Abstract
A 62-year-old man who had undergone a primary knee arthroplasty 3 years earlier, presented to the emergency department with an infected prosthesis. He underwent prosthesis resection. All cultures failed to identify the infecting organism. Analysis of the intraoperative samples by next-generation sequencing revealed Streptococcus canis (an organism that resides in the oral cavity of dogs). It was later discovered that the patient had sustained a dog scratch injury several days earlier. The patient reports that his dog had licked the scratch. Treatment was delivered based on the sensitivity of S. canis, and the patient has since undergone reimplantation arthroplasty.
Keywords: Periprosthetic joint infection, Hip arthroplasty, Knee arthroplasty, Culture negative, Next-generation sequencing
Introduction
The diagnosis of periprosthetic joint infection (PJI) continues to challenge the medical community. The lack of a “gold standard” test compels clinicians to rely on several tests, none of which have absolute accuracy. Despite efforts to improve the diagnosis of PJI, such as those proposed by the International Consensus Meeting [1], some challenges have proven insurmountable. Culture-negative PJI (CN-PJI) in particular, is one such issue, as the inability to isolate the infecting organism using conventional culture may cast doubt over the diagnosis, and cause uncertainty regarding optimal treatment [2]. The inability to isolate an organism leaves patients at the mercy of empiric antimicrobial therapy, and the potential failure to cover the infecting organism, thereby jeopardizing the outcome of treatment. There is currently no clear protocol for the management of patients with CN-PJI. Current recommendations state that patients with CN-PJI should receive antimicrobials covering the most common pathogens for PJI without committing to a specific protocol [3]. The fact that PJI can be caused by fungi and atypical organisms leaves infectious disease specialists to “best guess” the antimicrobial regimen for CN-PJI patients. This can lead to the true infecting organisms not being covered or indeed the administration of unnecessary antimicrobials, thus compromising the outcome of treatment and imparting adverse effects on patients. Considering the fact that the incidence of CN-PJI can reach 50% in some studies [4], [5], [6], [7], [8], this clinical situation is encountered commonly. Without knowing the infective organism, clinicians are unable to effectively monitor patients' response to treatment and determine if the infection has been controlled. In addition, some patients could suffer the psychological trauma of whether a joint infection existed in the first place that necessitated multiple surgical procedures and long-term antimicrobial treatment. This situation is not acceptable in modern-day medicine and further innovations are needed to address this issue in orthopaedics and other medical fields involving the use of implants and biofilm formation.
This case highlights the challenge of CN-PJI and provides an encouraging endorsement for the potential of molecular diagnostics, such as next-generation sequencing (NGS), in identifying infecting organisms in PJI. It further corroborates the belief that organisms from sources such as pets can result in PJI.
Informed consent: The patient provided written consent that data concerning the case would be submitted for publication.
Case history
A 62-year-old male was transferred to our institution after presenting to an outside hospital with an infected left total knee arthroplasty and systemic sepsis. The patient had undergone an uncomplicated total knee arthroplasty 3 years earlier. On arrival at the outside institution, aspiration of the knee was performed. The analysis of the synovial fluid revealed a nucleated cell count of 63,700 cells/μL with a 90% neutrophil differential. The serological markers of infection were elevated with an erythrocyte sedimentation rate of 82 mm/h and a C-reactive protein of 6.2 mg/dL. The result of laboratory investigations and clinical examination led the physicians at the outside institution to reach a diagnosis of PJI. At the family's request, he was transferred to our institution for definitive care of the infected knee after stabilization. On his arrival at our institution, the left knee was found to be swollen, warm, with erythema of the overlying skin, and painful range of motion. Radiographs showed soft tissue swelling, with arthroplasty components in good position, and no evidence of fracture or loosening (Fig. 1). In light of the clinical evaluation and laboratory investigations, which met the International Consensus Meeting criteria for the diagnosis of PJI [1], the patient was scheduled to undergo resection of the infected knee. Subsequently, the patient developed hypotension, hypoxia, and atrial fibrillation, thus necessitating intubation. In view of the systemic illness, the patient was started on empiric therapy with intravenous piperacillin/tazobactam and vancomycin before surgery. Synovial fluid obtained intraoperatively was sent for analysis, including culture, in addition to performing the leukocyte esterase test in the operating room, which produced a trace result [9]. At the time of debridement, tissue samples were also obtained from the knee, and sent for routine culture and NGS at PathoGenius Laboratory (Lubbock, TX). Samples obtained for culture and NGS included synovial fluid, synovium, and tissue from the femur and tibia. All 4 samples were sent for aerobic and anaerobic bacterial, fungal, and acid-fast cultures. Bacterial cultures were held for 14 days, whereas fungal and acid-fast cultures were held for 29 and 44 days, respectively.
Figure 1.
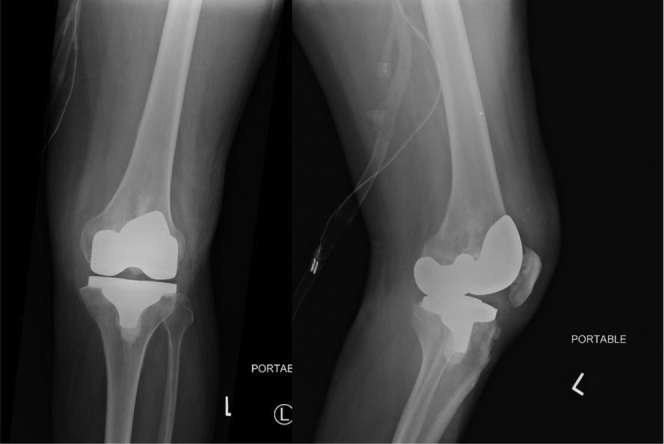
Preoperative anteroposterior and lateral radiographs.
Culture results of the synovial fluid at the outside institution, the synovial fluid obtained at our institution, and the periarticular tissues of the infected knee retrieved intraoperatively were all culture negative. Although the patient received systemic antibiotics on arrival at our institution, which is known to result in the inability to isolate the infective agent in PJI [10], the synovial fluid aspiration at the outside institution was obtained before administration of any antibiotics. Blood cultures obtained both at our institution and the referring institution, which were taken before antibiotic administration, were also negative.
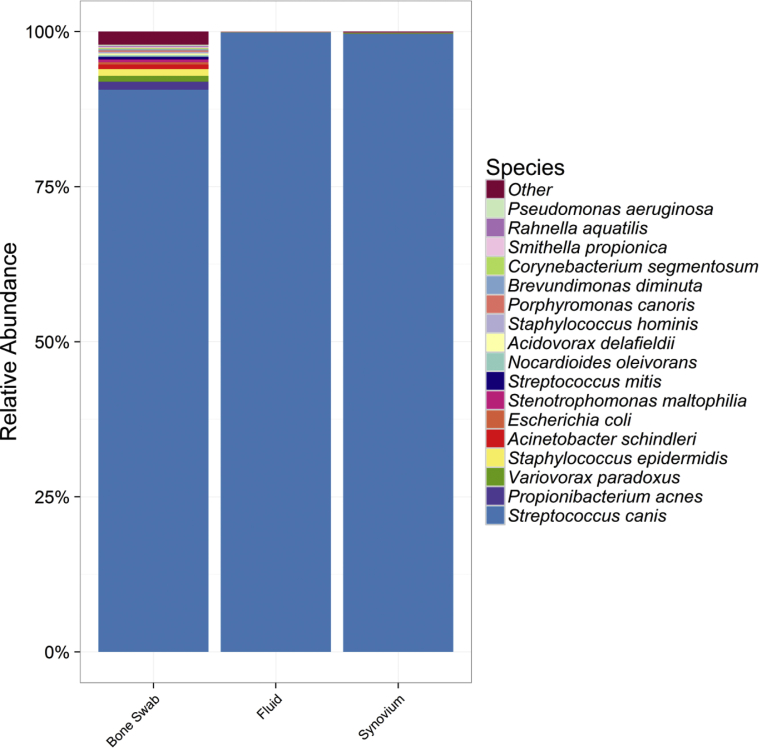
NGS analysis of the fluid and periarticular tissues all revealed Streptococcus canis, an oral pathogen in dogs, cats, and cattle (Fig. 2). It was later discovered that the patient had sustained a scratch from his pet dog, and was continuously licked by the dog, several days before developing the knee infection.
Figure 2.
DNA sequencing report detailing the microbes present in each sample.
NGS is a well-established molecular technique that involves amplification of microbial DNA using polymerase chain reaction (PCR) and subsequent sequencing of all the amplicons. The test is performed in 2 steps, with the first being a PCR to amplify the sequence of interest and subsequently sequencing the amplicons from that PCR. In this specific assay, the rRNA gene is amplified and sequenced. The 2 regions of interest for detection of bacterial and fungal species are the 16S and internal transcribed spacer sequences, respectively [11], [12]. These 2 sequences are both highly conserved and variable regions of the rRNA gene, allowing for specific microbial identification.
Based on NGS results, S. canis was found to be sensitive to vancomycin, and hence piperacillin/tazobactam was discontinued. It should be noted that the sensitivities provided by the results of NGS assay are projections based on the presence of antibacterial resistance genes and not a “true” sensitivity determined based on the growth of the organism. Because of his systemic sepsis, the patient had a difficult postoperative recovery, which was complicated by pneumonia, placement of a tracheostomy after failing to wean off ventilation, and was finally discharged to a rehabilitation facility. The patient completed a 6-week course of intravenous vancomycin followed by a 2-week antibiotic holiday, with monitoring of erythrocyte sedimentation rate and C-reactive protein. We do not routinely aspirate patients before reimplantation at our institution. At the time of reimplantation, synovial fluid and tissue samples were obtained and sent for culture and NGS in a similar fashion to that performed at the first stage. Both NGS and culture from the reimplantation procedure were negative. At his 6-month follow-up visit, the patient was walking with a cane and did not show any clinical signs of infection.
Discussion
Culture-negative infections in general, and PJI in particular, continue to challenge the medical community. Infections associated with implants, such as prosthetic joints, are known to exist as biofilms, which cannot easily be identified using conventional culture. Although numerous strategies for improving the yield of culture have been proposed, including withholding of antibiotics before taking culture samples [2], culturing synovial fluid in blood culture bottles [13] and holding cultures for longer periods, inability to isolate the infecting organism associated with implants is common. The incidence of CN-PJI at our institution is currently 28%. This is in line with the reported rate of CN-PJI, which can range between 27% and 55% [4], [5], [6], [7], [8].
There are numerous issues with the use of conventional cultures in modern medicine. Based on the recommendations of the Infectious Disease Society of America and the American Society for Microbiology [14], samples obtained for culture need to be transported in a specific fashion, and processed within 2 hours, which is difficult to implement in clinical practice. The conventional culture methods that were developed in the late 19th century also rely on a medium to grow the infecting organism. Although the latter may be possible with acute infections, most chronic infections, particularly those associated with biofilms, are difficult to grow using conventional culture [15]. In addition, the use of selective media may allow for preferential growth of one organism, that may not be the true pathogen, whereas suppressing the growth of other organisms. The shortfalls of conventional culture, particularly in relation to PJI, call for innovative techniques to overcome the high rate of culture-negative infections and also explore the possibility that infections in general, and PJI in particular, may be polymicrobial in nature.
Several culture-independent methods have been studied to address the aforementioned issue. The use of PCR for the diagnosis of PJI was first described at our institution in 1996 [16]. Numerous other studies have since evaluated the role of conventional and other PCR techniques [5], [7], [17], [18]. One of the issues related to the use of molecular techniques relates to the potential for isolation of a “contaminating” organism that is not a true pathogen [19]. A prior study raised concerns regarding the issue of false-positive results that seemed to occur commonly using multiplex PCR [20]. In that study, tissue samples obtained from 7 patients undergoing primary knee replacement with no prior surgery in the affected knee, 5 of which revealed an organism. The other issue with conventional or multiplex PCR is the need to know the profile of potential pathogens to allow for the design of primers for amplification. The technique, thus, fails to identify organisms that were not included in the amplification panel [21]. As demonstrated by this case, as well as those in the literature, any organism may be a potential pathogen for PJI, and hence may be missed by molecular techniques such as PCR.
One of the key issues facing PCR, as well as other molecular methods, is determining the material that contains pathogen DNA while minimizing amplification of DNA from contaminant organisms. The data regarding this is conflicting. A study by Rak et al., in which a multiplex PCR assay was used to compare the yield of tissue to sonication fluid, demonstrated that sonication fluid was superior to PCR in isolation of the “infecting” organism [22]. In contrast, a meta-analysis by Qu et al. concluded that tissue samples to be a better source for isolating the infecting organism and the least likely to be affected by previous treatment [23].
NGS is a well-established technique for amplification and sequencing of DNA material from any source. With the advances being made in the field of genomics and the declining expense associated with genomic analysis, NGS appears to be an appealing molecular technique in identifying infecting organisms. NGS is being applied with increasing frequency to determine the causative agents in infections. Perhaps isolation of leptospirosis using NGS, and the dramatic subsequent improvement in care, of a 14-year-old comatose boy in whom the infective agent could not be isolated by the culture is a testimony to the need for better methods of pathogen isolation [24]. An ongoing study to evaluate the role of NGS is being conducted at our institution, with encouraging preliminary results. Another potential application for NGS is polymicrobial PJI. Given the quantitative results the test can provide, better insight can be obtained into polymicrobial infections. Treatment of polymicrobial infection has been shown to have lower success rates when compared with monomicrobial infection [25], and a better understanding of these infections is needed to determine if they are truly polymicrobial in nature, or rather an infection with a dominant organism with other organisms acting in concert.
Summary
With all the challenges facing the medical community in isolating the infecting organism, we believe it is necessary to embrace and further refine molecular diagnostic techniques to improve their accuracy and better define their role. This case highlights the role that NGS may play a role in isolation of the infecting organism in a patient with PJI. This case report also highlights that a pathogen causing PJI may originate from a pet.
Footnotes
One or more of the authors of this paper have disclosed potential or pertinent conflicts of interest, which may include receipt of payment, either direct or indirect, institutional support, or association with an entity in the biomedical field which may be perceived to have potential conflict of interest with this work. For full disclosure statements refer to https://doi.org/10.1016/j.artd.2017.08.005.
Appendix A. Supplementary data
References
- 1.Parvizi J., Gehrke T. International Consensus Group on Periprosthetic Joint Infection. Definition of periprosthetic joint infection. J Arthroplasty. 2014;29:1331. doi: 10.1016/j.arth.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Parvizi J., Erkocak O.F., Della Valle C.J. Culture-negative periprosthetic joint infection. J Bone Joint Surg Am. 2014;96:430. doi: 10.2106/JBJS.L.01793. [DOI] [PubMed] [Google Scholar]
- 3.Restrepo C., Schmitt S., Backstein D. Antibiotic treatment and timing of reimplantation. J Arthroplasty. 2014;29:104. doi: 10.1016/j.arth.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 4.Baré J., MacDonald S.J., Bourne R.B. Preoperative evaluations in revision total knee arthroplasty. Clin Orthop. 2006;446:40. doi: 10.1097/01.blo.0000218727.14097.d5. [DOI] [PubMed] [Google Scholar]
- 5.Gallo J., Kolar M., Dendis M. Culture and PCR analysis of joint fluid in the diagnosis of prosthetic joint infection. New Microbiol. 2008;31:97. [PubMed] [Google Scholar]
- 6.Spangehl M.J., Masri B.A., O'Connell J.X., Duncan C.P. Prospective analysis of preoperative and intraoperative investigations for the diagnosis of infection at the sites of two hundred and two revision total hip arthroplasties. J Bone Joint Surg Am. 1999;81:672. doi: 10.2106/00004623-199905000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Gomez E., Cazanave C., Cunningham S.A. Prosthetic joint infection diagnosis using broad-range PCR of biofilms dislodged from knee and hip arthroplasty surfaces using sonication. J Clin Microbiol. 2012;50:3501. doi: 10.1128/JCM.00834-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shanmugasundaram S., Ricciardi B.F., Briggs T.W.R., Sussmann P.S., Bostrom M.P. Evaluation and management of periprosthetic joint infection - an International, Multicenter Study. HSS J. 2014;10:36. doi: 10.1007/s11420-013-9366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parvizi J., Jacovides C., Antoci V., Ghanem E. Diagnosis of periprosthetic joint infection: the utility of a simple yet unappreciated enzyme. J Bone Joint Surg Am. 2011;93:2242. doi: 10.2106/JBJS.J.01413. [DOI] [PubMed] [Google Scholar]
- 10.Parvizi J., Ghanem E., Menashe S., Barrack R.L., Bauer T.W. Periprosthetic infection: what are the diagnostic challenges? J Bone Joint Surg Am. 2006;88(Suppl 4):138. doi: 10.2106/JBJS.F.00609. [DOI] [PubMed] [Google Scholar]
- 11.Clarridge J.E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840. doi: 10.1128/CMR.17.4.840-862.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khot P.D., Ko D.L., Fredricks D.N. Sequencing and analysis of fungal rRNA operons for development of broad-range fungal PCR assays. Appl Environ Microbiol. 2009;75:1559. doi: 10.1128/AEM.02383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Font-Vizcarra L., García S., Martínez-Pastor J.C., Sierra J.M., Soriano A. Blood culture flasks for culturing synovial fluid in prosthetic joint infections. Clin Orthop. 2010;468:2238. doi: 10.1007/s11999-010-1254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baron E.J., Miller J.M., Weinstein M.P. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM)(a) Clin Infect Dis. 2013;57:e22. doi: 10.1093/cid/cit278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costerton J.W. Biofilm theory can guide the treatment of device-related orthopaedic infections. Clin Orthop. 2005:7. doi: 10.1097/00003086-200508000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Mariani B.D., Martin D.S., Levine M.J., Booth R.E., Tuan R.S. The Coventry Award. Polymerase chain reaction detection of bacterial infection in total knee arthroplasty. Clin Orthop. 1996:11. doi: 10.1097/00003086-199610000-00003. [DOI] [PubMed] [Google Scholar]
- 17.De Man F.H.R., Graber P., Lüem M. Broad-range PCR in selected episodes of prosthetic joint infection. Infection. 2009;37:292. doi: 10.1007/s15010-008-8246-1. [DOI] [PubMed] [Google Scholar]
- 18.Holst H., Salling N., Andresen K., Christensen J.J., Kemp M. Detection of anaerobic prosthetic joint infection by PCR and DNA sequencing – a case report. Acta Orthop. 2008;79:568. doi: 10.1080/17453670710015599. [DOI] [PubMed] [Google Scholar]
- 19.Bergin P.F., Doppelt J.D., Hamilton W.G. Detection of periprosthetic infections with use of ribosomal RNA-based polymerase chain reaction. J Bone Joint Surg Am. 2010;92:654. doi: 10.2106/JBJS.I.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacovides C.L., Kreft R., Adeli B. Successful identification of pathogens by polymerase chain reaction (PCR)-based electron spray ionization time-of-flight mass spectrometry (ESI-TOF-MS) in culture-negative periprosthetic joint infection. J Bone Joint Surg Am. 2012;94:2247. doi: 10.2106/JBJS.L.00210. [DOI] [PubMed] [Google Scholar]
- 21.Hischebeth G.T.R., Randau T.M., Buhr J.K. Unyvero i60 implant and tissue infection (ITI) multiplex PCR system in diagnosing periprosthetic joint infection. J Microbiol Methods. 2016;121:27. doi: 10.1016/j.mimet.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Rak M., KavčIč M., Trebše R., Cő R.A. Detection of bacteria with molecular methods in prosthetic joint infection: sonication fluid better than periprosthetic tissue. Acta Orthop. 2016;87:339. doi: 10.3109/17453674.2016.1165558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu X., Zhai Z., Li H. PCR-based diagnosis of prosthetic joint infection. J Clin Microbiol. 2013;51:2742. doi: 10.1128/JCM.00657-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson M.R., Naccache S.N., Samayoa E. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wimmer M.D., Friedrich M.J., Randau T.M. Polymicrobial infections reduce the cure rate in prosthetic joint infections: outcome analysis with two-stage exchange and follow-up ≥two years. Int Orthop. 2016;40:1367. doi: 10.1007/s00264-015-2871-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.