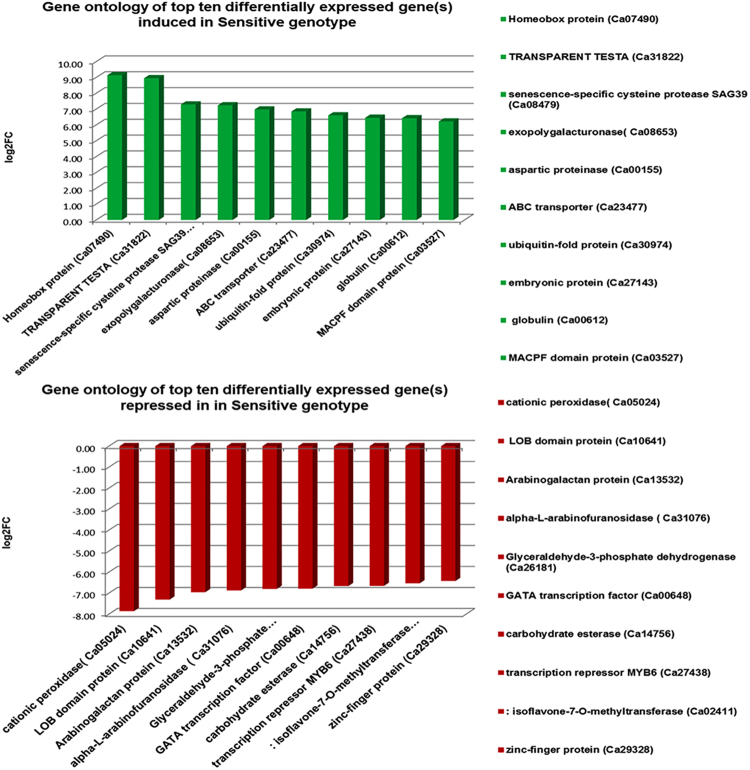
The genes that were significantly differentially expressed between the two genotypes at vegetative and reproductive stages were mainly associated with cell wall organisation, transmembrane transport, signal transduction, oxidative stress, transcriptional regulation, phytohormone signalling and root development (Fig. 8). In particular, majority of abiotic stress-related genes were significantly up-regulated in the tolerant genotype while down-regulated in the sensitive genotype under salt stress. These include lipid transfer proteins, NAC factors, sugar porters, cation/H+ exchangers, peroxidases, casparian strip membrane protein, root meristem development gene, cytochrome P450, Purple acid phosphatase, AP2-ERF factors, LEA proteins and auxin efflux carrier proteins. On the contrary, other genes such as cysteine-rich repeat secretory protein, pectin esterase, bidirectional sugar transporter, senescence-specific cysteine protease, abscisic acid hydroxylase and glutamate receptors were down-regulated in the tolerant genotype but up-regulated in the sensitive genotype (Fig. 9).
Figure 8.
Top ten differentially expressed genes in the tolerant genotype in response to salt stress. The most important genes like dirigent proteins are up-regulated in the tolerant genotype while glucose-6-phospahte genes were highly down-regulated in response to the salt stress.
Figure 9.
Top ten differentially expressed genes in the sensitive genotype in response to the salt stress. The most important genes like cationic peroxidase were highly down-regulated in response to the salt stress.
DEGs Involved In Sensory Mechanism for Cell Detoxification
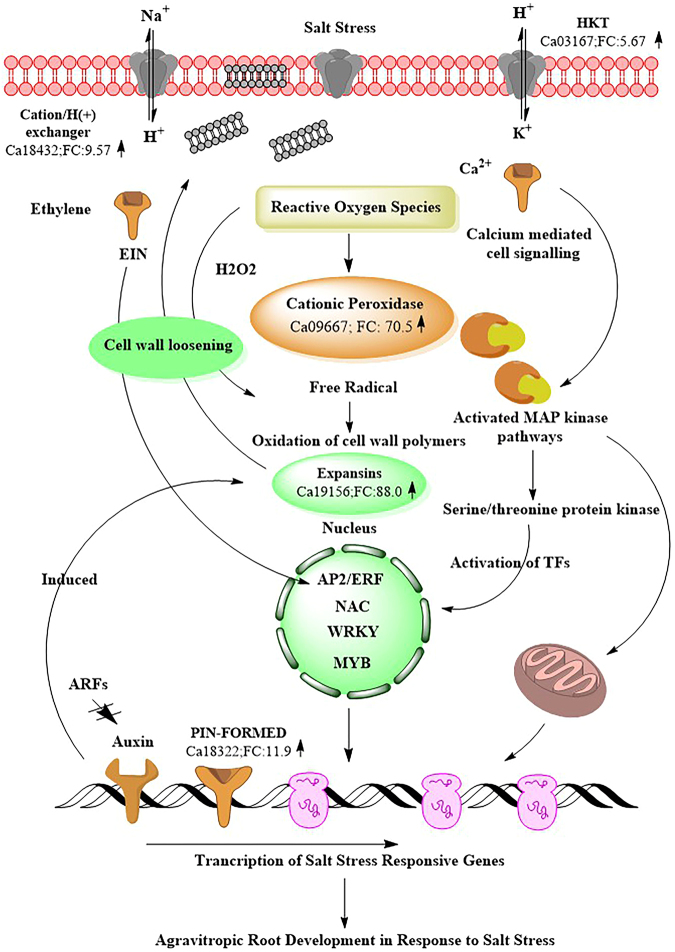
Plants are sessile organisms and have developed various sensory mechanisms to respond to abiotic stresses25. The first sensory mechanism when stress is perceived by the cell is an outburst of reactive oxygen species (ROS). These ROS instigate the H2O2 mediated detoxification and calcium-mediated activation of stress signalling pathways26. Further, the ROS activate the plasma membrane permeable calcium ion channels such as calcium-transporting ATPase which catalyse the Ca2+ induced cell signalling during salt stress27,28. Interestingly, calcium-transporting ATPase gene was highly induced in the tolerant genotype (Ca04200; FC: 18.50 ↑) while repressed in the sensitive genotype (Ca04200; FC: −3.89 ↓), which suggests that an efflux of Ca2+ is important for signalling sensory pathways (Fig. 10). Another calcium binding gene was significantly induced in the tolerant genotype (Ca07521, FC: 9.84 ↑) while repressed in the sensitive genotype (Ca07521, FC: −3.29 ↓).
Figure 10.
A schematic cell signalling cascade based on differential gene expression pattern in roots in response to the salt stress.
The calcium ions in turn activate the protein kinase genes such as mitogen activated protein kinases (MAPK) and calcium dependent protein kinases (CDPK) which help in signal relay mechanism (Wang et al.25). The MAP kinase gene (Ca01482; FC: 5.46 ↑) expression, followed by serine/threonine protein kinases (Ca09102; FC: 16.5 ↑) were induced in the tolerant genotype while repressed in the sensitive genotype (Ca09102; FC: −2.69 ↓). Importantly, these protein kinases cause the phosphorylation/dephosphorylation of signal cascade proteins and activate various transcription factors which eventually regulate salt stress responsive gene expression29.
Upon successful signalling, hydrogen peroxide has to be disintegrated. This is mainly achieved through the activation of peroxidases. Peroxidases play a diverse role in plant physiology and defence response and apart from detoxification of H2O2, they are involved in lignin biosynthesis30. Several antioxidant genes such as cationic peroxidases were significantly induced in the tolerant genotype (Ca09667; FC: 70.5 ↑) while being highly repressed in the sensitive genotype (Ca05024; FC: −232.32 ↓) in response to salt stress (Table 2). Importantly, lignin-forming anionic peroxidase that detoxifies H2O2 and has a putative role in combating environmental oxidative stress was significantly up-regulated in the tolerant genotype (Ca02966: FC: 14.12 ↑).
Table 2.
List of important differentially expressed genes in tolerant and sensitive genotypes in response to salt stress.
| Gene ID | Gene Name | Tolerant genotype (Fold change) | Sensitive genotype (Fold change) |
|---|---|---|---|
| Ca19118 | Acid beta-fructo furanosidase | 150.59 | −21.97 |
| Ca31032 | Pectate lyase Medicago truncatula | 56.60 | −12.91 |
| Ca05594 | 2-oxoglutarate dependent dioxygenase AOP1 | 43.47 | −7.83 |
| Ca17883 | Pectinesterase/pectinesterase inhibitor | 40.57 | −18.81 |
| Ca15441 | G2/mitotic specific cyclin | 38.22 | −9.01 |
| Ca07832 | Polygalacturonase | 36.52 | −6.23 |
| Ca05687 | DNA damage repair/toleration protein DRT | 29.45 | −4.69 |
| Ca27317 | Rho-GTPase | 28.98 | −63.27 |
| Ca15305 | Salicylate carboxymethyltransferase | 27.82 | −74.14 |
| Ca04516 | Non-specific lipid transfer protein | 23.02 | −3.40 |
| Ca11089 | Kinesin | 22.67 | −5.22 |
| Ca05419 | Leucine rich repeat receptor protein kinase | 21.57 | −6.81 |
| Ca05132 | Peroxidase | 20.54 | −11.21 |
| Ca15364 | Chaperone protein dnaJ | 20.45 | −5.32 |
| Ca31146 | Protein trichome birefringence | 19.11 | −15.57 |
| Ca28965 | Receptor protein kinase | 17.67 | −9.51 |
| Ca08233 | Basic leucine zipper | 17.56 | −25.54 |
| Ca22188 | Terpene synthase | 16.17 | −12.85 |
| Ca13532 | Arabinogalactan peptide | 15.97 | −123.97 |
| Ca05547 | Phospholipase D | 14.44 | −4.64 |
| Ca11315 | Endoglucanase | 13.39 | −5.21 |
| Ca18322 | Auxin efflux carrier | 12.78 | −7.22 |
| Ca27258 | ABC transporter | 11.53 | −14.25 |
| Ca06284 | Glucanase | 11.18 | −2.83 |
| Ca05458 | Auxin induced protein IAA6 | 11.02 | −9.41 |
| Ca12822 | Pectate lyase | 10.46 | −6.11 |
| Ca04109 | Condensin | 10.25 | −8.20 |
| Ca17712 | ASPARTIC PROTEASE IN GUARD CELL | 9.93 | −6.34 |
| Ca26863 | Gibberellin regulated family protein | 9.74 | −8.39 |
| Ca29191 | Alpha-glucosidase | 9.70 | −9.21 |
| Ca19842 | Histone H4 | 9.67 | −6.77 |
| Ca21266 | Glucan endo 1,3-betaglucosidase | 8.64 | −5.74 |
| Ca18622 | Sugar porter (SP) family MFS transporter | 7.38 | −5.64 |
| Ca20846 | Zinc finger | 7.12 | −4.21 |
| Ca18708 | Proline rich protein | 7.03 | −7.02 |
| Ca13369 | Allene oxide synthase | 6.64 | −9.66 |
| Ca12046 | alpha-L-fucosidase 2 | 6.41 | −18.78 |
| Ca05618 | Cdc2MsD protein | 6.36 | −4.50 |
| Ca31064 | ()isopiperitenol/()carveol dehydrogenase | 6.08 | −6.91 |
| Ca29950 | Protein SRG1 | 6.07 | −11.03 |
| Ca25807 | GDSL lipase/acylhydrolase | 5.98 | −11.36 |
| Ca30964 | Lamin protein | 5.66 | −12.08 |
| Ca27357 | Cellulose synthase | 5.51 | −3.42 |
| Ca12582 | Glucomannan 4- beta-mannosyltransferase | 5.41 | −28.43 |
| Ca14197 | SIEVE ELEMENT OCCLUSION B | 5.40 | −4.59 |
| Ca26452 | Alkaline alphagalactosidase | 5.16 | −2.55 |
| Ca14732 | Betagalactosidase | 4.56 | −13.29 |
| Ca13624 | Histone lysine N-methyltransferase | 4.32 | −9.36 |
| Ca12414 | Cytochrome P450 | 4.22 | −3.65 |
| Ca28514 | Protease inhibitor/seed storage/ltp | 4.12 | −2.78 |
| Ca26510 | ARM REPEAT PROTEIN INTERACTING WITH ABF2 | 4.09 | −7.24 |
| Ca06506 | Cellulose synthase A | 3.51 | −9.82 |
Transcription Factors (TFs)
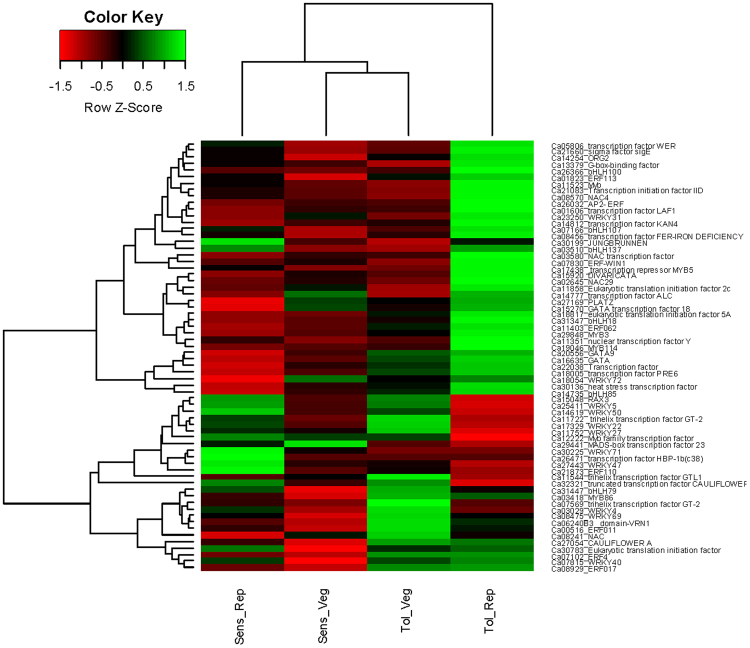
Transcription factors are special proteins that control the transcription of genes, and a lot of them are therefore expressed in a genotype, tissue, and stress-specific manner. A total of 151 TFs were differentially expressed, with 91 up-regulated and 60 down-regulated in response to salt stress. The most expressed TF families were NAC, ERF, WRKY, bHLH, MYB, trihelix-factor, HSF and GATA. It is important to note that the NAC, ERF, WRKY, and HSF were highly induced while trihelix factor was repressed in the tolerant genotype suggesting their role in stress response and plant growth processes (Fig. 11).
Figure 11.
Heatmap showing differential expression of different transcription factor families amongst the tolerant and sensitive genotypes at developmental stages in response to salt stress. Both row and columns are clustered based on the gene expression values.
AP2/ERF transcription factors: A suite of ERFs has been reported to be induced in response to high salinity, osmotic stress, and drought31. Interestingly, ethylene-responsive transcription factor (ERF-WIN1) was highly induced in the tolerant genotype (Ca07830; FC: 36.25 ↑) while being repressed in the sensitive genotype (Ca07830; FC: −1.03 ↓). Another important ethylene factor is AP2/ERF which plays a major role in plant development via cell proliferation and hormonal responses during the stress32. An AP2/ERF TF was 10-fold more induced in the tolerant genotype (Ca26032; FC: 11.71 ↑) as compared to the sensitive genotype (Ca26032; FC: 1.10 ↑). The AP2/ERF factors are thought to bind dehydration responsive elements at the promoter of target genes and instigate salt stress response33 (Sakuma et al.33). Interestingly, dehydration responsive element (DRE/CRT) was significantly induced in the tolerant genotype (Ca13645; FC: 15.56 ↑) while being repressed in the sensitive genotypes (Ca19176; FC: −6.86 ↓).
NAC transcription factors: In addition to ethylene factors, NACs are important TFs which have been reported to impart salt tolerance in Arabidopsis thaliana34, Oryza sativa35, and Glycine max36. The NAC TFs are involved in flower development, cell division, shoot apical meristem formation and secondary wall synthesis37. Many NAC factor transcripts were significantly induced in the tolerant genotype (Ca03580; FC: 77.70 ↑) while being repressed in the sensitive genotype (Ca15352; FC: −28.64 ↓). Significant differential expression of NAC factors amongst the two genotypes suggests they may contribute to salt tolerance/sensitivity in chickpea.
MYB transcription factors: MYBs are an important class of TFs which are widely distributed in plants and play a role in cell development, cell cycle and primary/secondary metabolism38. The MYB TFs can regulate both, positive and negative gene expression by silencing the transcription39. This is accomplished by the expression of MYB-repressor genes that control the synthesis of lignin and modulation of secondary cell wall formation in plants during the stress40. The MYB TFs were induced in the tolerant genotype (Ca24358; FC: 19.56 ↑) while being repressed in the sensitive genotype (Ca24358; FC: −6.96 ↓). Similarly, MYB-repressor (MYBH) was highly induced in the tolerant genotype (Ca17438; FC: 65.34 ↑) and repressed in the sensitive genotype (Ca27438; FC: −6.65 ↓). The effect of both these genes is essential to maintain auxin homoeostasis and their up-regulation in the tolerant genotype suggests major role in cell wall modification in order to protect the cell from osmotic pressure exerted by the salt stress41.
WRKY transcription factors: Another most important family of TFs extensively distributed in plant genome is WRKY transcription factors. They comprise of several diverse WRKY members which regulate the gene response to abiotic stress, root development, leaf senescence, seed germination and phytohormone signalling25,42,43. Different WRKY factors are highly tissue specific and reported to be critical for salt stress tolerance44,45. The WRKY72 factor (Ca18054; FC: 26.90 ↑) was highly induced while WRKY73 was highly repressed in the tolerant genotype (Ca21854; FC: −177.29 ↓). On the contrary, WRKY50 factor was induced in the sensitive genotype (Ca14619; FC: 5.09 ↑) while repressed in the tolerant genotype (Ca14619; FC: −1.67 ↓). Importantly, the stress responsive genes regulated by WRKY72 are known to be non-reponsive to salicylic acid while WRKY50 regulates the genes which are responsive to jasmonic acid46,47. This suggests that various WRKY genes have specific role in transcriptional re-programming48 and the two genotypes employ different mechanisms to combat the salt stress.
Heat stress transcription factors (HSF): Other important differentially regulated TFs in response to salt stress are heat stress transcription factor (HSF). They have been suggested to regulate signalling pathways during salinity, oxidative and osmotic stress in seedlings49. Over-expression of HSFs in Arabidopsis was demonstrated to improve salt tolerance50. An HSF gene was highly induced in the tolerant genotype (Ca30136; FC: 124.49 ↑) but significantly repressed in the sensitive genotype (Ca30136; FC: −13.8 ↓). The significant differential expression of this particular HSF needs to be validated further to investigate its role during salt stress in chickpea.
Truncated transcription factor CAULIFLOWER D: The truncated transcription factors are known to prevent early flora and meristem development. A truncated CAULIFLOWER D TF was significantly down-regulated in the tolerant genotype (Ca32321; FC: −206.50 ↓) while being up-regulated in the sensitive genotype (Ca32321; FC: 20.82 ↑). Several physiological studies in chickpea have indicated that salinity delays flowering time (Turner et al.8; Khan et al.5). The down-regulation of this gene in the tolerant genotype suggests that delaying flowering time could be an important mechanism to avoid the effect of salt stress during the reproductive phase to restore pod filling process.
DEGs Involved In Cell Wall Modification
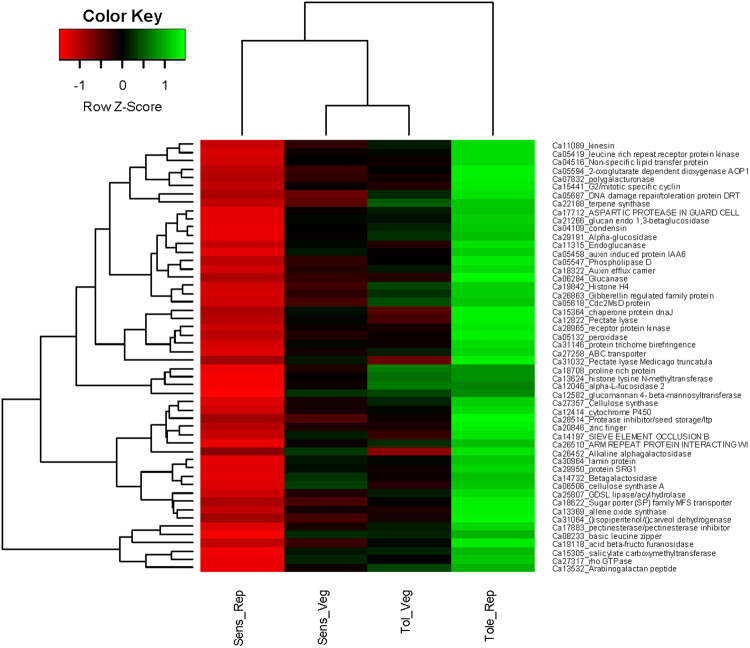
The plant cell wall is the first site to perceive and respond to abiotic stress51. The stress signal triggers cell wall remodelling so as to maintain the flexibility and protect the cell against ionic imbalance52. Major genes involved in cell wall organisation are dirigent protein, phosphatidylinositol phosphate kinase and lipid transfer proteins. The dirigent proteins (DIRs) are glycoproteins with an important role in lignan and lignin biosynthesis and are involved in secondary metabolism53. Interestingly, dirigent protein (Ca12786; FC: 1024 ↑), lipid-transfer protein (Ca19748; FC: 142.02 ↑), phosphatidylinositol phosphate kinase (Ca17709; FC: 77.7 ↑) and pectate lyase (Ca31032; FC: 60.96 ↑) were highly induced in the tolerant genotype but repressed at the same time in the sensitive genotype (Table 2). Among these genes, lipid transfer proteins are most important for stabilisation of membranes, cell wall organisation and signal transduction in response to biotic and abiotic stress. The lipid-transfer protein was highly induced in the tolerant genotype (Ca19748; FC: 142.02 ↑) but repressed in the sensitive genotype (Ca19748; FC: −1.42 ↓) indicating that the sensitive genotype lacks cell membrane stabilization during the adverse effect of ionic stress (Fig. 12).
Figure 12.
Heatmap showing differential expression of important genes amongst the tolerant and sensitive genotypes at developmental stages in response to salt stress. Both row and columns are clustered based on the gene expression values.
In a related observation, cell surface glycoproteins such as arabinogalactan peptide protein (AGPs) were differentially expressed in response to salt stress. These genes are wall-associated proteins and help in cell elongation, cell development and signalling54. Two such genes namely, arabinogalactan peptide protein (Ca13532; FC: 25.10 ↑) and proline-rich protein (Ca17660; FC: 34.77 ↑) which are involved in cell wall re-shaping were highly induced in the tolerant genotype but significantly repressed in the sensitive genotype (Ca13532; FC: −123.6 ↓).
Plant cell wall is composed of different biomolecules including cellulose, pectins and lignin. The cell wall modification under abiotic stress is accomplished by modulation in cross-linking between these polymers. Genes such as expansin and xyloglucan are involved in modulating these cross-links and regulate cell wall metabolism process. Importantly, expansin gene was highly induced in the tolerant genotype (Ca19156, FC: 88.03 ↑) but repressed in the sensitive genotype (Ca19156, FC: −2.44 ↓). Expansin and xyloglucan modifying enzymes have been reported to impart salt and drought tolerance when over-expressed in Capsicum and wheat in response to osmotic stress and salt stress, respectively55. Another important gene which is involved in cell wall metabolism is xyloglucan galactosyltransferase56. The xyloglucan galactosyltransferase was significantly up-regulated in the tolerant genotype (Ca00249, FC: 68.59 ↑) but down-regulated in the sensitive genotype (Ca00249, FC: −25.99 ↓). According to57 knock-out of xylogucan XHTs gene imparted more aluminium tolerance and reduced root elongation in Arabidopsis and they are regarded as important stress marker gene in salt, cold and drought stress58,59. Xyloglucans are linked to at least two cellulose molecules and are reported to remodel stomata cell walls to prevent excess water loss during the osmotic stress56. Cell wall remodelling promotes the plant growth and the up-regulation of expansin and xyloglucan genes in the tolerant genotype clearly suggests their activity in ‘cell wall loosening’ may impart salt stress tolerance in chickpea.
DEGs Involved In Hormonal Crosstalk in Response to Salt
Plants can continuously change their growth and development patterns by the action of plant hormones60. Hormones build up a signalling network and regulate several metabolic and transcriptional processes thus altering the physiological parameters in response to stress61. Differential regulation of classic hormones like auxin and ethylene suggest their major role in combating the stress. Many genes related to classic hormones like ethylene, auxin, gibberellin, salicylate and jasmonate were significantly up-regulated during the salt stress.
Ethylene-related genes: Auxin and ethylene hormones work as antagonists to regulate lateral root growth whereas they synergistically regulate root hair development and root cell elongation62. Recent studies have focussed on ethylene involvement in modulation of cell membrane receptors and transcription factors in salt response63. One such important ethylene precursor aminocyclopropane carboxylate (ACC) oxidase suppresses salt sensitivity in Arabidopsis64,65. Ethylene receptors like aminocyclopropane-1-carboxylate synthase were up-regulated in the tolerant genotype (Ca17276; FC: 29.65 ↑) while being highly down-regulated in the sensitive genotype (Ca17276; FC: −20.96 ↓). This indicates that ethylene-mediated downstream signalling is critically involved in stress response and is an important hormone playing a major role in salt tolerance.
ABA-related genes: It has been well characterised that the salt stress induces ABA-related genes. The accumulation of abscisic acid (ABA) hormone during salt stress helps to close the stomata and help plant acclimatise under low water availability by accumulating proteins which act as osmoprotectants for osmotic adjustment66,67. Interestingly, genes such as zeaxanthin (Ca20588; FC: 6.72 ↑) and ABF2 (Ca26510; FC: 4.22 ↑) were induced in the tolerant genotypes as shown while repressed in the sensitive genotype (Ca20588; FC: −1.74 ↓). Concomitantly, an aspartic protease protein found in guard cells plays important role in osmotic adjustment during stress by signalling increased ABA sensitivity and preventing water loss from the cells68. This gene was significantly up-regulated in the tolerant genotype (Ca17712; FC: 10.41 ↑) while highly repressed in the sensitive genotype (Ca17712; FC: −6.32 ↓) suggesting its important role in osmotic stress regulation. One of the important ABA-responsive protein is late embryonic abundant-like protein which was 82-fold more induced in the tolerant genotype (Ca14458; FC: 252.47 ↑) as compared to the sensitive genotype (Ca14458; FC: 3.05 ↑). This gene is known to prevent protein aggregation thus protecting the plants against high salinity, desiccation and temperature69,70.
Auxin-related genes: Root remodelling and plant development is a major challenge during the salt stress71. Auxins have a major function in root remodelling, tissue development, cell elongation and apical dominance which aid in plant adaptation72. Several studies report auxin as the main hormone responsible for altering the growth developmental pattern and inducing genes for stress tolerance73,74. Several auxin-related genes such as auxin-responsive protein (Ca11225; FC: 15.4 ↑), SAUR-like auxin-responsive family protein (Ca15351; FC: 7.51 ↑) and auxin-induced protein (Ca05458; FC: 10.55 ↑) were induced in the tolerant genotype as shown while repressed in the sensitive genotype (Ca11225; FC: −5.02 ↓). Gradients of auxin are important for root cell differentiation, lateral growth pattering and cell elongation75. This is regulated by number of membrane localised proteins such as PIN-FORMED family of auxin efflux carrier (Ca18322; FC: 11.95 ↑), multidrug resistance P-glycoprotein (Ca30923; FC: 5.81 ↑), and ABC transporters (Ca27258; FC: 11.71 ↑) (Lewis et al., 2007; Wu et al., 2007) that were significantly induced in the tolerant genotype as shown. On the contrary, auxin efflux carrier was repressed in the sensitive genotype (Ca18322; FC: −7.21 ↓). Another important auxin gene, indole-3-acetic acid-amido synthetase (GH3) was up-regulated in the tolerant genotype (Ca08527; FC: 40.22 ↑) while being down-regulated in the sensitive genotype (Ca08527; FC: 1.61 ↓). GH3 gene is thought to regulate expansin gene which loosens the cell wall and induces salicylate- and jasmonate-mediated disease resistance pathways76.
Jasmonic acid-related genes: Jasmonic acid (JA) is reported to impart salt tolerance in many different plants77,78. Jasmonates have important role in developmental and defense mechanisms, root growth inhibition, vegetative storage proteins (VSPs) accumulation, induced systemic resistance (ISR) and tolerance to ozone O379,80. Up-regulation of jasmonic acid-amido synthetase (JAR1) gene in the tolerant genotype (Ca33318; FC: 41.64 ↑) indicates its role in JA-mediated defence response pathways while this gene was not expressed in the sensitive genotype. Plant hormones are crucial endogenous molecules which regulate several plant developmental processes and response to biotic and abiotic stresses63. The differential expression of different hormone-related genes suggests their critical role in shaping chickpea plant morphology that enables it to adapt to salt stress.
DEGs Involved In Root Morphogenesis
Plants employ different sensory mechanisms to deal with high soil salinity and re-shaping the root architecture is one such important mechanism (Galvan-Ampudia and Testerink24). Crosstalk between classic phytohormones like auxin, ethylene, abscisic acid (ABA), salicylic acid (SA) and jasmonic acid (JA) coordinate the root growth and radial pattering81. Patterning in root and lateral root hair formation is important for plant adaptation during the stress82,83. Root endodermis is the membrane which functions in nutrient uptake, diffusion and stress resistance. Several root development genes were induced in the tolerant genotype in reponse to salt stress. One of the important root development gene such as casparian strip membrane protein was highly induced in the tolerant genotype (Ca08395; FC: 190.09 ↑) while being repressed in the sensitive genotype (Ca08395; FC: −3.68 ↓). In addition, important genes like SCARECROW protein (Ca25792; FC: 4.72 ↑), root meristem growth factor (Ca16049; FC: 29.85 ↑), LONGIFOLIA protein (Ca27082; FC: 44.94 ↑) and RADIALIS protein (Ca11227; FC: 4.08 ↑) which lead to formation of root endodermis were highly induced in the tolerant genotype but repressed in the sensitive genotype (Fig. 13). Further, the COBRA gene was highly repressed in the sensitive genotype (Ca27034; FC: −7.46 ↓). The COBRA gene helps in polar root expansion and its down-regulation in the sensitive genotype suggests that this important response was inhibited84. During salt stress, the root grows in agravitropic manner and cells expand radially85. These changes in lateral root development and agravitropism is mediated by auxin redistribution86,87. Importantly, this finding is supported by our physiological data (Figs 1 and 2), where the root volume increased in the tolerant genotype rather than root length. On the other hand, Transparent TESTA gene which has a role in the development of root hair in the epidermal cell layer was highly induced in the sensitive genotype (Ca31822; FC: 494.55 ↑)88. The bulging of root hair cells increases the influx of Ca2+/Na+ ions and up-regulation of this gene in the sensitive genotype could possibly be the reason for enhanced sensitivity to salt stress in chickpea88,89.
Figure 13.
Proposed root development modulation instigated by differentially expressed genes up-regulated in tolerant genotype. Integration of phenomics and transcriptomics data showing the auxin flow controlled by PIN-FORMED genes regulate root agravitropism in response to salt stress.
DEGs Involved In Network of Transmembrane Transporters
The plant’s first response to salinity is either ion-exclusion or sequestration of ions in the vacuoles leading to tissue tolerance9,90. Several ion-regulation pathways are integrally active in plant roots as the absorption of nutrients from soil solution requires constant inclusion/exclusion of ions. Many cation exchanger proteins were up-regulated in the tolerant genotype while down-regulated or not differentially expressed in the sensitive genotype. Cation/H+ antiporters are expressed in endodermis of the root and act as a channel to transport Na+ from endodermis to stele cells of the root eventually loading these ions in the xylem and aerial plant tissues90,91. One important cation/H(+) antiporter (CHX19) involved in ion-exclusion pathway was 7.3-fold more induced in the roots of the tolerant genotype (Ca18432; FC: 9.57 ↑) as compared to the sensitive genotype (Ca18432; FC: 1.37 ↑)92.
Plant High-Affinity Potassium Transporters (HKTs) are important ion channels that were recently demonstrated to selectively allow the Na+ and/or K+ transport and are therefore potential candidates for salt tolerance93,94. Potassium transporters are also known to unload the Na+ ions from xylem sap thus preventing the toxic ions from reaching and accumulating in the leaves95. Potassium channels also regulate import of K+ ions thereby balancing the K+/Na+ ratio in tissues and preventing salt stress96. One potassium transporter was 1.55-fold more induced in the tolerant genotype (Ca03167; FC: 5.27 ↑) as compared to the sensitive genotype (Ca03167; FC: 3.48 ↑) during the reproductive stage. This may prevent toxic Na+ ions from reaching the aerial parts of chickpea thereby ensuring successful seed setting and crop yield.
Another important key mechanism for ion-detoxification is vacuolar sequestration of toxic ions in the vacuole. We found a number of genes associated with vacuolar compartmentalisation of ions were differentially expressed indicating tissue tolerance through ion sequestration is possibly a main mechanism of salt tolerance in chickpea. The NRT1-PTR gene family (NPF) are identified as nitrate or di/tri-peptide transporters and involved in vacuolar compartmentalisation97. It was recently demonstrated to be expressed in root tissues and localised at the vacuolar membrane. An NRT1-PTR family gene was significantly induced in the tolerant genotype (Ca00319; FC: 95.67 ↑) while being repressed in the sensitive genotype (Ca00319; FC: −4.85 ↓). It is also known to transport phytohormones, nitrogen and secondary metabolites97. The ion-channels help to maintain low levels of cytosolic Na+ in the roots but if this mechanism fails, salinity will induce osmotic stress similar to drought88.
Apart from this, transmembrane water channels called aquaporins regulate the transport of fluids and are reported to play a major role during salt induced osmotic stress responses. Importantly, transmembrane major intrinsic protein (MIP) channels were induced in the tolerant genotype (Ca03453; FC: 54.5 ↑) while repressed in the sensitive genotype (Ca03453; FC: −2.73 ↓). They are major class of intrinsic proteins which regulate the transport of ion, water and carbohydrates. In addition to water, plants accumulate soluble osmolytes such as sugars, proline, betaine to help protect the cell against the osmotic stress98,99.
Other important transporters with ATPase activity to allow selective transport across the transmembrane were highly induced in the tolerant genotype. Examples are cyclic nucleotide-gated ion channel (Ca16646; FC: 37.53 ↑), metal transporter Nramp (Ca21750; FC: 5.57 ↑), extracellular ligand-gated ion channel protein (Ca13670; FC: 33.82 ↑), and white-brown complex protein (Ca26763; FC: 294.06 ↑). These were either not expressed or repressed in the sensitive genotype; example, cyclic nucleotide-gated ion channel (Ca16646; FC: −4.14 ↓). These results indicate that the two genotypes employ different approaches to deal with salt stress. Further, there exists strong evidence that ion-exclusion mechanism may confer salt tolerance in chickpea. This would be further validated and utilised to improve salt tolerance in chickpea.
Cellular Ion Homeostasis Through Metallozymes
Metallothioneins are an important class of intracellular metal binding enzymes involved in detoxification of heavy metals and maintaining ion-homeostasis. They often have a high affinity towards heavy metals100,101 and are known to provide tolerance against heavy metal ion toxicity102. Several metallozymes were found to be induced in the tolerant genotype suggesting their major role during the salt stress response. For instance, Type1 metallothionein (MT1) was induced in the tolerant genotype (Ca04758; FC: 4.69 ↑) while the expression of this gene class was not observed in the sensitive genotype. Apart from this, several metal proteins were highly induced only in the tolerant genotype and these include monothiol glutaredoxin (Ca27338; FC: 891.44 ↑), purple acid phosphatase (Ca01925; FC: 23.42 ↑), and metal tolerance protein (Ca08940; FC: 6.32 ↑). On the contrary, metal ion-binding gene (Ca27452; FC: −4.02 ↓) and heavy metal-detoxification gene (Ca29520; FC: −13.08 ↓) were highly repressed in the sensitive genotype. Another important metal enzyme is multi-copper oxidase protein which was induced in the tolerant genotype (Ca15190; FC: 4.37 ↑) while repressed in the sensitive genotype (Ca15190; FC: 3.89 ↓). Multi-copper oxidase gene is thought to oxidise copper and similar ions which could be toxic to cells. Its up-regulation in the tolerant genotype suggests a role in reducing metal toxicity during the salt stress. The functions of these metal enzymes in metal detoxification are well demonstrated in Arabidopsis103. However their role in plant metal detoxification during salt stress still remains to be established. Nevertheless, their differential expression in this study suggests that they play an important role in metal metabolism or detoxification during the salt stress103,104.
Methods
Plant Material
Two chickpea genotypes, desi JG 11 (salt tolerant) and kabuli ICCV 2 (salt sensitive), were subjected to salt stress in the glasshouse1. The physiological experiment was set-up in a Random Complete Block Design (RCBD) comprising three biological replicates of each genotype. The seeds were surface sterilised using 70% ethanol followed by three rinses with MilliQ water. The seeds were sprouted in Petri dishes in dark and moist conditions until radicle emergence. The etiolated seedlings were sown in 10.5 inches diameter pots filled with 9.5 kg of pasteurized sand soil mix. In the treatment pots, two adaptive doses of salt at the rate of 40 mM NaCl were added twice during the life cycle of plant; one at 10 days after sowing and another at the flowering stage. Initial salt treatment included mixing sodium chloride in the soil (1.75 g per kg of soil) before filling the pots6,8. Pots were sealed with sturdy tape to prevent the leakage of salt. To avoid water retention in pots, the soil field capacity was measured gravimetrically and pots were maintained at 80% field capacity throughout the experiment. To monitor the salt concentration, soil electrical conductivity was checked and maintained below ~1 dS/m considering the intrinsic salt sensitivity of chickpea. Control plants were grown normally in 9.5 kg of pasteurized sand soil mix and maintained at 80% field capacity. Plants were routinely watered and root tissues were harvested at the vegetative and reproductive stages and immediately frozen in liquid nitrogen.
RNA Isolation and Library Preparation
Total RNA was isolated using Qiagen RNeasy kit (GmBH, Germany). The frozen root tissue was ground to fine powder and weighed to add calibrated volumes of lysis buffer. Finally, RNA was eluted in 60 µl of RNase-free water. RNA was quantified using Nanodrop (NanoDrop™ Lite Spectrophotometer, Thermo Fisher Scientific) and qualitatively analysed on Bioanalyzer 2100 (Agilent Technologies). Only RNA with RIN values > 7 were chosen for mRNA enrichment.
mRNA Enrichment
From 1 µg of total RNA as starting material, poly(A+) mRNA was isolated using Dynabeads mRNA purification kit (Thermo Fisher Scientific, USA).
RNA-Seq Library Preparation
A total of 24 RNA-Seq libraries (2 genotypes x 2 time-points x 2 conditions x 3 biological replicates) were prepared from 100 ng of poly(A+) mRNA as starting material using Ion total RNA seq kit v2 (Thermo Fisher Scientific, USA). The mRNA was chemically fragmented followed by cDNA synthesis and adapter ligation. The unstranded library molecules were analysed on Bioanalyzer 2100 (Agilent Technologies) for peak size selection and region molarity calculations. Four uniquely barcoded libraries were pooled to an equimolar concentration of 0.8 nM and enriched with ion sphere particles (ISPs) using streptavidin beads and Ion One-Touch (Thermo Fisher Scientific, USA). The charged ISPs were loaded on to the Ion-Chip and ~20 million single-end 100 bp reads were sequenced per library.
RNA-Seq Data Processing And Differential Gene Expression Analysis
The raw fastq sequences were trimmed for adapters and read length using the q-trimming program. All reads less than 30 bp and poor quality bases were discarded. The reference genome was indexed using bowtie2-intel/2.1.0105. The structural annotation file (gff3) was indexed using tophat to generate a known transcript file. Ninety-two per cent clean reads were mapped against the chickpea (CDC Frontier) reference genome using tophat-gcc/2.0.13106. The gene counts were extracted from accepted hit bam files using HTSeq (Anders et al., 2014). The gene counts were used to identify differentially expressed genes using the EdgeR tool (Robinson et al., 2010), from Blast2GO PRO software. A gene was considered to be differentially expressed if log2 fold change was >1 and false discovery rate (FDR) was <0.05 (padj –BH method)107.
Data availability
The raw fastq files and gene count files for all samples including three biological replicates are submitted to the GEO databases under the accession GSE110127.
Conclusions
The availability of chickpea genome has provided a new opportunity to study the candidate genes involved in salt tolerance mechanisms in chickpea. The integration of phenotypic and genomic data in this study has provided a better understanding of salt tolerance mechanism in chickpea. A suite of genes important for salt tolerance were differentially expressed between the two genotypes suggesting that the tolerant genotype has evolved a more robust mechanism of salt tolerance in comparison to the sensitive genotype. Important candidate genes related to salt tolerance such as Cationic peroxidases, Expansins, Lipid transfer proteins, Cation/H + exchangers, Casparian strip membrane proteins, PIN-FORMED and Type1 metallothionein are related to physiological processes including cell wall modification, root growth modulation, ion-exclusion and phytohormone signalling. The pattern of gene expression suggested that salt induced osmotic stress followed by oxidative and ionic stress in the genotypes. However, the tolerant genotype utilised a number of genes to detoxify the effect of salt stress through highly induced actions of the above mentioned candidate genes. In conclusion, both the genotypes possess transcriptomic variation and deploy different molecular mechanisms through either stress-avoidance or stress-tolerance in response to salt stress. The candidate genes should be functionally validated using modern gene-knock out techniques such as CRISPR/Cas9 system before being employed in molecular breeding or genetic engineering for salt tolerance.
Acknowledgements
Authors are also grateful to VLSCI (Melbourne Bioinformatics) for providing some of the bioinformatics tools used for analysis. This work was financially supported by Australia-India Strategic Research Fund (AISRF) Grant number GCF010013, Commonwealth of Australia.
Author Contributions
M.K. conducted experiments and data acquisition. N.M., R.F. planned the research along with collaborators R.K.V., H.K., M.J. M.K. wrote the manuscript closely with N.M. and R.F. All the authors were involved in many discussions and revised the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rajeev Varshney, Email: r.k.varshney@cgiar.org.
Nitin Mantri, Email: nitin.mantri@rmit.edu.au.
References
- 1.Pushpavalli R, et al. Two key genomic regions harbour QTLs for salinity tolerance in ICCV 2 × JG 11 derived chickpea (Cicer arietinum L.) recombinant inbred lines. BMC plant biology. 2015;15:124. doi: 10.1186/s12870-015-0491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang X, Mu X, Shao H, Wang H, Brestic M. Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Critical reviews in biotechnology. 2015;35:425–437. doi: 10.3109/07388551.2014.889080. [DOI] [PubMed] [Google Scholar]
- 3.Kalaji HM, et al. Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta physiologiae plantarum. 2016;38:102. doi: 10.1007/s11738-016-2113-y. [DOI] [Google Scholar]
- 4.Kotula L, et al. Salt sensitivity in chickpea (Cicer arietinum L.): ions in reproductive tissues and yield components in contrasting genotypes. Plant, cell & environment. 2015;38:1565–1577. doi: 10.1111/pce.12506. [DOI] [PubMed] [Google Scholar]
- 5.Khan HA, Siddique KH, Munir R, Colmer TD. Salt sensitivity in chickpea: Growth, photosynthesis, seed yield components and tissue ion regulation in contrasting genotypes. Journal of plant physiology. 2015;182:1–12. doi: 10.1016/j.jplph.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Vadez V, et al. Large variation in salinity tolerance in chickpea is explained by differences in sensitivity at the reproductive stage. Field crops research. 2007;104:123–129. doi: 10.1016/j.fcr.2007.05.014. [DOI] [Google Scholar]
- 7.Varshney RK, et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnology advances. 2013;31:1120–1134. doi: 10.1016/j.biotechadv.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Turner NC, et al. Salinity tolerance and ion accumulation in chickpea (Cicer arietinum L.) subjected to salt stress. Plant and Soil. 2013;365:347–361. doi: 10.1007/s11104-012-1387-0. [DOI] [Google Scholar]
- 9.Munns, R. Comparative physiology of salt and water stress. Plant, cell & environment 25 (2002). [DOI] [PubMed]
- 10.Pan YQ, et al. The Photosynthesis, N(+)/K(+) Homeostasis and Osmotic Adjustment of Atriplex canescens in Response to Salinity. Frontiers in plant science. 2016;7:848. doi: 10.3389/fpls.2016.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flowers TJ, Colmer TD. Plant salt tolerance: adaptations in halophytes. Annals of botany. 2015;115:327–331. doi: 10.1093/aob/mcu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, D. et al. A Novel Soybean Intrinsic Protein Gene, GmTIP2;3, Involved in Responding to OsmoticStress. Frontiers in plant science 6, 10.3389/fpls.2015.01237 (2016). [DOI] [PMC free article] [PubMed]
- 13.Krishnamurthy L, et al. Consistent Variation Across Soil Types in Salinity Resistance of a Diverse Range of Chickpea (Cicer arietinum L.) Genotypes. Journal of Agronomy and Crop Science. 2011;197:214–227. doi: 10.1111/j.1439-037X.2010.00456.x. [DOI] [Google Scholar]
- 14.Yan K, et al. Physiological adaptive mechanisms of plants grown in saline soil and implications for sustainable saline agriculture in coastal zone. Acta physiologiae plantarum. 2013;35:2867–2878. doi: 10.1007/s11738-013-1325-7. [DOI] [Google Scholar]
- 15.Ding F, Yang J-C, Yuan F, Wang B-S. Progress in mechanism of salt excretion in recretohalopytes. Frontiers in Biology. 2010;5:164–170. doi: 10.1007/s11515-010-0032-7. [DOI] [Google Scholar]
- 16.Shabala S. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Annals of botany. 2013;112:1209–1221. doi: 10.1093/aob/mct205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shabala S, Bose J, Hedrich R. Salt bladders: do they matter? Trends in plant science. 2014;19:687–691. doi: 10.1016/j.tplants.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Mantri NL, Ford R, Coram TE, Pang ECK. Evidence of unique and shared responses to major biotic and abiotic stresses in chickpea. Environmental and experimental botany. 2010;69:286–292. doi: 10.1016/j.envexpbot.2010.05.003. [DOI] [Google Scholar]
- 19.Jain M, et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.) The Plant journal: for cell and molecular biology. 2013;74:715–729. doi: 10.1111/tpj.12173. [DOI] [PubMed] [Google Scholar]
- 20.Deokar AA, et al. Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly. BMC genomics. 2014;15:708. doi: 10.1186/1471-2164-15-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg R, et al. Transcriptome analyses reveal genotype- and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Scientific reports. 2016;6:19228. doi: 10.1038/srep19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantri NL, Ford R, Coram TE, Pang EC. Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC genomics. 2007;8:303. doi: 10.1186/1471-2164-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molina C, et al. The salt-responsive transcriptome of chickpea roots and nodules via deepSuperSAGE. BMC plant biology. 2011;11:31. doi: 10.1186/1471-2229-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galvan-Ampudia CS, Testerink C. Salt stress signals shape the plant root. Current opinion in plant biology. 2011;14:296–302. doi: 10.1016/j.pbi.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 25.Wang, H., Wang, H., Shao, H. & Tang, X. Recent Advances in Utilizing Transcription Factors to Improve Plant Abiotic Stress Tolerance by TransgenicTechnology. Frontiers in plant science 7, 10.3389/fpls.2016.00067 (2016). [DOI] [PMC free article] [PubMed]
- 26.Demidchik V. Mechanisms of oxidative stress in plants: From classical chemistry to cell biology. Environmental and experimental botany. 2015;109:212–228. doi: 10.1016/j.envexpbot.2014.06.021. [DOI] [Google Scholar]
- 27.Kudla J, Batistič O, Hashimoto K. Calcium Signals: The Lead Currency of Plant Information Processing. The Plant cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng, H. et al. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Frontiers in plant science6, 10.3389/fpls.2015.00600 (2015). [DOI] [PMC free article] [PubMed]
- 29.Danquah A, de Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnology advances. 2014;32:40–52. doi: 10.1016/j.biotechadv.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Novo-Uzal E, Gutiérrez J, Martínez-Cortés T, Pomar F. Molecular cloning of two novel peroxidases and their response to salt stress and salicylic acid in the living fossil Ginkgo biloba. Annals of botany. 2014;114:923–936. doi: 10.1093/aob/mcu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z-S, Chen M, Li L-C, Ma Y-Z. Functions and Application of the AP2/ERF Transcription Factor Family in Crop ImprovementF. Journal of integrative plant biology. 2011;53:570–585. doi: 10.1111/j.1744-7909.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- 32.Licausi, F. et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature479 (2011). [DOI] [PubMed]
- 33.Sakuma, Y. et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant cell18 (2006). [DOI] [PMC free article] [PubMed]
- 34.Jiang Y, Deyholos MK. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant molecular biology. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- 35.Fang Y, et al. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. Journal of experimental botany. 2015;66:6803–6817. doi: 10.1093/jxb/erv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussain RM, Ali M, Feng X, Li X. The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC plant biology. 2017;17:55. doi: 10.1186/s12870-017-1001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Frontiers in microbiology. 2013;4:248. doi: 10.3389/fmicb.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiology and Molecular Biology of Plants. 2013;19:307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H, et al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant biotechnology journal. 2017;15:107–121. doi: 10.1111/pbi.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vélez-Bermúdez I-C, et al. A MYB/ZML Complex Regulates Wound-Induced Lignin Genes in Maize. The Plant cell. 2015;27:3245–3259. doi: 10.1105/tpc.15.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C-K, et al. A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant molecular biology. 2015;88:269–286. doi: 10.1007/s11103-015-0321-2. [DOI] [PubMed] [Google Scholar]
- 42.Yu F, et al. GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC plant biology. 2012;12:144. doi: 10.1186/1471-2229-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rushton DL, et al. WRKY transcription factors: key components in abscisic acid signalling. Plant biotechnology journal. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- 44.Konda, A. K., Farmer, R., Soren, K. R., Shanmugavadivel, P. S. & Setti, A. Structural modelling and molecular dynamics of a multi-stress responsive WRKY TF-DNA complex, towards elucidating its role in stress signalling mechanisms in chickpea. Journal of biomolecular structure & dynamics, 1–25, 10.1080/07391102.2017.1349690 (2017). [DOI] [PubMed]
- 45.Kumar K, et al. WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum): comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene(s) DNA research: an international journal for rapid publication of reports on genes and genomes. 2016;23:225–239. doi: 10.1093/dnares/dsw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao QM, Venugopal S, Navarre D, Kachroo A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant physiology. 2011;155:464–476. doi: 10.1104/pp.110.166876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan Y, et al. PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant cell reports. 2015;34:831–841. doi: 10.1007/s00299-015-1745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T. WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. The Plant journal: for cell and molecular biology. 2010;63:229–240. doi: 10.1111/j.1365-313X.2010.04232.x. [DOI] [PubMed] [Google Scholar]
- 49.Bita CE, Gerats T. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Frontiers in plant science. 2013;4:273. doi: 10.3389/fpls.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu HC, Charng YY. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant physiology. 2013;163:276–290. doi: 10.1104/pp.113.221168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wimmer MA, Eichert T. Review: Mechanisms for boron deficiency-mediated changes in plant water relations. Plant Science. 2013;203:25–32. doi: 10.1016/j.plantsci.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Hofmann NR. A Functional Link between Mitochondria and the Cell Wall in Stress Responses. The Plant cell. 2016;28:1996–1996. doi: 10.1105/tpc.16.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pickel B, Schaller A. Dirigent proteins: molecular characteristics and potential biotechnological applications. Applied microbiology and biotechnology. 2013;97:8427–8438. doi: 10.1007/s00253-013-5167-4. [DOI] [PubMed] [Google Scholar]
- 54.Knoch, E., Dilokpimol, A. & Geshi, N. Arabinogalactan proteins: focus on carbohydrate active enzymes. Frontiers in plant science5, 10.3389/fpls.2014.00198 (2014). [DOI] [PMC free article] [PubMed]
- 55.Abuqamar S, Ajeb S, Sham A, Enan MR, Iratni R. A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Molecular plant pathology. 2013;14:813–827. doi: 10.1111/mpp.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tenhaken, R. Cell wall remodeling under abiotic stress. Frontiers in plant science5, 10.3389/fpls.2014.00771 (2015). [DOI] [PMC free article] [PubMed]
- 57.Zhu XF, et al. XTH31, Encoding an in Vitro XEH/XET-Active Enzyme, Regulates Aluminum Sensitivity by Modulating in Vivo XET Action, Cell Wall Xyloglucan Content, and Aluminum Binding Capacity in Arabidopsis. The Plant cell. 2012;24:4731–4747. doi: 10.1105/tpc.112.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hara Y, Yokoyama R, Osakabe K, Toki S, Nishitani K. Function of xyloglucan endotransglucosylase/hydrolases in rice. Annals of botany. 2014;114:1309–1318. doi: 10.1093/aob/mct292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong JL, Jiang YY, Chen RJ, Xu ZJ, Gao XL. Isolation of a novel xyloglucan endotransglucosylase (OsXET9) gene from rice and analysis of the response of this gene to abiotic stresses. Afr J Biotechnol. 2011;10:17424–17434. [Google Scholar]
- 60.Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant molecular biology. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 61.Nakashima K, Yamaguchi-Shinozaki K. ABA signaling in stress-response and seed development. Plant cell reports. 2013;32:959–970. doi: 10.1007/s00299-013-1418-1. [DOI] [PubMed] [Google Scholar]
- 62.Muday GK, Rahman A, Binder BM. Auxin and ethylene: collaborators or competitors? Trends in plant science. 2012;17:181–195. doi: 10.1016/j.tplants.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 63.Ryu H, Cho Y-G. Plant hormones in salt stress tolerance. Journal of Plant Biology. 2015;58:147–155. doi: 10.1007/s12374-015-0103-z. [DOI] [Google Scholar]
- 64.Cao W-H, et al. Modulation of Ethylene Responses Affects Plant Salt-Stress Responses. Plant physiology. 2007;143:707. doi: 10.1104/pp.106.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Datta R, et al. Glutathione Regulates 1-Aminocyclopropane-1-Carboxylate Synthase Transcription via WRKY33 and 1-Aminocyclopropane-1-Carboxylate Oxidase by Modulating Messenger RNA Stability to Induce Ethylene Synthesis during Stress. Plant physiology. 2015;169:2963–2981. doi: 10.1104/pp.15.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC plant biology. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cabot C, Sibole JV, Barcelo J, Poschenrieder C. Lessons from crop plants struggling with salinity. Plant science: an international journal of experimental plant biology. 2014;226:2–13. doi: 10.1016/j.plantsci.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 68.Yao X, Xiong W, Ye T, Wu Y. Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. Journal of experimental botany. 2012;63:2579–2593. doi: 10.1093/jxb/err433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu G, Xu H, Zhang L, Zheng Y. Fe Binding Properties of Two Soybean (Glycine max L.) LEA4 Proteins Associated with Antioxidant Activity. Plant and Cell Physiology. 2011;52:994–1002. doi: 10.1093/pcp/pcr052. [DOI] [PubMed] [Google Scholar]
- 70.Battaglia M, Covarrubias AA. Late Embryogenesis Abundant (LEA) proteins in legumes. Frontiers in plant science. 2013;4:190. doi: 10.3389/fpls.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chapman N, Whalley WR, Lindsey K, Miller AJ. Water supply and not nitrate concentration determines primary root growth in Arabidopsis. Plant, cell & environment. 2011;34:1630–1638. doi: 10.1111/j.1365-3040.2011.02358.x. [DOI] [PubMed] [Google Scholar]
- 72.Aquea F, et al. A molecular framework for the inhibition of Arabidopsis root growth in response to boron toxicity. Plant, cell & environment. 2012;35:719–734. doi: 10.1111/j.1365-3040.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- 73.Su, Y. H., Liu, Y. B., Bai, B. & Zhang, X. S. Establishment of embryonic shoot–root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Frontiers in plant science5, 10.3389/fpls.2014.00792 (2015). [DOI] [PMC free article] [PubMed]
- 74.Mockaitis K, Estelle M. Auxin Receptors and Plant Development: A New Signaling Paradigm. Annual Review of Cell and Developmental Biology. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 75.Petrášek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 76.Ding X, et al. Activation of the Indole-3-Acetic Acid–Amido Synthetase GH3-8 Suppresses Expansin Expression and Promotes Salicylate- and Jasmonate-Independent Basal Immunity in Rice. The Plant cell. 2008;20:228–240. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guerreiro, A., Figueiredo, J., Sousa Silva, M. & Figueiredo, A. Linking Jasmonic Acid to Grapevine Resistance against the Biotrophic Oomycete Plasmopara viticola. Frontiers in plant science7, 10.3389/fpls.2016.00565 (2016). [DOI] [PMC free article] [PubMed]
- 78.Ahmad P, et al. Jasmonates: Multifunctional Roles in Stress Tolerance. Frontiers in plant science. 2016;7:813. doi: 10.3389/fpls.2016.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan C, Xie D. Jasmonate in plant defence: sentinel or double agent? Plant biotechnology journal. 2015;13:1233–1240. doi: 10.1111/pbi.12417. [DOI] [PubMed] [Google Scholar]
- 80.Kazan K, Manners JM. JAZ repressors and the orchestration of phytohormone crosstalk. Trends in plant science. 2012;17:22–31. doi: 10.1016/j.tplants.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 81.Vanneste S, Friml J. Auxin: A Trigger for Change in Plant Development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 82.Krupinski P, Jönsson H. Modeling Auxin-regulated Development. Cold Spring Harbor Perspectives in Biology. 2010;2:a001560. doi: 10.1101/cshperspect.a001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mironova VV, et al. A plausible mechanism for auxin patterning along the developing root. BMC systems biology. 2010;4:98. doi: 10.1186/1752-0509-4-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN. Combining Expression and Comparative Evolutionary Analysis. The COBRA Gene Family. Plant physiology. 2007;143:172–187. doi: 10.1104/pp.106.087262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burssens S, et al. Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta. 2000;211:632–640. doi: 10.1007/s004250000334. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y, Li K, Li X. Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. Journal of plant physiology. 2009;166:1637–1645. doi: 10.1016/j.jplph.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 87.Zolla G, Heimer YM, Barak S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. Journal of experimental botany. 2010;61:211–224. doi: 10.1093/jxb/erp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Petricka JJ, Winter CM, Benfey PN. Control of Arabidopsis Root Development. Annual Review of Plant Biology. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pietra S, Lang P, Grebe M. SABRE is required for stabilization of root hair patterning in Arabidopsis thaliana. Physiologia plantarum. 2015;153:440–453. doi: 10.1111/ppl.12257. [DOI] [PubMed] [Google Scholar]
- 90.Deinlein U, et al. Plant salt-tolerance mechanisms. Trends in plant science. 2014;19:371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Craig Plett D, MØLler IS. Na+ transport in glycophytic plants: what we know and would like to know. Plant, cell & environment. 2010;33:612–626. doi: 10.1111/j.1365-3040.2009.02086.x. [DOI] [PubMed] [Google Scholar]
- 92.Qi, X. et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. 5, 4340, 10.1038/ncomms5340https://www.nature.com/articles/ncomms5340 (2014). [DOI] [PMC free article] [PubMed]
- 93.Cuéllar T, et al. A grapevine Shaker inward K+ channel activated by the calcineurin B-like calcium sensor 1–protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. The Plant Journal. 2010;61:58–69. doi: 10.1111/j.1365-313X.2009.04029.x. [DOI] [PubMed] [Google Scholar]
- 94.Li J, et al. The Os-AKT1 Channel Is Critical for K(+) Uptake in Rice Roots and Is Modulated by the Rice CBL1-CIPK23 Complex. The Plant cell. 2014;26:3387–3402. doi: 10.1105/tpc.114.123455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ren, Z.-H. et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet37, 1141–1146, http://www.nature.com/ng/journal/v37/n10/suppinfo/ng1643_S1.html (2005). [DOI] [PubMed]
- 96.Sunarpi, et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. The Plant Journal. 2005;44:928–938. doi: 10.1111/j.1365-313X.2005.02595.x. [DOI] [PubMed] [Google Scholar]
- 97.Chiba Y, et al. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. Journal of plant research. 2015;128:679–686. doi: 10.1007/s10265-015-0710-2. [DOI] [PubMed] [Google Scholar]
- 98.Munns, R. & Tester, M. Mechanisms of salinity tolerance. Annu Rev Plant Biol59 (2008). [DOI] [PubMed]
- 99.Flowers TJ, et al. Salt sensitivity in chickpea. Plant, cell & environment. 2010;33:490–509. doi: 10.1111/j.1365-3040.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 100.Vašák M. Advances in metallothionein structure and functions. Journal of Trace Elements in Medicine and Biology. 2005;19:13–17. doi: 10.1016/j.jtemb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 101.Klaassen CD, Liu J, Diwan BA. Metallothionein Protection of Cadmium Toxicity. Toxicology and applied pharmacology. 2009;238:215–220. doi: 10.1016/j.taap.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ruttkay-Nedecky B, et al. The Role of Metallothionein in Oxidative Stress. International journal of molecular sciences. 2013;14:6044–6066. doi: 10.3390/ijms14036044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jia, H. et al. Hydrogen sulfide - cysteine cycle system enhances cadmium tolerance through alleviating cadmium-induced oxidative stress and ion toxicity in Arabidopsis roots. 6, 39702, 10.1038/srep39702http://dharmasastra.live.cf.private.springer.com/articles/srep39702 (2016). [DOI] [PMC free article] [PubMed]
- 104.Hall JL. Cellular mechanisms for heavy metal detoxification and tolerance. Journal of experimental botany. 2002;53:1–11. doi: 10.1093/jexbot/53.366.1. [DOI] [PubMed] [Google Scholar]
- 105.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25–R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw fastq files and gene count files for all samples including three biological replicates are submitted to the GEO databases under the accession GSE110127.