Abstract
When Salmonella is grown in the nutrient-rich lysogeny broth (LB), the AraC-like transcriptional regulator HilD positively controls the expression of genes required for Salmonella invasion of host cells, such as the Salmonella pathogenicity island 1 (SPI-1) genes. However, in minimal media, the two-component system PhoP/Q activates the expression of genes necessary for Salmonella replication inside host cells, such as the SPI-2 genes. Recently, we found that the SL1344_1872 hypothetical gene, located in a S. Typhimurium genomic island, is co-expressed with the SPI-1 genes. In this study we demonstrate that HilD induces indirectly the expression of SL1344_1872 when S. Typhimurium is grown in LB; therefore, we named SL1344_1872 as grhD1 for gene regulated by HilD. Furthermore, we found that PhoP positively controls the expression of grhD1, independently of HilD, when S. Typhimurium is grown in LB or N-minimal medium. Moreover, we demonstrate that the grhD1 gene is required for the invasion of S. Typhimurium into epithelial cells, macrophages and fibroblasts, as well as for the intestinal inflammatory response caused by S. Typhimurium in mice. Thus, our results reveal a novel virulence factor of Salmonella, whose expression is positively and independently controlled by the HilD and PhoP transcriptional regulators.
Introduction
The acquisition of DNA fragments by horizontal transfer events has played a major role in the evolution of pathogenic bacteria. The acquired DNA may encode different factors that confer the ability to survive and replicate in distinct biological niches within an animal or human host, which leads to bacterial infection and disease1,2. To take advantage of the information contained in the acquired DNA, bacteria adapt regulatory mechanisms that allow the expression of the gained genes in those conditions where it is beneficial3.
Salmonella enterica is an important pathogen of humans and animals, causing a mild self-limiting gastroenteritis or a severe systemic infection4. Salmonella enterica serotype Typhimurium (S. Typhimurium) is a major cause of gastroenteritis in humans and several animals; but can also produce a systemic infection in laboratory mice, similar to the typhoid fever produced by S. Typhi in humans4,5. Therefore, S. Typhimurium is widely used as a model in infections to mice, cattle or eukaryotic cell cultures, to investigate the molecular mechanisms governing Salmonella virulence. Most of the virulence genes of Salmonella are grouped in acquired genomic regions called Salmonella pathogenicity islands (SPIs)6–8. SPI-1 and SPI-2 are major determinants for the Salmonella intestinal and systemic infection, respectively8. SPI-1 is present in the two Salmonella species, S. enterica and S. bongori, whereas SPI-2 is only conserved in the S. enterica species, suggesting that SPI-1 was acquired before SPI-2 during Salmonella evolution7,9. SPI-1 and SPI-2 both encode type III secretion systems (T3SSs), their cognate effector proteins, chaperones and transcriptional regulators controlling the expression of the respective genes within each island8,10. During initial infection, Salmonella invades host intestinal epithelium using the SPI-1-encoded T3SS (T3SS-1) and cognate effector proteins, which leads to gastroenteritis; by contrast, the SPI-2-encoded T3SS (T3SS-2) and cognate effector proteins provide to Salmonella the ability to survive and replicate inside epithelial cells and macrophages; within a membrane-bound compartment called Salmonella-containing vacuole (SCV), which leads to the systemic disease4,8. The SPI-2 genes also mediate a Salmonella non-proliferative stage inside phagocytes and non-phagocytic cells11,12 and contribute to the development of the intestinal inflammatory response13–15.
The SPI-1 and SPI-2 genes are expressed in different in vivo niches; the SPI-1 genes are activated when Salmonella is in the intestinal lumen and also in the cytosol of epithelial cells16,17; whereas the SPI-2 genes are activated within the SCV of host cells, such as macrophages, epithelial cells and fibroblasts12,16,18–20. The SPI-2 genes are also expressed in the intestinal lumen21, in the lamina propria or in the underlying mucosa17. In vitro, the SPI-1 genes are induced when Salmonella is grown at early stationary phase in the nutrient-rich lysogeny broth (LB)22–24; in contrast, the SPI-2 genes are induced when Salmonella is grown at late stationary phase in nutrient-rich media, as well as in minimal media containing low concentrations of phosphate, calcium and magnesium19,23–25.
The expression of the SPI-1 genes is controlled by the HilD, InvF and HilA regulators encoded in SPI-1, in a cascade fashion. HilD, an AraC-like transcriptional regulator, induces the expression of HilA, a regulator with an OmpR-ToxR-like DNA binding domain, which in turn activates the expression of InvF, another AraC-like regulator26–31. HilA and InvF activate the expression of the SPI-1 genes encoding the T3SS-1 components and effector proteins, respectively8. HilD also induces the expression of HilA through a feed-forward regulatory loop that it forms with HilC and RtsA30,32, which are AraC-like regulators that bind the DNA sites recognized by HilD33,34; HilC and RtsA are encoded within and outside SPI-1, respectively8. Furthermore, HilD induces, directly or through HilA, InvF or several other regulators, the expression of many horizontally acquired virulence genes located in different islands, as well as ancestral genes including those for flagella biosynthesis and chemotaxis8,23,35–43. Interestingly, HilD is involved in the expression of the ssrAB operon encoding the SsrA/B two-component system, the central positive regulator for the SPI-2 genes, but only when Salmonella is grown in LB23,44. When Salmonella is grown in minimal media, the expression of the ssrAB operon, and thus the SPI-2 genes, is induced by other regulators such as the MarR-like regulator SlyA and the two-component systems OmpR/EnvZ and PhoP/PhoQ, independently of HilD8,45.
The PhoP/PhoQ two-component system is formed by the sensor kinase protein PhoQ and its cognate response regulator PhoP46–48. In response to environmental signals such as acidic pH, low concentration of magnesium and antimicrobial peptides, PhoQ autophosphorylates and then phosphorylates PhoP, which binds to target genes48–53. Orthologous of PhoP/PhoQ are present in several bacteria, controlling the expression of genes for different cellular functions, including virulence, Mg2+ homeostasis, modification of lipopolysaccharides and resistance to antimicrobial peptides and acidic pH19,48,54–59. PhoP directly or indirectly regulates the expression of ∼9% of the S. Typhimurium genome, including the SPI-2 genes, thus having a fundamental role in phisiology and virulence19,41,60–62.
In this study, we show that the transcriptional regulator HilD indirectly induces the expression of the SL1344_1872 hypothetical gene, when S. Typhimurium is grown in LB. Furthermore, we demonstrate that SL1344_1872, here named as grhD1, ‘gene regulated by HilD’, is required for the invasion of S. Typhimurium into host cells and for the intestinal inflammatory response caused by S. Typhimurium in mice. In addition, we found that the response regulator PhoP also positively regulates the expression of grhD1, directly and independently of HilD, in response to different growth conditions. Therefore, our results from this study reveal a novel Salmonella virulence factor, GrhD1, whose expression is controlled by two major transcriptional regulators of Salmonella pathogenicity, HilD and PhoP.
Results
HilD positively regulates the expression of the SL1344_1872 (grhD1) gene
In a previous study, we identified a set of novel genes that are co-expressed with the SPI-1 genes in S. Typhimurium, by an in silico global expression analysis42. The characterization of some of these genes revealed a strong link between the co-expression with SPI-1 and the regulation by HilD. One uncharacterized gene co-expressed with SPI-1 is SL1344_1872, which is located in a S. Typhimurium acquired genomic island flanked by the yecA gene of unknown function and the tRNA-encoding leuZ gene (Fig. 1A). This island contains two additional genes, SL1344_1873 (ecgA) and SL1344_1874, as well as the pseudogene SL1344_1874A; SL1344_1873 and SL1344_1874 encode a peptidoglycan enzyme (EcgA) with L-endopeptidase activity, involved in S. Typhimurium virulence, and a hypothetical membrane protein, respectively (Fig. 1A)63. The SL1344_1872 gene encodes a hypothetical protein of 101 amino acids, predicted to form four α-helices and four β-strands (Fig. 1B). SL1344_1872 has no orthologs in other bacteria and does not present any conserved domain. Recent transcriptomic analysis supports that HilD positively regulates the expression of SL1344_187241.
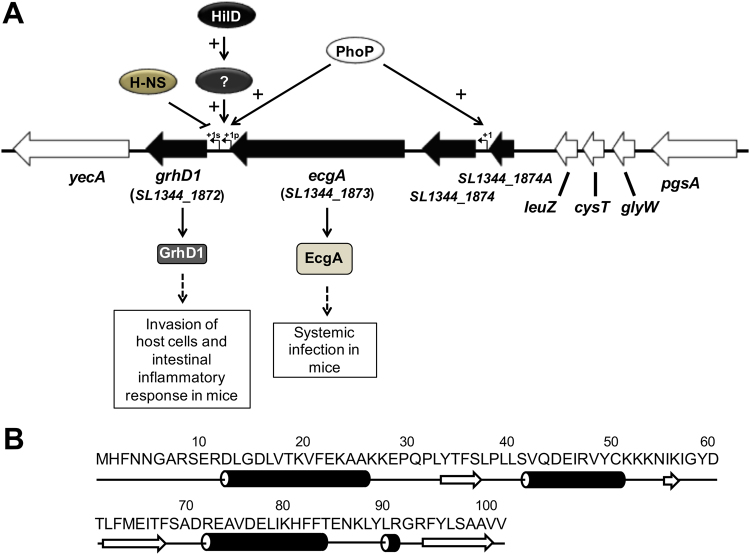
Figure 1.
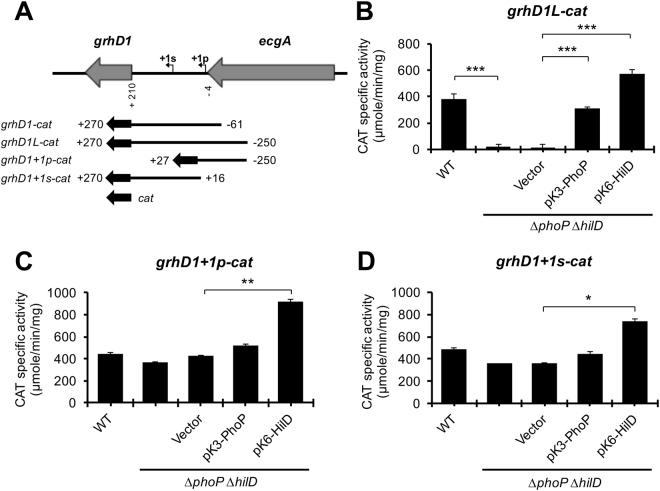
Genetic context, regulatory model and role in virulence of the grhD1 (SL1344_1872) gene of S. Typhimurium, as well as sequence and predictive secondary structure of its product. (A) Schematic representation of the S. Typhimurium SL1344 genome region harbouring grhD1. Arrows indicate coding sequences and lines represent intergenic regions. Black arrows indicate all the genes that are located in the genomic island containing grhD1. Bent arrows represent the primary (+1p) and secondary (+1s) transcription start sites reported in a previous study24. The virulence role for GrhD1 and EcgA, as well as the regulation by HilD, PhoP and H-NS, are also indicated. (B) Amino acid sequence and prediction of the secondary structure prediction of GrhD1. Arrows and cylinders indicate predicted β-strands and α-helices, respectively.
To determine whether SL1344_1872 indeed codes for a protein and to confirm whether its expression is controlled by HilD, the SL1344_1872 chromosomal gene was tagged in the wild-type (WT) S. Typhimurium SL1344 strain and its isogenic ∆hilD mutant, with the sequence encoding a 3XFLAG epitope. Total protein extracts were obtained from culture samples of these strains grown in LB at 37 °C, conditions that induce the expression of the SPI-1 genes, which were analyzed by Western blotting using anti-FLAG antibodies. An expression signal was detected in the WT strain, with the expected size for the SL1344_1872-FLAG protein (Fig. 2A). The expression of SL1344_1872-FLAG was drastically reduced in the ∆hilD mutant; the pK6-HilD plasmid expressing HilD restored the expression of SL1344_1872-FLAG in the ∆hilD mutant to WT levels (Fig. 2A). To investigate whether HilD regulates SL1344_1872 at transcriptional level, a transcriptional fusion of the intergenic region upstream of SL1344_1872 to the cat reporter gene was constructed in the pKK232-8 plasmid. The chloramphenicol acetyl transferase (CAT)-specific activity from this fusion was determined in the WT S. Typhimurium strain and its isogenic ∆hilD mutant, grown in LB. The activity of the SL1344_1872-cat fusion showed a 2-fold decrease in the ∆hilD mutant with respect to the WT strain, and was induced 3-fold in the ∆hilD mutant by the expression of HilD from the pK6-HilD plasmid (Fig. 2B). Together, these results demonstrate that HilD positively regulates the expression of SL1344_1872, herein named grhD1 for gene regulated by HilD.
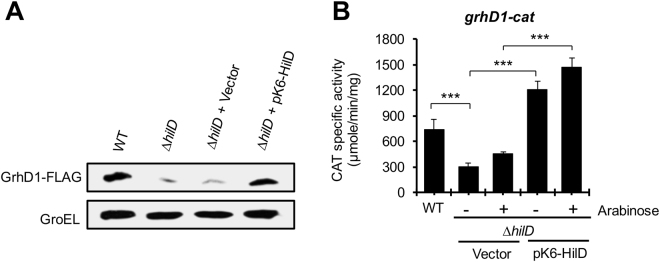
Figure 2.
HilD positively regulates the expression of grhD1 (SL1344_1872). (A) Expression of GrhD1-FLAG in the WT S. Typhimurium SL1344 strain and its derivative ∆hilD mutant containing or not the pK6-HilD plasmid, which expresses HilD from an arabinose-inducible promoter, or containing the pMPM-K6Ω vector. The expression of GrhD1-FLAG was analyzed from samples of bacterial cultures grown in LB at 37 °C by Western blotting, using monoclonal anti-FLAG antibodies. The expression of GroEL was also determined using polyclonal anti-GroEL antibodies, as a loading control. (B) Expression of the grhD1-cat transcriptional fusion contained in the pgrhD1-cat plasmid was determined in the WT S. Typhimurium strain and its derivative ∆hilD mutant carrying or not pK6-HilD or pMPM-K6Ω. Expression of HilD from pK6-HilD was activated by adding 0.001% L-arabinose to the medium. CAT specific activity was determined from samples of bacterial cultures grown in LB at 37 °C. Data are the average of three independent experiments done in duplicate. Bars represent the standard deviations. Statistically different values are indicated (***p < 0.001).
We next investigated whether the control of grhD1 expression by HilD is direct or indirect. Hence, electrophoretic mobility shift assays (EMSAs) were performed using affinity-purified maltose-binding protein (MBP)-HilD and the DNA fragment carrying the intergenic region upstream of grhD1. DNA fragments containing the regulatory regions of hilA or sigD were also tested as positive and negative controls, respectively. As expected, MBP-HilD specifically bound the DNA fragment of hilA, at concentrations of 0.5 to 1 µM; in contrast, at the same concentrations it did not shift the DNA fragment of grhD1, or that of the negative control, sigD (Fig. S1A and B). These results support that HilD regulates grhD1 indirectly; alternatively, an additional factor could be required for HilD binding on grhD1.
Escherichia coli K-12 lacks hilD, hilA and grhD1, as well as around 1400 other genes present in S. Typhimurium. The expression of genes known to be directly controlled by HiD, such as hilA, can be induced in E. coli K-12 when HilD is present42. Therefore, to further test if HilD regulates grhD1 indirectly, the activity of the grhD1-cat fusion was determined in the E. coli MC4100 strain carrying the pK6-HilD plasmid, grown in LB. As a positive control, a hilA-cat transcriptional fusion was also examined. As expected, the activity of both grhD1-cat and hilA-cat fusions was decreased in E. coli MC4100 with respect to the WT S. Typhimurium strain (Fig. S1C and D). Expression of HilD from the pK6-HilD plasmid activated the hilA-cat fusion, but not the grhD1-cat fusion, in E. coli MC4100 (Fig. S1C and D), indicating that an additional Salmonella factor is required for the HilD-mediated expression of grhD1.
HilD induces the expression of several transcriptional regulators, including HilC, HilA and InvF, encoded in SPI-1, as well as RtsA, SsrB and FlhDC, encoded outside SPI-18,23,31,38,40. To investigate whether HilD regulates grhD1 through any of these regulators, the expression of the grhD1-cat fusion was determined in the WT S. Typhimurium strain and its derivative ∆SPI-1 ∆rtsA mutant, as well as in ∆ssrB, ∆flhDC, ∆hilA, and ∆invF mutants, grown in LB. As expected, the grhD1-cat fusion presented a 2-fold-reduced expression in the ∆SPI-1 ∆rtsA mutant, since it lacks HilD; nevertheless, its expression was restored to WT levels in the presence of the pK6-HilD plasmid (Fig. S2A). On the other hand, the grhD1-cat fusion showed similar expression levels in the WT strain and its isogenic ∆ssrB, ∆flhDC, ∆hilA, and ∆invF mutants (Fig. S2B). These results indicate that the expression of grhD1 induced by HilD does not require any other regulator encoded in SPI-1 (HilC, HilA, InvF, SprB), neither RtsA, SsrB or FlhDC, in the growth condition tested.
Collectively, these results show that HilD indirectly induces the expression of grhD1, through a yet-unknown regulator controlled by HilD, found in S. Typhimurium but not in E. coli MC4100.
PhoP positively regulates the expression of grhD1
Recently, we reported that the response regulator PhoP positively and directly controls the expression of the operon containing the ecgA and SL1344_1874 genes, which is located close to grhD1, in the same S. Typhimurium genomic island (Fig. 1A)63. Therefore, we thought that PhoP could also be involved in the expression of grhD1, which is supported by recent transcriptomic analyses41. In order to determine this, we examined the activity of the grhD1-cat transcriptional fusion in the WT S. Typhimurium strain and its derivative ∆phoP mutant, grown in LB or N-minimal medium (N-MM). PhoP is known to be active when S. Typhimurium is grown in the nutrient-rich LB or in minimal media containing low concentrations of magnesium63. As positive and negative controls, the activity of transcriptional fusions to cat reporter of pagK, a gene positively regulated by PhoP, and sirA, a gene not regulated by PhoP, was also tested. The activity of the grhD1-cat, pagK-cat and sirA-cat fusions was higher in N-MM than in LB; however, the expression pattern for each fusion in the different genetic backgrounds tested was similar in both growth conditions (Fig. 3). The activity of the grhD1-cat and pagK-cat fusions was reduced in the ∆phoP mutant, with respect to the WT strain; expression of PhoP from pK3-PhoP recovered the activity of both fusions in the ∆phoP mutant to WT levels (Fig. 3A,B,D and E). In contrast, the activity of the sirA-cat fusion was not affected by the absence or overexpression of PhoP (Fig. 3C and F). To further support these results, we monitored the expression of GrhD1-FLAG in the WT strain and the ∆phoP mutant, grown in LB, intracellular salt medium (ISM) or acidified PCN (phosphate-carbon-nitrogen) medium. In all the conditions tested, the expression of GrhD1-FLAG was severely reduced in the ∆phoP mutant with respect to the WT strain (Fig. S3A and B). As expected, the presence of the pK3-PhoP plasmid restored the expression of GrhD1-FLAG in the ∆phoP mutant to WT levels (Fig. S3B). These results indicate that PhoP positively regulates the expression of grhD1 in S. Typhimurium growing in LB or minimal media.
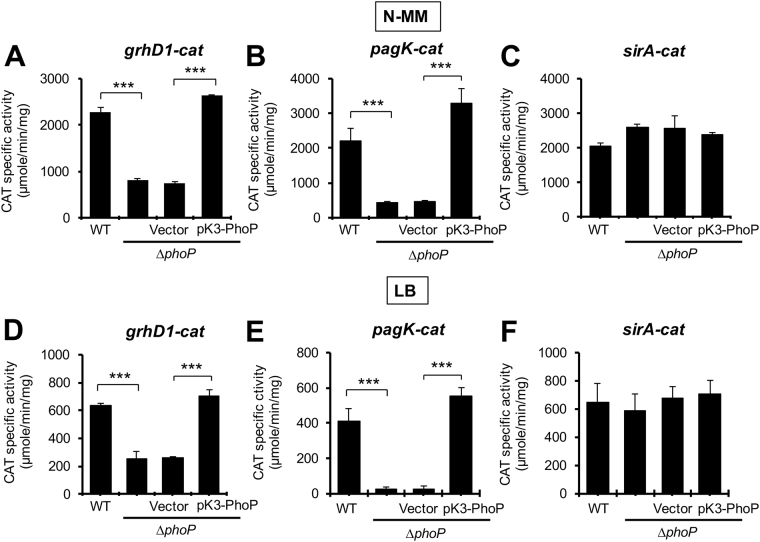
Figure 3.
PhoP actives the expression of grhD1. Expression of the grhD1-cat (A,D), pagK-cat (B,E) and sirA-cat (C,F) transcriptional fusions contained in the pgrhD1-cat, ppagK-cat and psirA-cat plasmids, respectively, was determined in the WT S. Typhimurium strain and its derivative ∆phoP mutant containing or not the pK3-PhoP plasmid or the pMPM-K3 vector, grown in N-MM (A–C) or LB (D–F) at 37 °C. pK3-PhoP constitutively expresses PhoP from a lac derivative promoter. Data are the average of three independent experiments done in duplicate. Bars represent the standard deviations. Statistically different values are indicated (***p < 0.001).
To determine whether PhoP regulates grhD1 directly or indirectly, we performed EMSAs with phosphorylated affinity-purified PhoP-6XHis (PhoP-H6) fusion protein and a labelled DNA fragment containing the regulatory region of grhD1. DNA fragments carrying the regulatory region of the orgB56 or ges64 genes were also tested as positive and negative controls, respectively. PhoP-H6 shifted the grhD1 and orgB fragments starting at a concentration of 3 µM; in contrast, the ges fragment was not shifted even at a concentration of 6 µM (Figs 4A and S4). Specific binding of PhoP-H6 to grhD1 was confirmed by competitive EMSAs (Fig. 4B). Together with the results from expression analyses, these binding assays indicate that PhoP directly regulates the expression of the grhD1 gene.
Figure 4.
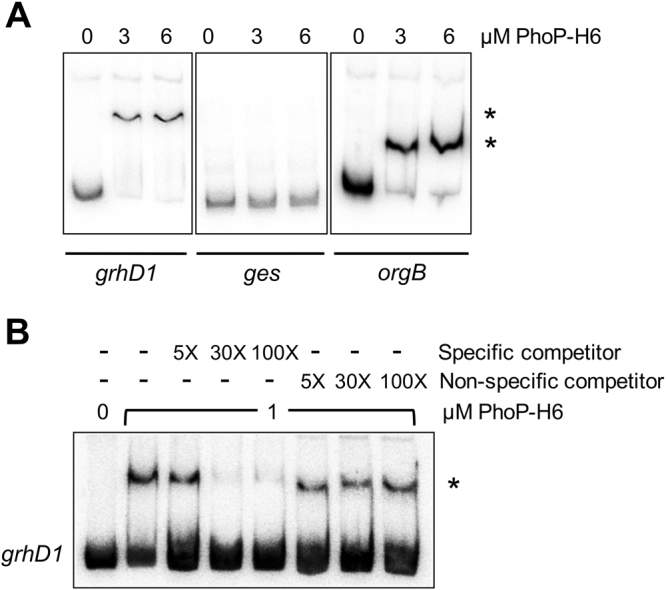
PhoP specifically binds to the grhD1 regulatory region. EMSAs with PhoP-H6 and DNA fragments containing the regulatory regions of the grhD1, ges or orgB genes (A). 32P-5′-end-labelled DNA fragments of the respective gene were incubated with increasing concentrations of purified and phosphorylated PhoP-H6 (0, 3 and 6 µM). The ges and orgB genes were used as negative and positive controls, respectively. PhoP binding to grhD1 was further tested by competitive EMSAs (B). The 32P-5′-end-labelled DNA fragment of grhD1 was incubate with 1 µM of purified and phosphorylated PhoP-H6 in the absence or presence of 5-, 30- and 100-fold excess of unlabelled specific (grhD1) or non-specific (nucA) competitors. The DNA-protein complexes, which are indicated by an asterisk, were resolved in a nondenaturing 8% Tris-borate-EDTA-polyacrylamide gel. After electrophoresis, the gel was dried and analyzed in a Typhoon FLA 7000 IP laser scanner.
HilD and PhoP independently control the expression of grhD1
Our results indicate that both HilD and PhoP positively control the expression of grhD1, which could act independently of each other or one through the other; for instance, since HilD regulates grhD1 indirectly, it could act through PhoP. To investigate this, firstly, the expression of the grhD1-cat fusion was compared in the WT S. Typhimurium strain and its isogenic ∆hilD, ∆phoP and ∆phoP ∆hilD mutants, grown in LB or N-MM. In LB, the grhD1-cat fusion showed a similar 2-fold reduction of its activity in the ∆hilD and ∆phoP mutants with respect to the WT strain, whereas in the ∆phoP ∆hilD double mutant its activity was even 4-fold lower than in the single mutants (Fig. 5A), indicating that HilD and PhoP have an additive effect on grhD1. In contrast, in N-MM, the activity of the grhD1-cat fusion was not affected in the ∆hilD mutant and presented a similar 2.5-fold decrease in the ∆phoP and ∆phoP ∆hilD mutants (Fig. 5B), showing that PhoP regulate grhD1 independently of HilD in these growth conditions. Then, we determined the activity of the grhD1-cat fusion in the ∆phoP ∆hilD double mutant expressing PhoP, HilD and SirA from the pK3-PhoP, pK6-HilD or pK3-SirA plasmids, respectively, grown in LB. SirA is a transcriptional regulator expected to be not involved in the expression of grhD1. Expression of PhoP or HilD, but not SirA, induced the activity of the grhD1-cat fusion in the ∆phoP ∆hilD mutant (Fig. 5C). Together, these results show that PhoP and HilD regulate grhD1 independently of each other; interestingly, the overexpression of one of these regulators compensates the absence of the other for grhD1 expression.
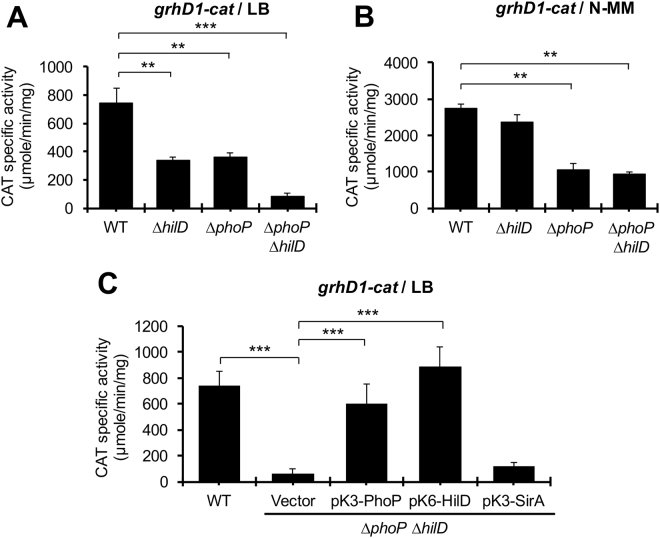
Figure 5.
HilD and PhoP induce the expression of grhD1 independently. Expression of the grhD1-cat transcriptional fusion contained in the pgrhD1-cat plasmid was determined in the WT S. Typhimurium and its derivative ∆hilD, ∆phoP and ∆phoP ∆hilD mutants grown in LB (A) or N-MM (B) at 37 °C, as well as in the WT S. Typhimurium strain and its isogenic ∆phoP ∆hilD mutant carrying or not the pMPM-K3 vector or the pK3-PhoP, pK6-HilD and pK3-SirA plasmids, grown in LB at 37 °C (C). Expression of HilD from pK6-HilD was induced by adding 0.001% L-arabinose to the medium at the beginning of the bacterial cultures. pK3-PhoP and pK3-SirA constitutively express PhoP and SirA, respectively. Data are the average of three independent experiments done in duplicate. Bars represent the standard deviations. Statistically different values are indicated (**p < 0.01; ***p < 0.001).
Previous RNA-sequencing analyses revealed a primary and a secondary transcriptional start site (TSS) in the intergenic region upstream of grhD1, located at 230 and 148 bp from the start codon of grhD1, respectively (Fig. 6A)24. The grhD1-cat fusion tested in the experiments described above carries a DNA region containing these two TSSs. Therefore, to define whether the expression of grhD1 is sustained by two promoters and in that case, whether HilD and PhoP each affects one or both promoters, we constructed three additional grhD1-cat transcriptional fusions. The grhD1L-cat fusion carries an extended 3′ grhD1 upstream region with respect to that contained in the initial assessed grhD1-cat fusion, whereas the grhD1 + 1p-cat and grhD1 + 1s-cat fusions carry segments of the grhD1 upstream region containing only the primary or secondary TSS, respectively (Fig. 6A). The activity of these new constructed fusions was monitored in the WT S. Typhimurium strain and its isogenic ∆phoP ∆hilD double mutant, grown in LB. The grhD1L-cat, grhD1 + 1p-cat and grhD1 + 1s-cat fusions were similarly expressed in the WT strain (Fig. 6B–D), indicating that two independent promoters sustain the expression of grhD1 in these growth conditions. The grhD1L-cat fusion, carrying both promoters, showed a 17-fold-reduced activity in the ΔphoP ΔhilD mutant, which was restored to WT levels or even higher by the expression of PhoP or HilD from the pK3-PhoP or pK6-HilD plasmids, respectively (Fig. 6B). Thus, these results, together with those from Fig. 5C, support that HilD and PhoP independently induce the transcription of both grhD1 promoters. Surprisingly, the activity of the grhD1 + 1p-cat and grhD1 + 1s-cat fusions was not significantly affected in the ∆phoP ∆hilD mutant containing or not the pK3-PhoP plasmid, with respect to the WT strain; only the presence of pK6-HilD further increased the activity of these fusions (Fig. 6C and D). These results suggest that proper control of grhD1 expression requires negative regulatory sequences located around the promoters; in the absence of these sequences the expression of grhD1 becomes independent of HilD and PhoP.
Figure 6.
Two different promoters regulated by HilD and PhoP induce the expression of grhD1. Schematic representation of the intergenic region upstream grhD1 (A). The primary (+1p) and secondary (+1s) transcription start sites of grhD1, previously reported24, are indicated by a bent arrow. The DNA fragments carried by the grhD1-cat, grhD1L-cat, grhD1 + 1p-cat and grhD1 + 1s-cat transcriptional fusions, are shown; positions are indicated with respect to the primary transcriptional start site of grhD1. Expression of the grhD1L-cat (B), grhD1 + 1p-cat (C), and grhD1 + 1s-cat (D) transcriptional fusions contained in the pgrhD1L-cat, pgrhD1 + 1p-cat and pgrhD1 + 1s-cat plasmids, respectively, was tested in the WT S. Typhimurium strain and its isogenic ∆phoP ∆hilD mutant carrying or not the pK3-PhoP or pK6-HilD plasmids, or the pMPM-K3 vector. CAT specific activity was determined from samples of bacterial cultures grown in LB at 37 °C. Expression of HilD from pK6-HilD was induced by adding 0.001% L-arabinose to the medium at the beginning of the bacterial cultures. pK3-PhoP constitutively expresses PhoP. Data are the average of three independent experiments done in duplicate. Bars represent the standard deviations. Statistically different values are indicated (*p < 0.05; **p < 0.01; ***p < 0.001).
H-NS represses the expression of grhD1
The histone-like protein H-NS works as a global transcriptional regulator that silences the expression of genes acquired by Salmonella65,66. We investigated whether the negative regulatory sequences on grhD1 could mediate repression by H-NS. Given that the hns null mutation generates severe growth defects in Salmonella66, we analyzed the effect of H-NS on grhD1 by overexpressing the H-NSG113D mutant, which does not have DNA-binding activity but still forms heterodimers with WT H-NS monomers67 and thus acts as a dominant negative mutant68. The activity of the grhD1-cat fusion was tested in the ∆hilD and ∆phoP mutants containing the pT6-HNS-G113D plasmid expressing H-NSG113D, as well as in the WT strain. The expression of H-NSG113D induced the activity of the grhD1-cat fusion in the ∆hilD mutant, but not in the ∆phoP mutant (Fig. S5A). These results indicate that with the inactivation of H-NS the expression of grhD1 becomes independent of HilD, thus revealing that H-NS represses grhD1.
To determine whether H-NS regulates grhD1 directly or indirectly, we performed EMSAs with the affinity-purified H-NS-3XFLAG-6XHis (H-NS-FH) protein and a fragment containing the grhD1 regulatory region. DNA fragments containing the regulatory regions of ssrAB or sigD were also tested as positive and negative controls, respectively. H-NS-FH bound the DNA fragments of grhD1 and ssrAB, starting at a concentration of 0.5 and 0.25 µM, respectively; as expected, it did not bind the negative control sigD (Fig. S5B and C). Together with the expression analyses, these binding assays demonstrate that H-NS directly represses the expression of grhD1.
GrhD1 is required for invasion of S. Typhimurium into host cells
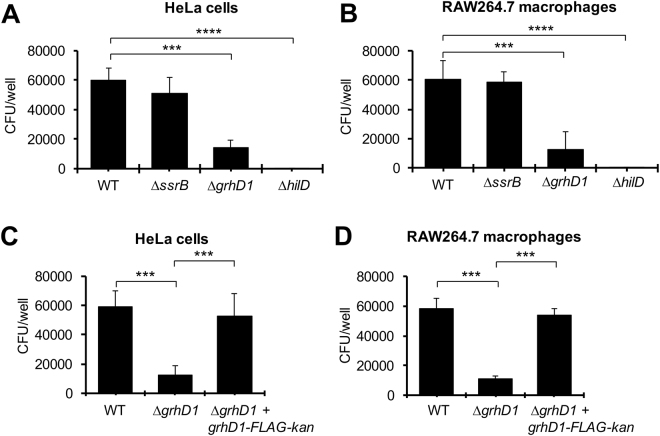
HilD controls the expression of a high number of genes mainly required for Salmonella invasion of host cells8,40–43. Therefore, we investigated whether the grhD1 gene, also regulated by HilD, is involved in this virulence phenotype. Gentamicin protection assays were used to analyze the bacterial invasion of the WT S. Typhimurium strain and its derivative ∆grhD1 mutant into HeLa cells and RAW264.7 mouse macrophages. The ∆hilD and ∆ssrB mutants were also assessed as positive and negative controls, respectively. The ∆grhD1 mutant showed a 4-fold reduction in the invasion of both HeLa cells and macrophages in comparison to the WT strain (Fig. 7A and B). As expected, the ∆ssrB mutant was not affected in the invasion phenotype and the ∆hilD mutant was unable to invade the HeLa cells and macrophages (Fig. 7A and B). The invasion defect of the ∆grhD1 mutant was also observed in NRK-49F fibroblasts using two different initial infection times (10 and 20 min) (Fig. S6). A centrifugation step can be used in the invasion assays to accelerate the contact between bacteria and eukaryotic cells, which overcomes an invasion deficiency by a motility defect69. Even with the centrifugation step the ΔgrhD1 mutant showed a reduced invasion phenotype in HeLa cells, compared to the WT strain (Fig. S7A); in contrast, the ΔflhDC mutant, which lacks of motility, greatly recovered its ability to invade HeLa cells with the centrifugation step (Fig. S7B). Next, we sought to complement the invasion phenotype of the ∆grhD1 mutant with a plasmid expressing GrhD1. For this, we constructed the low-copy number pK3-GrhD1 and pK3-GrhD1-FLAG plasmids, which constitutively express GrhD1 and GrhD1-FLAG proteins, respectively, from a constitutive lac promoter. Unexpectedly, the pK3-GrhD1 and pK3-GrhD1-FLAG plasmids further decreased the invasion of the ∆grhD1 mutant to HeLa cells; moreover, these plasmids also drastically inhibited the invasion of the WT strain (Fig. S8A). To further explore this phenomenon, we monitored by Western blot the expression of the GrhD1-FLAG protein from the grhD1::3XFLAG gene located in the chromosome and that carried by the pK3-GrhD1-FLAG plasmid. As shown in Fig. S8B, GrhD1-FLAG reached much higher levels from the pK3-GrhD1-FLAG plasmid that from the chromosomal gene. Together, these results support that both the absence and overexpression of GrhD1 negatively affects the S. Typhimurium invasion of host cells independently of motility.
Figure 7.
GrhD is required for invasion of S. Typhimurium into HeLa cells and macrophages. Epithelial HeLa cells (A) and murine RAW 264.7 macrophages (B) were infected with the WT S. Typhimurium strain or its isogenic ∆ssrB, ∆grhD1 and ∆hilD mutants. Epithelial HeLa cells (C) and murine RAW 264.7 macrophages (D) were infected with the WT S. Typhimurium strain and its isogenic ∆grhD1 and ∆grhD1 + grhD1-FLAG-kan mutants. Invasion was measured by enumerating the intracellular CFUs at 1 h post-infection, using a gentamicin protection assay. Data are the average of three independent experiments done in triplicate. Bars represent the standard deviations. Statistically different values are indicated (***p < 0.001; ****p < 0.0001).
Since a specific concentration of GrhD1 seems to be required for its role in the S. Typhimurium invasion of host cells, we decided to complement the ∆grhD1 mutant by inserting the grhD1::3XFLAG allele into the chromosome (Fig. S9). Expression of GrhD1-FLAG was similar in the complemented ∆grhD1 + grhD1-FLAG-kan strain and the WT strain carrying the grhD1::3XFLAG allele (Fig. S8C). Accordingly, the complemented ∆grhD1 + grhD1-FLAG-kan strain presented an invasion phenotype similar to that of the WT strain, in both HeLa cells and macrophages (Fig. 7C and D). These results confirm that GrhD1 is an additional S. Typhimurium factor required for invasion of host cells.
We also investigated whether grhD1 have a role during survival/replication of Salmonella inside host cells. For this, we infected RAW264.7 macrophages and HeLa cells with the WT S. Typhimurium strain and its isogenic ∆grhD1 mutant, as well as with the ∆ssrB mutant, used as a positive control. As expected, the ∆ssrB mutant presented an affected replication/survival ability, which was more evident in macrophages that in HeLa cells; in contrast, the replication/survival phenotype of the ∆grhD1 mutant was very similar to that showed by the WT strain, in both macrophages and HeLa cells (Fig. S10A and B).
In all, these results indicate that grhD1 is necessary for invasion of S. Typhimurium into host cells, but not for its intracellular replication/survival.
GrhD1 is required for the intestinal inflammatory response induced by S. Typhimurium in mice
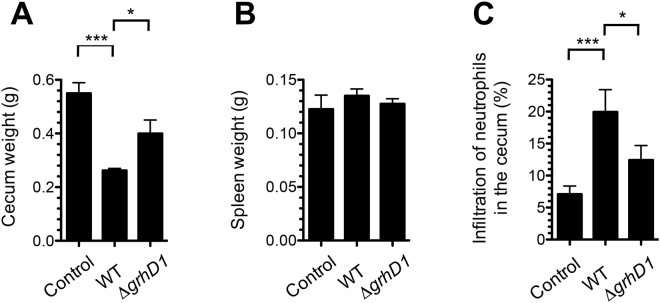
Salmonella invasion of intestinal epithelial cells eventually leads to the development of enterocolitis8. To determine whether GrhD1 is necessary for the induction of intestinal inflammation, we analyzed the infection caused by the WT S. Typhimurium strain and the ∆grhD1 mutant in streptomycin-pretreated mice, which is used as a S. Typhimurium colitis model70. As expected, the WT strain was able to colonize the intestine (cecum and ileum) and the spleen of mice (Fig. S11). Furthermore, the mice infected with the WT strain showed a reduced cecum weight and an increased infiltration of neutrophils in the cecum content (Fig. 8A and C), two hallmarks of the intestinal inflammatory response induced by S. Typhimurium in streptomycin-pretreated mice70. Interestingly, the ∆grhD1 mutant colonized the cecum similarly to the WT strain (Fig. S11); however, the mice infected with the ∆grhD1 mutant showed a significant higher weight of the cecum and a significant lower infiltration of neutrophils in the cecum content, than those infected with the WT strain (Fig. 8A and C). Additionally, the ∆grhD1 mutant showed a 3-fold reduction in the colonization of the ileum, with respect to the WT strain (Fig. S11). In contrast, the ∆grhD1 mutant and the WT strain colonized similarly the spleen (Fig. S11) and the mice infected with these strains showed a similar weight of the spleen (Fig. 8B). These results show that GrhD1 is involved in the intestinal infection, particularly in the intestinal inflammatory response induced by S. Typhimurium in the streptomycin mouse model.
Figure 8.
GrhD1 is involved in the intestinal inflammatory response induced by S. Typhimurium in mice. Mice pretreated with streptomycin were infected with the WT S. Typhimurium strain or the ΔgrhD1 mutant, or treated with sterile 1X PBS (Control). The mice were sacrificed two days post-infection and the total weight (g) of the cecum (A) and the spleen (B), as well as the infiltration of neutrophils in the cecum content (C), was measured for each mouse. Samples of the cecum content were analyzed by microcopy to determine the number of neutrophils with respect to the total eukaryotic cells, which is expressed as a percentage score. Data are the average from four separate animals. Bars represent the standard deviations. Statistically different values are indicated (*p < 0.05; ***p < 0.001).
Discussion
HilD and PhoP are two major transcriptional regulators controlling the expression of virulence genes in Salmonella8. Our results reveal one additional virulence gene, grhD1, which is regulated by HilD and PhoP. When S. Typhimurium proliferates in conditions that favor expression of the SPI-1 and other genes required for the Salmonella invasion of host cells, both HilD and PhoP are required for the expression of grhD1. We show that HilD and PhoP independently affect two promoters located upstream grhD1; PhoP directly and HilD through an additional factor that remains to be identified. Furthermore, our data also show that the histone-like protein H-NS directly represses the expression of grhD1; H-NS silences the expression of many other genes acquired by Salmonella65,66. A model for the regulation of the grhD1 expression is depicted in Fig. 1A.
HilD and PhoP also positively and independently regulate the expression of the orgBC SPI-1 operon, in SPI-1-inducing growth conditions, PhoP directly and HilD through HilA8,56. This operon codes for a cytoplasmic protein required for invasion and an effector protein secreted through the T3SS-1, OrgB and OrgC, respectively71,72. Opposite regulation mediated by HilD and PhoP on the hilA gene, encoding a master regulator for the SPI-1 genes, has also been reported26,29,73,74. Additionally, HilD and PhoP positively and independently regulate the expression of the slrP gene, in SPI-1-inducing growth conditions, PhoP directly and HilD by an unknown mechanism37. The slrP gene encodes a virulence effector protein that is translocated into macrophages through both T3SS-1 and T3SS-225. The overlap between the HilD and PhoP regulons extends to several other genes, as revealed by recent transcriptomic analyses41 and by our results (unpublished).
We found that GrhD1 is required for the invasion of S. Typhimurium to host cells; interestingly, both the absence or overexpression of GrhD1 inhibit the invasion phenotype. Consistently, we show that GrhD1 is involved in the intestinal inflammatory response induced by S. Typhimurium in streptomycin-pretreated mice. Many other genes regulated by HilD, such as the SPI-1 genes, required for the invasion of host cells, also contribute to the induction of intestinal inflammatory response8,14,15,75. Our results indicate that GrhD1 is not secreted when S. Typhimurium is grown in SPI-1-inducing growth conditions (Fig. S12); on the other hand, the absence of GrhD1 does not affect the typical SPI-1-mediated protein secretion profile or has a significant effect on motility (Fig. S13), which are all factors involved in the Salmonella invasion of host cells. Therefore, the specific role of GrhD1 for invasion remains to be defined, which is a matter of our current investigation.
During the growth of S. Typhimurium in minimal media, which favor expression of the SPI-2 and other genes required for the Salmonella replication inside host cells, the expression of grhD1 requires PhoP, but not HilD. In SPI-2-inducing growth conditions, PhoP also positively controls transcription of the orgBC operon and the slrP gene independently of HilD37,56. In addition to PhoP, SlyA and possibly other regulators are also involved in the expression of grhD1 in SPI-2-inducing growth conditions, as revealed by transcriptomic analyses41 and confirmed by our results (Fig. S3A). PhoP and SlyA induce expression of virulence genes when Salmonella is within host cells8; consistently, the grhD1 gene is expressed inside macrophages20,76. Our results indicate that the grhD1 gene is not necessary for the replication of S. Typhimurium inside HeLa cells and RAW264.7 mouse macrophages. Furthermore, our results show that grhD1 is not required for the colonization of the spleen of streptomycin-pretreated mice, which suggests that it is not involved in the systemic infection caused by S. Typhimurium. PhoP positively regulates the ecgA-SL1344_1874 operon, located close to the grhD1 gene, in the same genomic island; this operon codes for EcgA, a peptidoglycan D,L-endopeptidase that contributes to systemic infection in mice, but it is not required for the invasion of or replication within HeLa cells of S. Typhimurium63.
Our results indicate that PhoP and SlyA are required for the expression of the grhD1 gene in both SPI-1-inducing and SPI-2-inducing growth conditions, which could be explained by the reciprocal positive regulation between PhoP and SlyA that has been demonstrated in Salmonella77–80.
Our findings further expand the HilD, PhoP and SlyA regulons, provide additional evidence on the overlap between these virulence regulons, and reveal a novel virulence factor of Salmonella.
Methods
Bacterial strains and growth conditions
Bacterial strains used in this work are listed in Table S1. Bacterial cultures were grown at 37 °C in nutrient-rich lysogeny broth (LB), N-minimal medium (N-MM), phosphate-carbon-nitrogen (PCN) minimal medium or intracellular salts medium (ISM) as described previously81,82. When needed, the antibiotics ampicillin (200 µg/ml), streptomycin (100 µg/ml), kanamycin (20 µg/ml) or tetracycline (10 µg/ml) were added to the media. The chloramphenicol acetyltransferase (CAT) assays were performed as we described previously23,83.
Construction of plasmids
Tables S1 and S2 indicate the plasmids and primers used in this study, respectively. To construct the plasmids containing the transcriptional fusions pgrhD1-cat, pgrhD1L-cat, pgrhD1 + 1p-cat and pgrhD1 + 1s-cat, different segments of the upstream region of grhD1 were amplified by PCR with the combination of primers 1872FW-1/1872RV-2, 1872FW-1/1872Rv-3, 1872FW-1/1872Rv + 1s, and 1872Fw + 1p/1872Rv-3, respectively. The generated PCR products were digested with SalI and HindIII restriction enzymes, purified and cloned into the vector pKK232-8, which carries a cat reporter gene lacking the promoter (Amersham Pharmacia LKB Biotechnology), digested with the same restriction enzymes. To construct the ppagK-cat plasmid, the upstream region of pagK was amplified by PCR with the primers pagKyM-Fw and pagKyM-Rv. This PCR product was digested with BamHI and HindIII restriction enzymes, purified and cloned into the vector pKK232-8 digested with the same restriction enzymes. To construct the pK3-PhoP plasmid, the phoP gene was amplified by PCR using the primers PhoP-RV11 and PhoPFW22. This PCR product was digested with BamHI and HindIII restriction enzymes, purified and ligated into the pMPM-K3 vector84 digested with the same restriction enzymes. The pK3-PhoP plasmid expresses PhoP from the vector lac promoter. To construct the pBAD-H-NS-FH plasmid, the hns gene was amplified by PCR using the primers HNS-NcoI and Flag-His, and chromosomal DNA from the EPEC E2348/69 hns::3xFLAG-kan strain (V.H. Bustamante, unpublished) as template. This PCR product was digested with NcoI and HindIII restriction enzymes, purified and cloned into the vector pBADMycHisA digested with the same restriction enzymes. The pBAD-H-NS-FH plasmid expresses H-NS fused to 3XFLAG and 6XHis (H-NS-FH) from an arabinose-inducible promoter. To construct the pT6-H-NS-G113D plasmid, the G113D hns mutant allele was amplified by PCR using the primers hns-Nco and hns-22R and chromosomal DNA from the E. coli HM52 strain67 as template. The resulting PCR product was digested with NcoI and HindIII, purified and ligated into the pMPM-T6Ω vector84 digested with the same restriction enzymes. The pT6-HNS-G113D plasmid expresses H-NSG113D under an arabinose-inducible promoter. To construct the pK3-GrhD1 and pK3-GrhD1-FLAG plasmids, the grhD1 gene was amplified by PCR using the primers 1872Fw-K3 and 1872Rv-K3, and chromosomal DNA from the WT and DTM106 (grhD1::3XFLAG) S. Typhimurium strains, respectively, as template. The PCR products were digested with KpnI and SacI restriction enzymes, purified and ligated into the pMPM-K3 vector digested with the same restriction enzymes. Plasmids pK3-GrhD1 and pK3-GrhD1-FLAG express GrhD1 and GrhD1-FLAG, respectively, from the vector lac promoter. To construct the p2795-GrhD1-FLAG plasmid, the grhD1 gene was amplified by PCR using the primers 1872-SalIFw and 1872Rv-3, and chromosomal DNA from the S. Typhimurium DTM106 strain (grhD1::3XFLAG) as template. The PCR products were digested with BamHI and SalI restriction enzymes, purified and ligated into the p2795 vector85 digested with the same restriction enzymes.
Chloramphenicol acetyltransferase (CAT) assays
The CAT specific activity was determined as described previously86.
Construction of deletion mutant strains and strains expressing FLAG-tagged proteins
The grhD1 and phoP genes were replaced with a selectable kanamycin resistance cassette in the S. Typhimurium SL1344 strain by the λRed recombinase system, as reported previously87, thus generating the DTM101 (∆grhD1::kan) and DTM103 (∆phoP::kan) strains. The chromosomal grhD1 and avrA genes were FLAG-tagged in the S. Typhimurium SL1344 strain, using a previously reported method based on the λRed recombinase system88, generating the DTM107 (grhD1::3XFLAG-kan) and DTM113 (avrA::3XFLAG-kan) strains. P22 transduction was used to transfer the ∆hilD::kan allele from JPTM5 into DTM104, generating the DTM105 strain, and to transfer the grhD1::3XFLAG-kan allele from DTM107 into JPTM25, DTM104, SV4198 and SV4235, generating the DTM109, DTM111, MD3883 and MD3870 strains, respectively. The kanamycin resistance cassette was excised from the DTM101, DTM103, DTM105, DTM107, DTM109 and DTM111 strains, by using helper plasmid pCP20, expressing the FLP recombinase, as described previously87, generating the DTM102, DTM104, DTM106, DTM108, DTM110 and DTM112 strains, respectively. The complemented DTM114 strain (∆grhD1 + grhD1::3XFLAG-kan) was generated by inserting in cis the grhD1::3XFLAG-kan into the chromosome of the DTM102 strain (∆grhD1), using a previously reported method based on the λRed recombinase system85 and the p2795-GrhD1-FLAG plasmid. All modified strains were verified by PCR amplification and sequencing.
Western blotting
Whole-cell extracts were prepared from samples of bacterial cultures and analyzed by Western blot as described previously82. Antibodies anti-FLAG M2 (Sigma) or anti-GroEL (StressGen) were used at 1:2 000 and 1:100 000 dilutions, respectively. The secondary antibodies Horseradish peroxidase-conjugated anti-mouse or anti-rabbit (Pierce) were used at a dilution of 1:10 000. Reaction bands on membranes were developed with the Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer) and the exposition to Kodak X-Omat films.
Protein secretion analysis
Protein secretion assays were performed as described previously83.
Expression and purification of MBP-HilD
The maltose binding protein (MBP)-HilD was expressed and purified from E. coli BL21/DE3 containing the pMAL-HilD1 plasmid, using an amylose affinity column, as described previously23.
Expression and purification of PhoP-H6
The His-tagged fusion protein PhoP-H6 was expressed in E. coli BL21/DE3 carrying the pPB1020 plasmid and purified by using a Ni2+-NTA-agarose affinity column, as described previously89.
Expression and purification of H-NS-FH
The His-tagged fusion protein H-NS-FH was expressed in E. coli BL21/DE3 containing the pBAD-H-NS-FH plasmid and purified by using a Ni2+-NTA-agarose affinity column, as described previously23.
Electrophoretic mobility shift assays (EMSAs)
For EMSAs with MBP-HilD or H-NS-FH, the upstream regions of hilA, sigD, grhD1 and ssrAB were amplified by PCR using the combination of primers hilA1FBamHI/hilA2RHindIII, SigDBHIF/SigDH3R, 1872Fw-1/1872Rv-2 (or 1872Fw-1/1872Rv-3) and SsaBFBglII/SsrBRS6E, respectively. The generated PCR products were purified with the QIAquick PCR purification kit (Qiagen). Binding reactions were performed by mixing each PCR product (≈100 ng) with increasing concentrations of purified MBP-HilD or H-NS-FH in binding buffer containing 10 mM Tris-HCl (pH 8), 50 mM KCl, 1 mM DTT, 0.5 mM EDTA, 5% glycerol and 10 µg/ml bovine serum albumin (BSA), in a total volume of 20 µl. These reactions were incubated at room temperature for 20 min and then analyzed by electrophoresis on 6% nondenaturing acrylamide gels ran with 0.5X Tris-borate-EDTA buffer, at room temperature. The DNA fragments were visualized by staining with ethidium bromide, in an Alpha-Imager UV transilluminator (Alpha Innotech Corp.).
For EMSAs with PhoP-H6, the primers that anneal to the coding strand of the promoters analyzed were labeled with T4 polynucleotide kinase and [γ-32P] ATP. The promoter regions of grhD1, orgB, ges, and nucA were amplified by PCR using primer pairs stm1939 Fwd/stm1939 rv, orgB PE 3/PROM 2869, ges1/ges2, and nucA FW/nuclease RV, respectively. Approximately 6 fmol of labeled promoter region DNA in a 20-µl volume was incubated at room temperature for 30 min with the indicated amounts of purified PhoP-H6 protein, which was previously phosphorylated by incubation for 3 h at 25 °C with 25 mM acetyl phosphate as reported90. The binding buffer used for protein-DNA incubations contained 20 mM Tris-HCl (pH 7.4), 50 mM KCl, 5 mM MgCl2, 10% glycerol, and 25 µg/ml BSA. Samples were separated in an 8% nondenaturing Tris-borate-EDTA–polyacrylamide gel at room temperature. After electrophoresis, the gel was dried and analyzed in a Typhoon FLA 7000 IP laser scanner.
Motility assays
Salmonella strains were grown overnight at 37 °C with appropriate antibiotics. Then, the strains were sub-cultured 1:100 in fresh LB and grown at 37 °C with shaking until an OD600 ∼ 1. At this point, 1 µl of each culture was spotted onto LB 0.3% agar plates and allowed to dry for 3 min at room temperature. Plates were incubated for 7 h at 37 °C and the diameter of the motility haloes was measured.
Cell infection assays
Invasion of HeLa cells or RAW264.7 macrophages was tested by gentamicin assays as previously described69,82. Monolayers of HeLa cells or RAW264.7 macrophages were infected at a multiplicity of infection (MOI) of 40:1 and 10:1 (bacteria to eukaryotic cells), respectively. In some experiments, monolayers were centrifuged at 1000 g for 10 min immediately after addition of the bacteria and then incubated for 10 min at 37 °C. To test the intracellular replication/survival, the monolayers of HeLa cells or RAW264.7 macrophages were further incubated with DMEM containing 10 µg/ml gentamicin up to the indicated times. After removing the DMEM, the cells were lysed at 1, 4, 8, and 16 h post-infection in 1 ml or 200 µl of 0.2% (w/v) sodium deoxycholate in 1X PBS for HeLa cells and RAW264.7 macrophages, respectively. Serial dilutions of the cell lysates were plated onto LB agar containing streptomycin at 100 µg/ml. To evaluate invasion, the CFUs were counted at 1 h post-infection; to test intracellular replication/survival, the CFUs were enumerated at 4, 8, or 16 h post-infection. Fold-replication represents the CFUs recovered at 4, 8, or 16 h relative to the CFUs at 1 h post-infection.
The fibroblast cell line NRK-49F (ATCC CRL-1570) of rat origin, were propagated in DMEM containing 10% (v/v) fetal bovine serum, as described previously91. For the invasion assay, bacteria were grown at 37 °C in static non-aerated cultures obtained upon inoculation of 2 ml of LB with a bacterial colony and subsequent overnight incubation (final OD600 ~ 1.0). Infection was carried out for either 10 or 20 min using a MOI of 10:1, as previously described91. After extensive washing, fibroblasts were incubated in fresh tissue culture medium containing 100 µg/ml gentamicin up to 2 h post-infection. At that time, fibroblasts were lysed in 1X PBS pH 7.4, 1% (v/v) Triton X-100. Number of viable intracellular bacteria was determined by plating.
Mouse infection experiments
Animal manipulation in this work was carried out according to the standard and operating protocols approved by the Internal Committee for Animal Care and Use from CICUAL-UNAM, and by the Official Mexican Norm NOM-062Z00-1999. Pathogen-free BALB/c mice (6- to 7-week-old) were obtained from the Experimental Medicine Research Unit, School of Medicine, UNAM, Mexico. Groups of four animals were maintained in different ventilated cages. Water and food were withdrawn 4 h before treatment of mice with 50 mg of streptomycin by orogastric administration; then, animals were supplied with water and food ad libitum. For infection, overnight cultures of the Salmonella strains were diluted 1:100 in 5 ml of fresh LB and incubated at 37 °C with shaking for 3 h. After 24 h of the streptomycin treatment, water and food were withdrawn again for 4 h before the infection of mice with 50 µl of a bacterial suspension containing 1 × 108 CFUs/ml in 1X PBS, or the administration of 50 µl of sterile 1X PBS (control). Thereafter, drinking water and food were offered to the animals ad libitum. At 48 h post-infection, the animals were euthanatized with an overdose of the anesthetics
Ketamine plus Xylazine administrated intraperitoneally, in a workstation hood (Thermo-Scientific). The spleen, cecum and terminal ileum were aseptically removed, weighed and homogenized in 1 ml of sterile and cold 1X PBS. To evaluate infiltration of neutrophils, samples of the cecum contents were analyzed by microscopy to determine the number of neutrophils and total eukaryotic cells from 15 different fields, using the Diff-Quick staining method and an inverted microscope Nikon TE300 (objective 60X). To analyze bacterial colonization, 2% Triton X-100 was added to the organ homogenizates and CFUs were determined by plating serial dilutions of the obtained cell lysates onto LB agar containing streptomycin at 100 µg/ml.
Statistical analysis
Data were analyzed with the GraphPad Prism 5.0 software (GraphPad Inc., San Diego, CA) using two-tailed Student’s t-test. P values of <0.05 were considered significant.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the Dirección General de Asuntos del Personal Académico de la UNAM/México (IN203415) and from the Consejo Nacional de Ciencia y Tecnología (CONACYT)/México (254531) to V.H.B., from the Spanish Ministry of Economy, Industry and Competitiveness and European Regional Development Funds (FEDER) [BIO2013-46281-P and BIO2016-77639-P-(MINECO-FEDER)] to F.G.-dP., and from CONACYT/México (256263) to M.A.dC., F.C.S. is a career investigator of CONICET and of the Rosario National University Research Council (CIUNR). M.M.B. was supported by a pre-doctoral fellowship from CONACYT (403748). R.M.D. was supported by a master fellowship from CONACYT (367095). C.L. is a fellow of the National Scientific and Technical Research Council (CONICET). We thank D. Pérez-Morales for constructing the pagK-cat plasmid and for help during cell infection assays; A. Vázquez for constructing the DTM103 strain; F.J. Santana for constructing the pK3-PhoP plasmid; C. Zavala-Alvarado for purifying the H-NS-FH protein; M. Hensel for providing the p2795 vector; F.J. Santana, M. Fernández-Mora, D. Vences-Vences, A. López-Huerta and M. Martínez-Parrilla for technical assistance; I. Martínez-Flores for critical reading of the manuscript; as well as J.C.D. Hinton and J.L. Puente for sharing results from transcriptomics before publication.
Author Contributions
V.H.B., F.G.-dP., F.C.S., and M.A.dC. designed research; M.M.B., C.L., R.M., G.R.-P., P.G., R.R.-R., and M.A.dC. performed experimental research; M.M.B., C.L., R.M., G.R.-P., P.G., R.R.-R., M.A.dC., F.C.S., F.G.-dP., and V.H.B. analyzed data; and M.M.B. and V.H.B. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-018-25327-6.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23068-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/11/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
- 1.Ochman, H., Lawrence, J. G. & Groisman, E. A. Lateral gene transfer and the nature of bacterial innovation. Nature405, 10.1038/35012500 (2000). [DOI] [PubMed]
- 2.Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ilyas B, Tsai CN, Coombes BK. Evolution of Salmonella-host cell interactions through a dynamic bacterial genome. Front Cell Infect Microbiol. 2017;7:428. doi: 10.3389/fcimb.2017.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nature Rev Microbiol. 2008;6:53. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 5.Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu Rev Med. 2001;52:259–274. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- 6.Marcus SL, Brumell JH, Pfeifer CG, Finlay BB. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2000;2:145–156. doi: 10.1016/S1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 7.Porwollik, S. & McClelland, M. Lateral gene transfer in Salmonella. Microbes Infect5, 977–989 (2003). [DOI] [PubMed]
- 8.Fàbrega A, Vila J. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groisman EA, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 10.Hansen-Wester I, Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–559. doi: 10.1016/S1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 11.Grant AJ, et al. Attenuated Salmonella Typhimurium lacking the pathogenicity island-2 type 3 secretion system grow to high bacterial numbers inside phagocytes in mice. PLoS Pathog. 2012;8:e1003070. doi: 10.1371/journal.ppat.1003070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Núñez-Hernández C, Alonso A, Pucciarelli MG, Casadesús J, García-del Portillo F. Dormant intracellular Salmonella enterica serovar Typhimurium discriminates among Salmonella pathogenicity island 2 effectors to persist inside fibroblasts. Infect Immun. 2014;82:221–232. doi: 10.1128/IAI.01304-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bispham J, Tripathi B, Watson P, Wallis T. Salmonella Pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect Immun. 2001;69:367–377. doi: 10.1128/IAI.69.1.367-377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun. 2005;73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombes BK, et al. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun. 2005;73:7161–7169. doi: 10.1128/IAI.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knodler LA, et al. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci USA. 2010;107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laughlin RC, et al. Spatial segregation of virulence gene expression during acute enteric infection with Salmonella enterica serovar Typhimurium. MBio. 2014;5:e00946–00913. doi: 10.1128/mBio.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage‐dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 19.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 20.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 21.Brown NF, et al. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathog. 2005;1:e32. doi: 10.1371/journal.ppat.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J Bacteriol. 1999;181:3433–3437. doi: 10.1128/jb.181.11.3433-3437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bustamante VH, et al. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci USA. 2008;105:14591–14596. doi: 10.1073/pnas.0801205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kröger C, et al. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Miao EA, Miller SI. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schechter LM, Damrauer SM, Lee CA. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 27.Lostroh CP, Bajaj V, Lee CA. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI‐1. Mol Microbiol. 2000;37:300–315. doi: 10.1046/j.1365-2958.2000.01991.x. [DOI] [PubMed] [Google Scholar]
- 28.Lostroh CP, Lee CA. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of P prgH from Salmonella pathogenicity island 1. J Bacteriol. 2001;183:4876–4885. doi: 10.1128/JB.183.16.4876-4885.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schechter LM, Lee CA. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol Microbiol. 2001;40:1289–1299. doi: 10.1046/j.1365-2958.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- 30.Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2005;57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 31.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics. 2012;190:79–90. doi: 10.1534/genetics.111.132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saini S, Ellermeier JR, Slauch JM, Rao CV. The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog. 2010;6:e1001025. doi: 10.1371/journal.ppat.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olekhnovich IN, Kadner RJ. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:4148–4160. doi: 10.1128/JB.184.15.4148-4160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olekhnovich IN, Kadner RJ. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol. 2007;189:6882–6890. doi: 10.1128/JB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thijs IM, et al. Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J Bacteriol. 2007;189:4587–4596. doi: 10.1128/JB.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Main-Hester KL, Colpitts KM, Thomas GA, Fang FC, Libby SJ. Coordinate regulation of Salmonella pathogenicity island 1 (SPI1) and SPI4 in Salmonella enterica serovar Typhimurium. Infec Immun. 2008;76:1024–1035. doi: 10.1128/IAI.01224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordero-Alba M, Ramos-Morales F. Patterns of expression and translocation of the ubiquitin ligase SlrP in Salmonella enterica serovar Typhimurium. J Bacteriol. 2014;196:3912–3922. doi: 10.1128/JB.02158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mouslim C, Hughes KT. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog. 2014;10:e1003987. doi: 10.1371/journal.ppat.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrone BL, Stringer AM, Wade JT. Identification of HilD-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2014;196:1094–1101. doi: 10.1128/JB.01449-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer HM, Kühne C, Deditius JA, Hughes KT, Erhardt M. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J Bacteriol. 2014;196:1448–1457. doi: 10.1128/JB.01438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colgan AM, et al. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 2016;12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Flores I, et al. In silico clustering of Salmonella global gene expression data reveals novel genes co-regulated with the SPI-1 virulence genes through HilD. Sci Rep. 2016;6:37858. doi: 10.1038/srep37858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith C, Stringer AM, Mao C, Palumbo MJ, Wade JT. Mapping the regulatory network for Salmonella enterica serovar Typhimurium invasion. MBio. 2016;7:e01024–01016. doi: 10.1128/mBio.01024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martínez LC, Banda MM, Fernández-Mora M, Santana FJ, Bustamante VH. HilD induces expression of Salmonella pathogenicity island 2 genes by displacing the global negative regulator H-NS from ssrAB. J Bacteriol. 2014;196:3746–3755. doi: 10.1128/JB.01799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fass E, Groisman EA. Control of Salmonella pathogenicity island-2 gene expression. Curr Opin Microbiol. 2009;12:199–204. doi: 10.1016/j.mib.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groisman EA, Chiao E, Lipps CJ, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci USA. 1989;86:7077–7081. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prost LR, et al. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Bader MW, et al. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol. 2003;50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 51.Bader MW, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 52.Prost LR, Miller SI. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Microbiol. 2008;10:576–582. doi: 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 53.Véscovi EG, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 54.Groisman EA. The ins and outs of virulence gene expression: Mg2+ as a regulatory signal. Bioessays. 1998;20:96–101. doi: 10.1002/(SICI)1521-1878(199801)20:1<96::AID-BIES13>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 55.Soncini FC, Vescovi EG, Solomon F, Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aguirre A, et al. PhoP-induced genes within Salmonella pathogenicity island 1. J Bacteriol. 2006;188:6889–6898. doi: 10.1128/JB.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2000;68:6139–6146. doi: 10.1128/IAI.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L, et al. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 59.Ernst RK, Guina T, Miller SI. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 2001;3:1327–1334. doi: 10.1016/S1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- 60.Adams P, et al. Proteomic detection of PhoPQ‐and acid‐mediated repression of Salmonella motility. Proteomics. 2001;1:597–607. doi: 10.1002/1615-9861(200104)1:4<597::AID-PROT597>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 61.Thompson JA, Liu M, Helaine S, Holden DW. Contribution of the PhoP/Q regulon to survival and replication of Salmonella enterica serovar Typhimurium in macrophages. Microbiology. 2011;157:2084–2093. doi: 10.1099/mic.0.048926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu J-L, Guo L. Quantitative proteomic analysis of Salmonella enterica serovar Typhimurium under PhoP/PhoQ activation conditions. J Proteome Res. 2011;10:2992–3002. doi: 10.1021/pr101177g. [DOI] [PubMed] [Google Scholar]
- 63.Rico‐Pérez G, et al. A novel peptidoglycan D, L‐endopeptidase induced by Salmonella inside eukaryotic cells contributes to virulence. Mol Microbiol. 2016;99:546–556. doi: 10.1111/mmi.13248. [DOI] [PubMed] [Google Scholar]
- 64.Pontel LB, Audero MEP, Espariz M, Checa SK, Soncini FC. GoIS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol Microbiol. 2007;66:814–825. doi: 10.1111/j.1365-2958.2007.05963.x. [DOI] [PubMed] [Google Scholar]
- 65.Lucchini S, et al. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navarre WW, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 67.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 68.De la Cruz, M. A. et al. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol66, 727–743 (2007). [DOI] [PubMed]
- 69.Ibarra JA, et al. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella–host cell interactions in vitro. Microbiology. 2010;156:1120–1133. doi: 10.1099/mic.0.032896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barthel M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klein JR, Fahlen TF, Jones BD. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect Immun. 2000;68:3368–3376. doi: 10.1128/IAI.68.6.3368-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Day JB, Lee CA. Secretion of the orgC gene product by Salmonella enterica serovar Typhimurium. Infect Immun. 2003;71:6680–6685. doi: 10.1128/IAI.71.11.6680-6685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 74.Baxter MA, Jones BD. Two-component regulators control hilA expression by controlling fimZ and hilE expression within Salmonella enterica serovar Typhimurium. Infect Immun. 2015;83:978–985. doi: 10.1128/IAI.02506-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hapfelmeier S, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol. 2005;174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 76.Srikumar S, et al. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 2015;11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Norte VA, Stapleton MR, Green J. PhoP-responsive expression of the Salmonella enterica serovar Typhimurium slyA gene. J Bacteriol. 2003;185:3508–3514. doi: 10.1128/JB.185.12.3508-3514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Navarre WW, et al. Co‐regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005;56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 79.Song H, et al. Modulation of the regulatory activity of bacterial two-component systems by SlyA. J Biol Chem. 2008;283:28158–28168. doi: 10.1074/jbc.M801058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao G, Weatherspoon N, Kong W, Curtiss R, Shi Y. A dual-signal regulatory circuit activates transcription of a set of divergent operons in Salmonella typhimurium. Proc Natl Acad Sci USA. 2008;105:20924–20929. doi: 10.1073/pnas.0807071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Headley VL, Payne SM. Differential protein expression by Shigella flexneri in intracellular and extracellular environments. Proc Natl Acad Sci USA. 1990;87:4179–4183. doi: 10.1073/pnas.87.11.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pérez-Morales D, et al. The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella. PLOS Pathog. 2017;13:e1006497. doi: 10.1371/journal.ppat.1006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez LC, et al. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol. 2011;80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mayer MP. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163:41–46. doi: 10.1016/0378-1119(95)00389-N. [DOI] [PubMed] [Google Scholar]
- 85.Husseiny MI, Hensel M. Rapid method for the construction of Salmonella enterica serovar Typhimurium vaccine carrier strains. Infect Immun. 2005;73:1598–1605. doi: 10.1128/IAI.73.3.1598-1605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puente JL, Bieber D, Ramer SW, Murray W, Schoolnik GK. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 87.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Castelli, Ma. E., Véscovi, E. G. A. & Soncini, F. C. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J Biol Chem275, 22948–22954, 10.1074/jbc.M909335199 (2000). [DOI] [PubMed]
- 90.Lejona S, et al. PhoP can activate its target genes in a PhoQ-independent manner. J Bacteriol. 2004;186:2476–2480. doi: 10.1128/JB.186.8.2476-2480.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aiastui A, Pucciarelli MG, García-del Portillo F. Salmonella enterica serovar Typhimurium invades fibroblasts by multiple routes differing from the entry into epithelial cells. Infect Immun. 2010;78:2700–2713. doi: 10.1128/IAI.01389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.