Abstract
Mutations in the human dynamin-related protein Drp1 cause mitochondria to form perinuclear clusters. We show here that these mitochondrial clusters consist of highly interconnected mitochondrial tubules. The increased connectivity between mitochondria indicates that the balance between mitochondrial division and fusion is shifted toward fusion. Such a shift is consistent with a block in mitochondrial division. Immunofluorescence and subcellular fractionation show that endogenous Drp1 is localized to mitochondria, which is also consistent with a role in mitochondrial division. A direct role in mitochondrial division is suggested by time-lapse photography of transfected cells, in which green fluorescent protein fused to Drp1 is concentrated in spots that mark actual mitochondrial division events. We find that purified human Drp1 can self-assemble into multimeric ring-like structures with dimensions similar to those of dynamin multimers. The structural and functional similarities between dynamin and Drp1 suggest that Drp1 wraps around the constriction points of dividing mitochondria, analogous to dynamin collars at the necks of budding vesicles. We conclude that Drp1 contributes to mitochondrial division in mammalian cells.
INTRODUCTION
Mitochondria are dynamic structures that frequently divide and fuse with one another (Bereiter-Hahn and Voth, 1994; Hermann and Shaw, 1998). Mitochondrial division is needed during cell division to distribute mitochondria to the daughter cells. Quiescent cells also exhibit mitochondrial division during cellular differentiation, during cell growth or in response to extracellular stimuli. The mitochondrial division and fusion processes must be tightly regulated, because cell survival depends on the preservation of an adequate number of mitochondria in each cell. The mechanisms of mitochondrial division and fusion are also likely to be complex, because mitochondria have double membranes, which present distinct topological and energetic barriers.
A first clue to the mechanism of mitochondrial division came from the recent discoveries that this process is controlled by a dynamin-related protein called DRP-1 in Caenorhabditis elegans (Labrousse et al., 1999) or Dnm1p in yeast (Bleazard et al., 1999; Sesaki and Jensen, 1999). Loss of Dnm1p/DRP-1 function causes an increase in mitochondrial connectivity both in C. elegans and in yeast, consistent with reduced frequencies of mitochondrial divisions (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999). In C. elegans, the mitochondrial inner membrane continues to divide, indicating that DRP-1 is only required for division of the mitochondrial outer membrane (Labrousse et al., 1999). Overexpression of wild-type DRP-1 increases the number of mitochondrial division events in C. elegans (Labrousse et al., 1999). Immunofluorescence and immuno-electron microscopy showed that epitope-tagged versions of yeast Dnm1p were localized in spots along mitochondria or at the tips of mitochondria (Bleazard et al., 1999). Time-lapse photography showed that green fluorescent protein (GFP)-tagged DRP-1 from C. elegans is localized in spots on mitochondria where fission is about to occur, consistent with a direct role in mitochondrial division (Labrousse et al., 1999). Taken together, these results provide strong evidence that C. elegans DRP-1 and yeast Dnm1p are essential for the final stages of the mitochondrial division process.
The mammalian homolog of C. elegans DRP-1 and yeast Dnm1p has alternatively been called Drp1, Dlp1, DVLP, or Dymple (Shin et al., 1997; Imoto et al., 1998; Kamimoto et al., 1998; Smirnova et al., 1998; Yoon et al., 1998). Here, we refer to this protein as Drp1. The function of mammalian Drp1 is still a matter of debate. It has been suggested that Drp1 helps form vesicles, playing a role similar to the role of dynamin in vesicle formation. One study suggested that Drp1 is required during an early stage of the secretory pathway (Imoto et al., 1998). Another study suggested that Drp1 contributes to a novel route of vesicular transport or direct fusion between endoplasmic reticulum (ER) and mitochondria (Pitts et al., 1999). This study showed no effect on the secretory or endocytic pathways, consistent with our own previous results that also showed no effect on vesicular transport (Smirnova et al., 1998). Instead, we found that mutant Drp1 causes mitochondria to collapse into perinuclear clusters, which indicates that Drp1 specifically affects mitochondrial morphology (Smirnova et al., 1998). The underlying defect that induces mitochondrial clustering has remained uncertain.
We now show that the mitochondrial clusters contain a highly interconnected network of mitochondria, as expected from a defect in mitochondrial division. We are also able to detect a fraction of endogenous Drp1 that is localized in spots along mitochondria and we can detect colocalization of GFP-tagged Drp1 with mitochondrial division events with the use of time-lapse photography. We conclude that Drp1 contributes to mitochondrial division in mammalian cells, as it does in C. elegans and in yeast.
MATERIALS AND METHODS
Transfection Constructs, Cell Culture, and Transfection Procedures
The pcDNA3-HA-Drp1 plasmid, which was used for expression of human Drp1 in mammalian cells, was described previously (Smirnova et al., 1998). The K38A, V41F, T59A, and G281D mutations were introduced into the Drp1 coding sequence with the use of standard polymerase chain reaction methods and Pfu polymerase (Stratagene, La Jolla, CA). New clones were checked by sequencing. The GFP::Drp1 construct was made by recloning GFP from pGreenLantern (Life Technologies, Rockville, MD) upstream of Drp1 in pcDNA3 (Invitrogen, San Diego, CA). Drp1::YFP and YFP::Drp1 were made by recloning Drp1 upstream or downstream of YFP coding sequence in pEYFP-C1 (CLONTECH, Palo Alto, CA). The pECFP-Mito plasmid (CLONTECH) was used as a mitochondrial matrix marker. The mitochondrial outer membrane marker was made by amplifying the N-terminal 73 amino acids of Tom20 from a human stromal cell cDNA library and cloning this fragment upstream of GFP in pcDNA3 (Invitrogen).
COS-7 and C2C12 cell lines were grown in DMEM supplemented with 10 or 20% fetal calf serum, respectively. COS-7 cells were transiently transfected with the use of FuGENE transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN), and C2C12 with SuperFect (Qiagen, Valencia, CA) according to the manufacturers' procedures. The transfected cells were seeded onto glass coverslips for analysis by immunofluorescence or in glass-bottom dishes (MatTek, Ashland, MA) for in vivo observation. The cells were grown for 16–26 h between transfection and analysis by fluorescence microscopy.
Antibody Production, Immunofluorescence Staining, and Fluorescence Imaging
Anti-Drp1 antibodies were made by immunizing rabbits with peptides synthesized on MAPS resin by the University of Southern California (Los Angeles, CA) microchemistry core facility. Two peptides (271-KKYPSLANRNGT-282 and 445-HCSNYSYQELLRFP-458) were used to immunize rabbits (Robert Sargeant, Ramona, CA). The antibodies were blot-purified with the use of a fragment of Drp1 that was expressed in Escherichia coli as described previously (Smirnova et al., 1998). The rabbit antibody that was used throughout this study was increased against the 445–458 peptide. For double labeling with other rabbit antibodies, we used a chicken anti-Drp1 antibody, which was described previously (Smirnova et al., 1998). The VSV-G KKTN construct for labeling ER was also described previously (Smirnova et al., 1998). VSV-G was detected with a monoclonal antibody from Sigma (St. Louis, MO). Rabbit antiribophorin-I antibody (Hortsch and Meyer, 1985) was kindly provided by from Dr. D.I. Meyer (Biological Chemistry, UCLA). Protein disulfide isomerase was detected with a mouse monoclonal antibody from Stressgene (Victoria, British Columbia, Canada).
Before fixation for immunofluorescence, transfected cells were incubated with 0.1 μM MitoTracker Red (Molecular Probes, Eugene, OR) for 30 min at 37°C. The cells were fixed by incubating with 3.7% paraformaldehyde in phosphate-buffered saline for 15 min at 37°C. The cells were permeabilized with a 10-min incubation in ice-cold acetone, stained with anti-Drp1 antibody and secondary antibody, and viewed with standard fluorescein isothiocyanate and rhodamine filter sets as described previously (Smirnova et al., 1998). Double labeling of endogenous Drp1 and the mitochondria was achieved by incubating COS-7 cells with MitoTracker and then fixing and permeabilizing the cells by incubating them with methanol acetone (1:1) for 3 min at −20°C. These cells were stained with anti-Drp1 antibody and further processed as described above.
GFP variants were viewed with filters from Chroma Technologies (Brattleboro, VT), a PXL charge-coupled device camera from Photometrics (Tuscon, AZ), and software from Inovision (Durham, NC). Images for time-lapse experiments were acquired at 5-s intervals in CFP and YFP channels. The lengths of mitochondria in the charge-coupled device images were measured by tracing them with NIH Image software.
Subcellular Fractionation and Western Blotting
All fractionation procedures were performed on ice. Fresh bovine brain (50 g) was minced in ice-cold buffer A (250 mM sucrose, 10 mM Tris-HCl, pH 7.5, 1 mM EGTA with protease inhibitor mix; Roche Diagnostics GmbH, Mannheim, Germany) and homogenized in a final volume of 250 ml with a Dounce homogenizer. The crude lysate was centrifuged for 15 min at 1000 × g. The resulting low-speed pellet, which contained nuclei and unbroken cells, was discarded. The low-speed supernatant (S1) was centrifuged for 15 min at 10,000 × g. This step yielded a medium-speed supernatant (S2) and pellet (P2). The P2 fraction, which contains mitochondria and lysosomes, was washed twice by resuspending and repelleting in buffer A. The S2 fraction, which contains cytosol and light membranes such as ER and Golgi, was recentrifuged for 15 min at 15,000 × g. The resulting supernatant was centrifuged for 1 h at 100,000 × g to separate ER and Golgi, which are in the P3 fraction, from cytosol, which is in the S3 fraction.
Mitochondria in the P2 fraction were further purified with a 45%Percoll gradient (Amersham Pharmacia Biotech AB, Uppsala, Sweden) established by centrifugation for 30 min at 30 krpm in a Ti70 rotor (Beckman Instruments, Palo Alto, CA). The mitochondrial band was diluted in a large volume of buffer A. The mitochondria were pelleted by centrifugation for 15 min at 10,000 × g and washed by resuspending and repelleting in buffer A. The pellet containing purified mitochondria was resuspended in a small volume of buffer A.
Protein (75 μg) from each fraction was precipitated with trichloroacetic acid and separated by SDS-PAGE. Western blots were probed with our anti-Drp1 antibody, with 20C11-B11-B11 from Molecular Probes, which recognizes the 39-kDa subunit of NADH-ubiquinol oxidoreductase, and an anti-tubulin antibody provided by A. Rajesekaran (Department of Pathology, UCLA). The blots were developed with horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence reagents (Amersham Pharmacia Biotech AB). The blots were quantified with a densitometer (Molecular Dynamics, Sunnyvale, CA) and the values were adjusted to achieve volume equivalents. Values obtained with the mitochondrial marker were used to correct for loss of mitochondria in the Percoll gradient and subsequent washes.
Protein Expression and Electron Microscopy
The methods for protein purification with the use of baculovirus are described in detail elsewhere (Smirnova et al., 2000). Briefly, a Drp1 expression construct with an N-terminal tag of six histidines was made in pBlueBac4 vector (Invitrogen) and expressed in Sf9 insect cells. The overexpressed protein was purified on TALON metal affinity resin (CLONTECH). Purified Drp1 was diluted to a concentration of 250 μg/ml in a HEPES buffer (20 mM HEPES pH 7.2, 2 mM MgCl2, 0.5 mM EGTA, 0.5 mM dithiothreitol, and 160 mM NaCl). The Drp1 solution was dialyzed for 16 h at 4°C in the same buffer with or without 200 μM GDP, 500 μM AlCl3, and 5 mM NaF as described (Carr and Hinshaw, 1997). The samples were diluted 1:10 in the HEPES buffer, adsorbed onto carbon-coated electron microscopy grids, negatively stained with 1% uranyl acetate, and air dried. Electron micrographs were obtained with a JEM 1200-EX electron microscope (JEOL, Tokyo, Japan).
RESULTS
Overexpression of Mutant Drp1 Protein Increases Mitochondrial Connectivity
To investigate how the Drp1 protein affects mitochondrial morphology, we transfected COS-7 cells with constructs containing dominant negative mutations in human Drp1. Four different mutations were tested to help distinguish the primary defect caused by disrupting Drp1 from possible side effects. The first mutation, K38A, changes the critical lysine in the G1 consensus motif of the GTPase domain into an alanine, thereby presumably inhibiting GTP binding by Drp1. This mutation was used in our previous study of mammalian Drp1 (Smirnova et al., 1998). The second mutation, V41F, was modeled after a mutation in C. elegans let-60/ras that confers temperature sensitivity to GTP hydrolysis (Eisenmann and Kim, 1997). The third mutation, T59A, is a mutation of a conserved threonine, which is most likely within the G2 consensus motif of GTP-binding proteins (van der Bliek, 1999). The analogous mutation in C. elegans DRP-1 was shown to have a strong dominant negative effect on mitochondrial morphology (Labrousse et al., 1999). The fourth mutation, G281D, was modeled after a temperature-sensitive mutation in the Drosophila shibire/dynamin gene (van der Bliek and Meyerowitz, 1991). The analogous mutation in human dynamin causes a temperature-sensitive block in endocytosis in transfected COS-7 cells (Damke et al., 1995). The Drp1 mutant constructs were transiently transfected into COS-7 cells. Transfected cells were identified by immunofluorescence with anti-Drp1 antibody and mitochondrial morphology was observed by staining the cells with the dye Mitotracker.
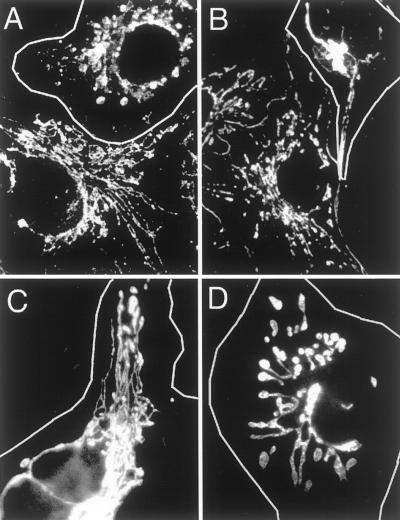
We first tested whether the V41F and G281D mutations confer temperature-sensitive dominant negative properties to human Drp1. Cells were transfected with the Drp1(V41F) or the Drp1(G281D) construct and grown at 30 or 40°C before staining with anti-Drp1 antibody, to identify transfected cells, and with MitoTracker, to monitor mitochondrial morphology. The mitochondria of cells transfected with mutant Drp1 were almost all aberrant (outlined cells in Figure 1), whereas the untransfected cells had wild-type mitochondrial morphologies at both lower and higher growth temperatures (Figure 1, A and B, bottom). There were also clear differences between the transfected cells grown at 30 or 40°C. The mitochondria of transfected cells grown at 30°C were distributed throughout the cell. In contrast, the mitochondria of transfected cells grown at 40°C were clustered around the nucleus (Figure 1B) and in some cases appeared to be much thicker than wild-type mitochondria (Figure 1D). It would thus appear that the V41F and G281D mutations confer temperature-sensitive properties to Drp1.
Figure 1.
Effect of temperature-sensitive mutations in Drp1 on mitochondrial morphology. (A and B) Cells transfected with Drp1(V41F) and grown at 30°C (A) or 40°C (B). (C and D) Cells transfected with Drp1(G281D) and grown at 30°C (C) or 40°C (D). The transfected cells were identified by immunofluorescence with anti-Drp1 antibody (outlined cells). Mitochondrial morphology was monitored by staining with Mitotracker. A and B each also show an untransfected cell, to illustrate that the growth temperature does not appreciably affect mitochondrial distribution in cells without mutant Drp1.
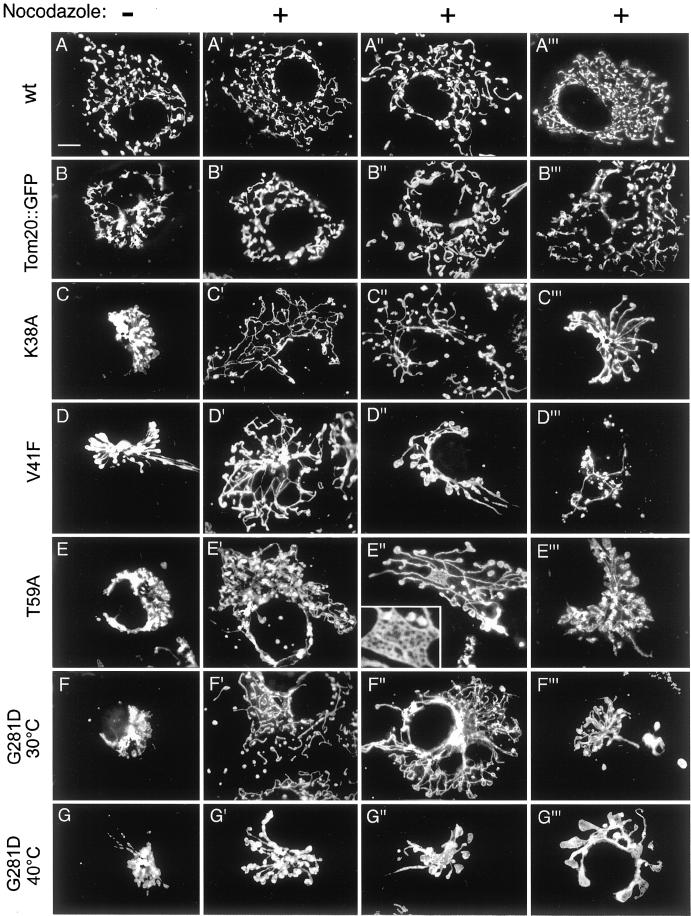
To analyze the effects of mutant Drp1 more systematically, we also transfected COS-7 cells with Drp1 containing the other two mutations (K38A and T59A). In these cells the mitochondrial network invariably collapsed into large perinuclear aggregates similar to the effects of the temperature-sensitive mutations at higher temperatures (compare wild-type in Figure 2A with affected cells in Figure 2, C–G). Occasionally, a few long tubules are retained (Figure 2D). These phenotypes are similar to the ones previously observed with Drp1(K38A) in transfected COS-7 cells (Smirnova et al., 1998). Electron microscopy of thin sections of those cells had shown that the mitochondrial aggregates consist of clustered mitochondrial tubules. The mitochondrial clusters were so compact that it was impossible to distinguish connected from unconnected mitochondria (Smirnova et al., 1998). To help determine whether these mitochondria were more connected than mitochondria in wild-type cells, we tested a variety of drug treatments for their ability to loosen the mitochondrial clusters. We found that treatment with nocodazole was most effective. Treating COS-7 cells for 1 h with 5 μM nocodazole depolymerizes all of the microtubules that can be detected by immunofluorescence with anti-tubulin antibody (our unpublished results) and it disperses many of the mitochondrial clusters that are induced by mutant Drp1.
Figure 2.
Range of mitochondrial distribution defects induced by different mutations in Drp1. The left column of images shows transfected cells that were not treated with nocodazole. The remaining three columns show different examples of transfected cells that were treated with nocodazole. (A–A‴) COS-7 cells transfected with a wild-type Drp1 construct. (B–B‴) Cells transfected with a Tom20::GFP construct, which was used as a negative a control, because it causes mitochondrial clustering without affecting connectivity. (C–C‴) Cells transfected with a Drp1(K38A) construct. (D–D‴) Cells transfected with a Drp1(V41F) construct. The cell shown in D′ was grown at 30°C, whereas the cells shown in D" and D‴ were grown at 40°C. (E–E‴) Cells transfected with a Drp1(T59A) construct. The inset in E" shows an enlargement of a net formed by the mitochondria in this cell. (F–F‴) Cells transfected with a Drp1(G281D) construct and grown at 30°C. (G–G‴) Cells transfected with a Drp1(G281D) construct and grown at 40°C.
Dispersion of the mitochondrial clusters with nocodazole made it possible to see how the mitochondria within clusters were affected by mutant Drp1. Examples of the mutant phenotypes are shown in Figure 2 and a quantitative evaluation is presented in Table 1. Cells transfected with wild-type Drp1 (Figure 2, A–A‴) and cells transfected with a fusion between the mitochondrial outer membrane protein Tom20 and GFP (Figure 2, B–B‴) were used as controls. As described previously, the Tom20::GFP fusion protein induces mitochondrial clustering (Yano et al., 1997). On treatment with nocodazole, the clusters induced by Tom20::GFP were dispersed, showing a degree of mitochondrial fragmentation similar to that observed in cells transfected with wild-type Drp1. This indicates that the collapse of mitochondria into a perinuclear cluster by Tom20::GFP and treatment with nocodazole do not affect the degree of mitochondrial connectivity.
Table 1.
Summary of mitochondrial morphologies observed in COS-7 cells transfected with mutant Drp1
| n | Weak
|
Strong
|
Average phenotype | |||
|---|---|---|---|---|---|---|
| Wild type | Loose net | Collapsed | Degenerate | |||
| WT | 103 | 100 | 0 | 0 | 0 | None |
| K38A | 203 | 0 | 18 | 64 | 18 | Strong |
| V41F at 30°C | 96 | 2 | 33 | 60 | 5 | Intermediate |
| V41F at 40°C | 108 | 0 | 8 | 44 | 48 | Strong |
| T59A | 112 | 0 | 12 | 61 | 27 | Strong |
| G281D at 30°C | 107 | 40 | 48 | 11 | 1 | Weak |
| G281D at 40°C | 120 | 6 | 13 | 46 | 35 | Strong |
The transfected cells were treated with nocodazole to disperse the perinuclear mitochondrial clusters. The phenotypes of individual cells were classified as described in the text and Figure 1. The number of cells that were counted is shown (n) followed by the percentage of cells in each phenotypic class. Staining with anti-Drp1 antibody showed that the overexpressed protein was mostly cytosolic with wild-type Drp1, aggregated with Drp1(K38A), cytosolic with Drp1(V41F) at both temperatures, sometimes aggregated with Drp1(T59A), cytosolic Drp1(G281D) at 30°C, and sometimes aggregated with Drp1(G281D) at 40°C.
In contrast, cells transfected with mutant Drp1 displayed a range of mitochondrial morphologies. Many transfected cells had loose networks of interconnected mitochondria (Figure 2, C′, D′, E", and F"; classified as a “loose net” phenotype in Table 1). In some cells, the loosened mitochondrial tubules revealed a tight internal net, as if the loose network that was observed in other cells had been drawn together like a piece of cloth (Figure 2E", insert; these were also classified as a loose net phenotype in Table 1). We also observed long mitochondrial filaments radiating from a central core (Figure 2, C‴, D", and E‴; classified as a “collapsed” phenotype in Table 1). Many of these filaments had club-shaped endings. Other cells contained a single short, thick mitochondrion with large club-shaped endings, suggesting that the mitochondria had fused and retracted into a single giant mitochondrion (Figure 2G‴; these were also classified as collapsed in Table 1). In some cells, the mitochondria started to degenerate, as judged by reduction of the number of mitochondria (Figure 2D‴; classified as a “degenerate” phenotype in Table 1) and their decreased ability to retain MitoTracker, which reflects a decrease in mitochondrial membrane potential (our unpublished results).
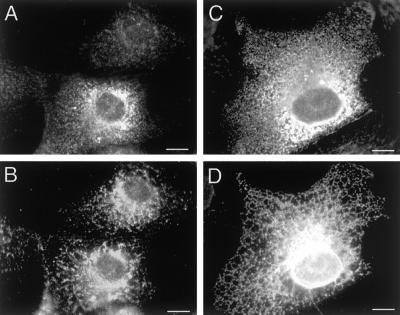
To verify that mutant Drp1 had no effect on ER, as shown previously with Drp1(K40A) (Smirnova et al., 1998), we also stained cells transfected with Drp1(T59A) with different ER markers (Figure 3). The markers that we used here were protein disulfide isomerase (Figure 3B) and a cotransfected ER marker (Figure 3D; VSV-G with a C-terminal ER retention signal, as described previously [Smirnova et al., 1998]). Transfected cells were identified by staining with anti-Drp1 antibody (Figure 3, A and C). As observed previously, the transfected cells yield a cytosolic staining pattern with anti-Drp1 antibody, presumably because the endogenous binding sites become saturated by overexpression of Drp1 (Smirnova et al., 1998). Staining with protein disulfide isomerase antibody showed no difference between transfected and untransfected cells (Figure 3, A and B). Staining with VSV-G antibody also showed ER that looked wild type (Figure 3, C and D). We conclude that ER staining intensity and morphology are unaffected by mutant Drp1.
Figure 3.
Morphology of the endoplasmic reticulum is not affected by Drp1(T59A). (A and B) Cells transfected with Drp1(T59A) alone. (C and D) A cell cotransfected with Drp1(T59A) and an exogenous ER-marker (VSV-G::KKTN), which we know from previous experiments helps to visualize the reticular nature of the ER (Smirnova et al., 1998). The transfected cells were identified by staining with anti-Drp1 antibody (A and C). The top cell in A shows endogenous levels of Drp1, whereas the bottom cell shows overexpression of Drp1, as does the cell in C. ER morphology was observed by staining with antiprotein disulfide isomerase antibody (B) or with anti-VSV-G antibody (D). The intensity and morphology of ER were unaffected by Drp1(T59A) (compare the staining pattern of the untransfected cell at the top of B with the staining pattern of the transfected cell at the bottom). The reticular nature of the ER also appears unaffected (compare the staining pattern in D with similar results obtained previously with wild-type and Drp1(K40A) transfected cells (Smirnova et al., 1998)).
To further assess the increased connectivity caused by mutant Drp1, we measured the lengths of mitochondria in transfected cells treated with nocodazole. Mitochondrial lengths could be measured by tracing them on images obtained with standard epifluorescence, because the transfected COS-7 cells were generally very flat and therefore captured within one focal plane. We verified that these mitochondria were indeed connected with the use of confocal microscopy (our unpublished results). We found that cells transfected with wild-type Drp1 had on average 96 mitochondria (SD = 16.5, n = 5) with average lengths of 5.4 μm (SD = 6.3, n = 5). In contrast, cells transfected with Drp1(K38A) usually had one large interconnected mitochondrion (average length of the large mitochondrion is 401 μm, SD = 86.5, n = 10) and a few small mitochondria (average number of small mitochondria per cell is 7, SD = 6.6; average length of the small mitochondria is 3.1 μm, SD = 3.8, n = 10). We conclude that the mitochondria of cells expressing mutant Drp1 were almost all more connected than mitochondria in wild-type cells, as would be expected when fission is blocked but fusion can still occur. Some of the cells transfected with Drp1(G281D) and grown at 30°C had mitochondria that were indistinguishable from wild type. Others had a loose net of mitochondria and there were others with a tight cluster that could not be dispersed by nocodazole. In contrast, all of the Drp1(G281D) cells grown at 40°C had tight clusters of mitochondria that could not be dispersed by nocodazole. Cells transfected with Drp1(V41F) were typically more severely affected than cells transfected with Drp1(G281D), even at 30°C, with many cells containing tight clusters of mitochondria (Table 1). The different effects obtained at lower and higher temperatures have enabled us to determine which of the mitochondrial phenotypes which stronger and which are weaker.
Differences in the severity of the mitochondrial defects can be used to arrange the phenotypes into the equivalent of an allelic series. The weakest phenotypes are usually the closest to the primary defect, whereas stronger phenotypes may be caused by secondary defects. The finding that Drp1(G281D) at 30°C induces a mixture of phenotypes, including mitochondria that appear wild type, suggests that these are the least severe phenotypes. Under these conditions, a loose, but highly interconnected, net of mitochondria is prevalent, suggesting that the loose net phenotype is the weakest phenotype and thus an immediate effect of mutant Drp1 (Table 1). The increased mitochondrial connectivity is consistent with a shift in the balance between mitochondrial division and fusion, when mitochondrial division is blocked. The collapsed or retracted mitochondria observed at 40°C and with stronger mutations may represent further progression from fused nets of mitochondrial tubules into more and more tightly knit structures. Together, these results indicate that the primary defect in cells with mutant Drp1 is a block in mitochondrial division.
Endogenous Drp1 Is Partly Cytosolic and Partly Localized to Mitochondria
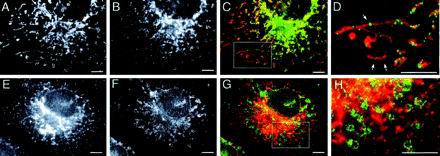
Previous attempts to localize mammalian Drp1 to specific organelles were hampered by the fact that most of the protein is cytosolic (Shin et al., 1997; Imoto et al., 1998; Kamimoto et al., 1998; Smirnova et al., 1998; Yoon et al., 1998). To help identify organelles with which Drp1 might associate, we tested a variety of fixation and staining methods and we tested newly developed antibodies directed against human Drp1. Immunofluorescence with the new antibodies showed diffuse cytosolic staining superimposed on a faint punctate staining pattern. The antibodies were specific for Drp1, because they detect a single band of the expected size (80 kDa) on Western blots of total cell lysates (our unpublished results). We sought to improve the signal of the punctate staining pattern, because this pattern might reveal a fraction of Drp1 that is associated with specific organelles. We found that cells fixed with a 1:1 mixture of methanol and acetone had reduced levels of cytosolic staining, thereby helping to uncover the punctate staining pattern. By double labeling with MitoTracker we found that the punctate staining pattern coincides with mitochondria (Figure 4, A–D). Double labeling with the ER marker ribophorin-I showed no colocalization with Drp1 spots (Figure 4, E–H), in contrast with previously published results, which had suggested that Drp1 is associated with ER (Yoon et al., 1998). Our own trials with various other ER markers suggest that these discrepancies may have arisen from cross-reactivity with mitochondria (our unpublished results). We conclude that a small but reproducible fraction of endogenous Drp1 colocalizes with mitochondria. The distribution of Drp1 between cytosol and membrane fractions is similar to that of dynamin, which is also found in a large cytosolic fraction and a smaller membrane-bound fraction. It seems likely that Drp1, like dynamin, cycles between the cytosol and its target membrane.
Figure 4.
Localization of endogenous Drp1 as determined by immunofluorescence. (A) Mitochondria stained with MitoTracker. (B) Endogenous Drp1 detected by immunofluorescence with anti-Drp1 antibody. (C) Merged image with Drp1 staining shown green and mitochondrial staining shown in red. (D) Enlargement of the boxed area in C. Arrows point to Drp1 spots that appear to transect the mitochondria. (E and F) Lack of colocalization of ER and Drp1. (E) ER staining pattern obtained with anti-ribophorin-I antibody. (F) Staining pattern obtained with anti-Drp1 antibody. (G) Merged image showing Drp1 staining in green and ER staining in red. (H) Enlargement of the boxed area in G. Bar, 5 μm.
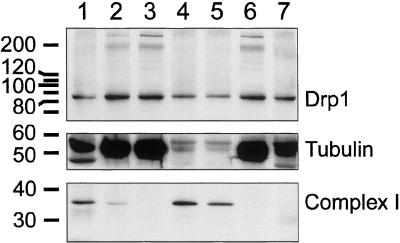
To determine what fraction of Drp1 is associated with mitochondria, we conducted subcellular fractionation experiments with bovine brain. Brain was selected because it has the highest level of Drp1 expression (Smirnova et al., 1998). The homogenized tissue was subjected to differential centrifugation. The medium-speed pellet, which contains mitochondria, was subjected to further fractionation on a Percoll gradient. The resulting fractions were analyzed by probing Western blots with anti-Drp1 antibody and antibodies for tubulin, to track cytosolic fractions, and Ox-Phos complex I, to track mitochondrial fractions. The bulk of Drp1 protein was in the cytosolic fraction. There was also a small but reproducible amount of Drp1 in the mitochondrial fraction. This mitochondrial Drp1 was barely detectable when volume equivalents were analyzed, but readily detectable when equivalent amounts of proteins from the different fractions were loaded on a gel and analyzed by Western blotting (Figure 5). Densitometry of the chemiluminescence bands, followed by corrections to obtain corresponding values for the volume equivalents, indicates that ∼3% of the total amount of Drp1 cofractionates with mitochondria in the medium-speed pellet and on the Percoll gradient.
Figure 5.
Localization of Drp1 determined by subcellular fractionation of bovine brain. Lane 1, crude extract; lane 2, postnuclear supernatant (S1); lane 3, medium speed supernatant (S2); lane 4, medium-speed pellet (P2); lane 5, mitochondrial fractions after further purification on a Percoll gradient; lane 6, high-speed supernatant (S3); and lane 7, high-speed pellet (P3). The subcellular fractions were probed with antibody for Drp1, for a cytosolic marker (tubulin), and for mitochondria (39-kDa subunit of OxPhos complex I). The number of volume equivalents needed to load equal amounts of protein (75 μg/lane) was 1× for lane 1, 3× for lane 2 and 3, 43× for lane 4, 21× for lane 5, 3× for lane 6, and 75× for lane 7. Densitometry and adjustment for volume equivalents shows that ∼3% of Drp1 is in the mitochondrial fractions (lanes 4 and 5), 2% is in the high-speed pellet and the rest is in the supernatant. This fractionation experiment was replicated five times, each time giving similar results.
A large part of Drp1 remains in the supernatant, even after high-speed centrifugation, consistent with the cycling of Drp1 on and off of mitochondria. However, ∼2% of Drp1 can be pelleted by high-speed centrifugation. This fraction of Drp1 may not be associated with membranous organelles, such as ER or vesicles, because it could not be solubilized by 1% Triton X-100, even though an ER marker, calnexin, is solubilized under these conditions (our unpublished results). It seems more likely that this fraction of Drp1 can be pelleted because Drp1 can form higher order multimers, like dynamin multimers, which are also pelleted by high-speed centrifugation. In contrast, the 3% of Drp1 that was detected in the medium speed pellet can be solubilized by Triton X-100, suggesting that this fraction is indeed associated with a membranous organelle such as mitochondria (our unpublished results).
GFP::Drp1 Fusion Protein Localizes to Sites of Mitochondrial Division
The movement of Drp1 was tracked in live cells with the use of GFP or YFP fused to Drp1. The expression constructs were transfected into COS-7. The transfected cells were examined within 24 h after transfection. At later time points protein aggregates were formed, presumably because of the exceedingly high expression levels reached in cells that contain large T antigen. Protein aggregates were never observed in transfected HeLa cells, which lack large T antigen. To avoid possible artifacts caused by protein aggregation, further observations were made with cells containing low levels of fusion protein. Cells that were expressing low levels of fusion protein showed a mixture of diffuse cytosolic staining and punctate staining on mitochondria (Figure 6A). The punctate staining pattern was detected with a variety of constructs, including GFP or YFP fused to the N terminus of Drp1 and with YFP fused to the C terminus of Drp1, but not with GFP or YFP alone. Similar distributions were previously observed in yeast and C. elegans (Otsuga et al., 1998; Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999). These fusion proteins are likely to be functional, because it was previously shown that the overexpression of GFP::DRP-1 in C. elegans muscle cells increases the rate of mitochondrial division (Labrousse et al., 1999), whereas a Dnm1::GFP fusion can rescue the mutant phenotype of a Dnm1 deletion in yeast (Sesaki and Jensen, 1999). The distributions of GFP::Drp1 fusions observed here are also consistent with the immunofluorescence results obtained with fixed cells.
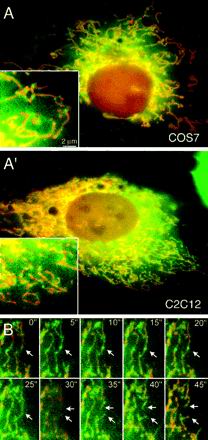
Figure 6.
(A) Localization of a yellow-fluorescent protein (YFP)::Drp1 fusion protein. COS-7 and C2C12 cells were transfected with a YFP::Drp1 construct (green) and a construct encoding cyan-fluorescent protein that was targeted to the mitochondrial matrix (red). Insets show enlargements of peripheral portions of the cells. The images show that Drp1 is largely diffuse throughout the cytosol or localized to spots that are on mitochondria. (B) Time-lapse photography of a C2C12 cell with YFP::Drp1 in spots on mitochondria. C2C12 cells were cotransfected with a YFP::Drp1 construct (green) and a construct encoding cyan-fluorescent protein that was targeted to the mitochondrial matrix (red) and photographed at 5-s intervals. The arrows point to a division event preceded by a YFP::Drp1 spot. The two arrows at later time points indicate the two mitochondrial ends that are formed by mitochondrial division.
To determine whether the subcellular localization of Drp1 is consistent with the proposed function in scission of the mitochondrial membrane, we conducted time-lapse experiments with the use of cells that express YFP::Drp1 (yellow fluorescent protein fused to the N terminus of Drp1) and mito::CFP (cyan fluorescent protein targeted to the mitochondrial matrix with a mitochondrial leader sequence). The time-lapse experiments were limited to ∼20 images before the signal became too weak due to photobleaching. This number was, however, enough to observe several mitochondrial scission events in a single cell. An example of a time-lapse series is shown in Figure 6B. As with the earlier single images, the time-lapse series show many spots of Drp1, evenly distributed along the mitochondria. Each series of time-lapse photographs revealed only a few actual mitochondrial division events. However, these division events were always in a location that had a fluorescent Drp1 spot in preceding images. In our experimental setup, division occurs in ∼5% of the fluorescent spots per hour, but these cells were kept at room temperature during the time lapse series. Cells kept at 37°C presumably have a higher mitochondrial division rate, suggesting that most, if not all, Drp1 spots are primed for division events. Although this has not been verified, we can draw the opposite conclusion, namely, that places of mitochondrial division coincide with Drp1 spots, which is consistent with a direct role of Drp1 in the mitochondrial division process. It therefore seems likely that Drp1 assembles into a multimeric complex on mitochondria before division can occur.
Purified Drp1 Oligomerizes In Vitro
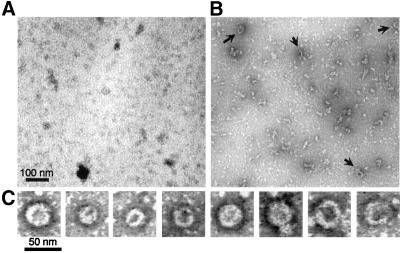
The similar arrangement of protein domains in Drp1 and dynamin suggests that Drp1 might be capable of assembling into a multimeric spiral similar to the spirals formed by dynamin (Hinshaw and Schmid, 1995; Takei et al., 1995). To determine whether Drp1 can form such spirals, full-length protein was made with the baculovirus expression system, purified by Ni-NTA chromatography with the use of a 6-his tag and examined by electron microscopy of negative-stained grids. High and low salt conditions were tested, because these had previously been shown to reversibly induce disassembly and assembly of dynamin spirals (Carr and Hinshaw, 1997). Because dynamin assembly is weakened by GTP and promoted by GDP and aluminum fluoride (Carr and Hinshaw, 1997), we also tested Drp1 assembly under these conditions.
Under high salt conditions (160 mM NaCl) almost all Drp1 was disassembled. The disassembled state was previously shown to be primarily tetrameric (Shin et al., 1999), similar to the soluble tetramers formed by dynamin under high salt conditions (Muhlberg et al., 1997). There was an occasional ring-like structure with a diameter of ∼30–50 nm, similar to the diameter of dynamin spirals (Figure 7A). Decreasing the salt concentration or adding GDP and aluminum fluoride by dialysis increased the numbers of larger structures (Figure 7B). We never observed large numbers of spirals nor did we observe stacks of spirals as previously observed with purified dynamin (Carr and Hinshaw, 1997). The sizes were also not as uniform as those observed with dynamin, even in control experiments in which the two proteins were treated identically (our unpublished results). Some of the structures were elongated as if they were opened up rings. A gallery of ring-like structures is shown in Figure 7C. We conclude that Drp1 can multimerize into ring-like structures with dimensions similar to those of dynamin rings. There must, however, be differences in the assembly properties of dynamin and Drp1, because the structures formed by purified Drp1 were never as uniform, abundant, or extensive as those formed by dynamin. Additional factors on the surface of mitochondria may be required to stimulate Drp1 assembly in vivo. The assemblies made in vitro nevertheless show that Drp1 can form rings or spirals. It seems likely that these spirals wrap around constricted parts of the mitochondrial outer membrane. Taken together, the evidence from this and previous studies suggests that Drp1 spirals contribute to a final stage in the division of the mitochondrial outer membrane (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999).
Figure 7.
In vitro formation of ring-like structures by purified Drp1. (A) Electron micrograph of Drp1 incubated with 150 mM NaCl. (B) Drp1 incubated with GDP and AlF4−. Arrows point to the ring-like structures. (C) Examples of ring-like structures that were observed with GDP and AlF4−. The proteins were detected by negative staining with uranyl acetate.
DISCUSSION
This article addresses the question whether Drp1 controls mitochondrial division in mammalian cells, as it does in yeast and C. elegans. We previously found that mutant mammalian Drp1 disrupts the distribution of mitochondria, but at the time we were unable to determine the underlying cause of this mitochondrial defect (Smirnova et al., 1998). A possible division defect was masked by the formation of tight perinuclear clusters of mitochondria in cells with mutant Drp1. Here we show that mitochondrial clustering can be reversed by nocodazole, thereby exposing the underlying defects caused by mutant Drp1.
Evidence Favoring a Role in Mitochondrial Division
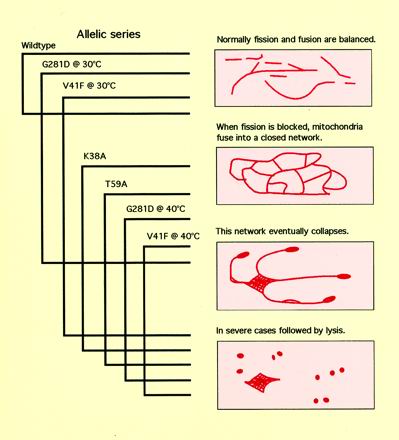
The primary defect caused by mutant Drp1 was investigated with a series of mutations that cause varied mitochondrial phenotypes. These phenotypes can be arranged into the equivalent of an allelic series, as depicted in Figure 8. The primary defect is likely to be the first defect to appear in the most weakly affected cells. The weakest phenotypes were observed with a mutation modeled after the ts1 mutation in the Drosophila shibire gene (van der Bliek and Meyerowitz, 1991). Some of the transfected cells that were grown at a relatively low temperature had wild-type mitochondria, whereas others had more interconnected mitochondria. Stronger effects are induced by increasing the growth temperature or with the use of other mutations in Drp1. In the allelic series, the defects begin with mitochondria becoming more interconnected. The mitochondrial network is likely to result from a shift in the balance between fission and fusion of mitochondria, similar to the formation of closed mitochondrial networks by blocking mitochondrial division in yeast and C. elegans (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999). The network of mitochondria is eventually drawn into a tight net, which is then converted to thick club-shaped mitochondria, before becoming lysed in the most severely affected cells. The fact that increased mitochondrial connectivity was the only defect observed in cells with the weakest mutation in Drp1 points to a primary defect in mitochondrial division.
Figure 8.
Phenotypes caused by mutant Drp1 arranged in an allelic series. The drawings on the right depict the different mitochondrial morphologies that were observed with mutant Drp1. The brackets on the left indicate which morphologies were observed with each mutation. The overlap of phenotypes obtained with different mutations in Drp1 made it possible to rank those phenotypes from weak (a closed network of mitochondria) through strong (a collapsed network) to very strong (cell lysis). The weakest phenotype is most directly linked to the primary defect, indicating that this defect is a block in mitochondrial division. The stronger phenotypes may be due to secondary defects.
Secondary defects, such as the collapse of mitochondria into a perinuclear cluster and their retraction into thick club-shaped structures, may reflect faulty distribution of a fused mitochondrial network throughout the cell. Loosening of the mitochondrial clusters by nocodazole suggests that the clustering defect is microtubule-dependent. There are several possible explanations for this phenomenon. For example, the fused mitochondrial networks might be too bulky to attach to the kinesins that are responsible for anterograde transport of mitochondria (Tanaka et al., 1998). It is even possible that the mitochondrial distribution process depends on a direct interaction with the division machinery. Alternatively, the mitochondrial networks might cluster as part of a general mechanism to dispose of errant organelles, because other defects of the mitochondrial outer membrane also lead to the formation of perinuclear clusters of mitochondria (Yano et al., 1997). However, each of these explanations is consistent with a primary defect in mitochondrial division.
The direct role of Drp1 in mitochondrial division is substantiated by the localization of Drp1 in spots on mitochondrial tubules. The Drp1 spots were detected by immunofluorescence of endogenous Drp1 and with GFP-tagged Drp1. The Drp1 spots were evenly spaced along the lengths of the mitochondrial tubules. Time-lapse photography shows that the Drp1 spots coincide with the locations of actual mitochondrial division events, consistent with a direct role in mitochondrial division. In C. elegans, these spots were shown to assemble before a division event, colocalize with the division event, and then stay attached to one of the newly formed mitochondrial tips. The spots eventually disappear, presumably by being released from the mitochondrial membrane into the cytosol (Labrousse et al., 1999). Unfortunately, the fluorescence in mammalian cells was too weak to detect the cycling of Drp1 between mitochondria and the cytosol. However, both the immunofluorescence and the subcellular fractionation data show a large pool of cytosolic Drp1 and a small fraction of Drp1 on mitochondria, consistent with the cycling of Drp1 onto and off of mitochondria. These localization results together with the mutant phenotypes provide strong evidence that wild-type Drp1 contributes to mitochondrial division in mammalian cells.
Can Our Results Be Reconciled with a Function in Vesicular Transport?
It was previously suggested that Drp1 affects the secretory pathway in mammalian cells (Imoto et al., 1998). However, mitochondrial morphology was not investigated in that study, increasing the possibility that secretion was affected as a secondary consequence of a mitochondrial division defect. Our own studies and those of others were unable to confirm an effect on the secretory or endocytic pathways of mammalian cells (Smirnova et al., 1998; Pitts et al., 1999). The yeast homolog of Drp1, Dnm1p, was also originally thought to affect vesicular traffic (Gammie et al., 1995), but later shown to be specific for mitochondrial division (Bleazard et al., 1999; Sesaki and Jensen, 1999), as was C. elegans DRP-1 (Labrousse et al., 1999). We therefore find it unlikely that Drp1 plays a direct role in the secretory or endocytic pathways.
It has also been suggested that Drp1 mediates membrane or protein flux between ER and mitochondria, either by vesicular transport or by fusion between these two organelles (Pitts et al., 1999). This suggestion was based on the observation that mutant Drp1 not only causes mitochondria to cluster, but also decreases the amount of ER proteins detected by immunofluorescence. As shown in Figure 3, we were not able to confirm this observation, nor is it immediately apparent how a block in transport between ER and mitochondria might affect the amount of resident ER proteins. Perhaps the synthesis of resident ER proteins is down-regulated in response to a mitochondrial defect. It is also possible that ER protein synthesis is indirectly affected by the clustering of mitochondria, because mitochondria may provide a localized source of ATP. The reduced levels of ER proteins may therefore have been a secondary defect arising from a severe block in mitochondrial division.
Along with the debate about the function of Drp1, there has also been disagreement on the localization of Drp1. Our own immunofluorescence results clearly show punctate staining on mitochondria and a background of diffuse cytosolic staining, consistent with the cycling of Drp1 between cytosol and mitochondria. It has, however, also been reported that Drp1 is localized in punctae that only partially colocalize with mitochondria (Yoon et al., 1998; Pitts et al., 1999). These differences in Drp1 localization could be due to differences in the fixation methods. Our determination that Drp1 is localized to mitochondria is confirmed by the results that we obtained with subcellular fractionation and with YFP::Drp1 fusion proteins. This localization is also consistent with the localization of Drp1 homologs in yeast and C. elegans, as determined with immunofluorescence, immuno-electron microscopy, and GFP::Drp1 fusions.
Our subcellular fractionation experiment confirms that Drp1 is largely cytosolic with only 3% of the protein is associated with mitochondria. In addition, ∼ 2% of Drp1 is in the high speed pellet. This latter fraction had previously been observed and was attributed to an association with ER (Yoon et al., 1998). We were, however, unable to solubilize Drp1 in the high speed pellet with Triton X-100, although Triton X-100 did solubilize resident ER proteins in the high speed pellet as well as the mitochondrial fraction of Drp1, which was in the medium speed pellet. The results indicate that the fraction of Drp1 in the high-speed pellet is not membrane bound. Instead, Drp1 might be pelleting as an oligomeric complex. We conclude that Drp1 is associated with mitochondria, but not with other membrane bound organelles. This association is inconsistent with a role in vesicular traffic. It is, however, consistent with a role in mitochondrial division, as was first proposed for yeast and C. elegans homologs and is now proposed for mammalian Drp1.
How Might Drp1 Affect Mitochondrial Division?
The ability of purified Drp1 to assemble into ring-like multimers with dimensions similar to the dimensions of dynamin rings (Hinshaw and Schmid, 1995), suggests that both proteins use similar mechanisms. Spiral-shaped assemblies have also been observed with yet another class of dynamin family members, the MX proteins, which are interferon-induced proteins that inhibit virus infection through an as yet unknown mechanism (Nakayama et al., 1993). The similar arrangement of protein domains in each of these proteins suggests that the ability to assemble into a multimeric spiral is a general property of dynamin family members (van der Bliek, 1999), because those domains that are conserved between dynamin family members may also be the ones that are responsible for spiral formation (Smirnova et al., 1999). The Drp1 spirals presumably wrap around constricted parts of mitochondria where they somehow control severing of the mitochondrial outer membrane. The precise mechanism used by Drp1 is likely to be similar to that of dynamin at the plasma membrane. That mechanism is, however, still a matter of debate. Some experiments suggesting that dynamin is a force-producing enzyme (Sweitzer and Hinshaw, 1998; Marks et al., 2001), whereas others suggest that dynamin is a regulatory GTPase (Sever et al., 1999, 2000), possibly activating another protein that finishes the scission process. It is, nevertheless, apparent that Drp1 and dynamin play critical roles in their respective scission processes.
The analogies between Drp1 and dynamin suggests that the mechanism of mitochondrial outer membrane division may have been derived from the endocytic mechanism upon entry of the first endosymbiont into the host cell (van der Bliek, 2000). Other components of the mitochondrial division apparatus were recently found in yeast. One protein called Mdv1/Fis2/Gag3 is a WD-repeat protein that may serve as an adapter within the Dnm1 protein complex (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000). There are no obvious homologs of this protein in mammals or in C. elegans, suggesting that other binding partners await discovery. The second protein, Fis1/Mdv2, is an integral component of the mitochondrial outer membrane, where it may help target Drp1 to the surface of mitochondria (Mozdy et al., 2000; Tieu and Nunnari, 2000). Other organisms, such as humans and C. elegans, contain possible homologs of Fis1/Mdv2, but their functions have not yet been studied.
Finally, there may also be a mitochondrial inner membrane division machinery waiting to be discovered. Perhaps for technical reasons, we did not observe uncoupling between inner and outer membrane division, as was previously observed when mitochondrial outer membrane division was blocked in C. elegans (Labrousse et al., 1999). There are, however, other instances in mammalian cells in which the mitochondrial inner membrane divides without division of the outer membrane, confirming the existence of a separate inner membrane division apparatus (Duncan et al., 1980). An inner membrane division apparatus may have been introduced by the first endosymbiotic bacteria, because algal mitochondria (Beech et al., 2000) and chloroplasts (Osteryoung et al., 1998) contain homologs of the bacterial division protein ftsZ. However, yeast, C. elegans and mammals do not contain ftsZ homologs, suggesting that other as yet unknown proteins have adopted this function. Once these novel inner membrane division proteins are identified, it will become possible to investigate how divisions of the mitochondrial inner and outer membranes are coordinated.
ACKNOWLEDGMENTS
We thank the other lab members for comments on the manuscript. We thank Dr. Sergey Ryazentsev for assistance with electron microscopy. E.S. was supported by fellowships from the Myasthenia Gravis Foundation and the American Heart Association of Greater Los Angeles. This work was supported by grants from the American Heart Association (965084) and the National Institutes of Health (GM-58166) to A.M.v.d.B.
REFERENCES
- Beech PL, Nheu T, Schultz T, Herbert S, Lithgow T, Gilson PR, McFadden GI. Mitochondrial FtsZ in a chromophyte alga. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr JF, Hinshaw JE. Dynamin assembles into spirals under physiological salt conditions upon the addition of GDP and gamma-phosphate analogues. J Biol Chem. 1997;272:28030–28035. doi: 10.1074/jbc.272.44.28030. [DOI] [PubMed] [Google Scholar]
- Damke H, Baba T, van der Bliek AM, Schmid SL. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CJ, Greenaway HC, Publicover SJ, Rudge MF, Smith JL. Experimental production of “septa”and apparent subdivision of muscle mitochondria. J Bioenerg Biomembr. 1980;12:13–33. doi: 10.1007/BF00745010. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Kim SK. Mechanism of activation of the Caenorhabditis elegans ras homologue let-60 by a novel, temperature-sensitive, gain-of-function mutation. Genetics. 1997;146:553–565. doi: 10.1093/genetics/146.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes P, Shepard KA, Yaffe MP. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Kurihara LJ, Vallee RB, Rose MD. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Shaw JM. Mitochondrial dynamics in yeast. Annu Rev Cell Dev Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Meyer DI. Immunochemical analysis of rough and smooth microsomes from rat liver. Segregation of docking protein in rough membranes. Eur J Biochem. 1985;150:559–564. doi: 10.1111/j.1432-1033.1985.tb09057.x. [DOI] [PubMed] [Google Scholar]
- Imoto M, Tachibana I, Urrutia R. Identification and functional characterization of a novel human protein highly related to the yeast dynamin-like GTPase vps1p. J Cell Sci. 1998;111:1341–1349. doi: 10.1242/jcs.111.10.1341. [DOI] [PubMed] [Google Scholar]
- Kamimoto T, Nagai Y, Onogi H, Muro Y, Wakabayashi T, Hagiwara M. Dymple, a novel dynamin-like high molecular weight GTPase lacking a proline-rich carboxyl-terminal domain in mammalian cells. J Biol Chem. 1998;273:1044–1051. doi: 10.1074/jbc.273.2.1044. [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zapaterra M, Rube DA, van der Bliek AM. C. elegans dynamin-related protein drp-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Marks B, Stowell MH, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Yazaki K, Kusano A, Nagata K, Hanai N, Ishihama A. Structure of mouse Mx1 protein. Molecular assembly and GTP-dependent conformational change. J Biol Chem. 1993;268:15033–15038. [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts KR, Yoon Y, Krueger EW, McNiven MA. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol Biol Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S, Damke H, Schmid SL. Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J Cell Biol. 2000;150:1137–1148. doi: 10.1083/jcb.150.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S, Muhlberg AB, Schmid SL. Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- Shin HW, Shinotsuka C, Torii S, Murakami K, Nakayama K. Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J Biochem. 1997;122:525–530. doi: 10.1093/oxfordjournals.jbchem.a021784. [DOI] [PubMed] [Google Scholar]
- Shin HW, Takatsu H, Mukai H, Munekata E, Murakami K, Nakayama K. Intermolecular and interdomain interactions of a dynamin-related GTP-binding protein, Dnm1p/Vps1p-like protein. J Biol Chem. 1999;274:2780–2785. doi: 10.1074/jbc.274.5.2780. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Newman-Smith ED, Pishvaee B, van der Bliek AM. A model for dynamin self-assembly based on binding between three different protein domains. J Biol Chem. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, van der Bliek AM. Mapping dynamin interdomain interactions through two-hybrid and GST-pulldowns. In: Balch WE, Der CJ, Hall A, editors. Regulators and Effectors of Small GTPases. San Diego: Academic Press; 2000. pp. 468–477. [Google Scholar]
- Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- van der Bliek AM. A mitochondrial division apparatus takes shape. J Cell Biol. 2000;151:F1–F4. doi: 10.1083/jcb.151.2.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- Yano M, Kanazawa M, Terada K, Namchai C, Yamaizumi M, Hanson B, Hoogenraad N, Mori M. Visualization of mitochondrial protein import in cultured mammalian cells with green fluorescent protein and effects of overexpression of the human import receptor Tom20. J Biol Chem. 1997;272:8459–8465. doi: 10.1074/jbc.272.13.8459. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, Dahan S, McNiven MA. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J Cell Biol. 1998;140:779–793. doi: 10.1083/jcb.140.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]