Summary
Improving health of the rapidly growing aging population is a critical medical, social, and economic goal. Identification of genes that modulate healthspan, the period of mid-life vigor that precedes significant functional decline, will be an essential part of the effort to design anti-aging therapies. Because locomotory decline in humans is a major contributor to frailty and loss of independence and because slowing of movement is a conserved feature of aging across phyla, we screened for genetic interventions that extend locomotory healthspan of Caenorhabditis elegans. From a group of 54 genes previously noted to encode secreted proteins similar in sequence to extracellular domains of insulin receptor, we identified two genes for which RNAi knockdown delayed age-associated locomotory decline, conferring a high performance in advanced age phenotype (Hpa). Unexpectedly, we found that hpa-1 and hpa-2 act through the EGF pathway, rather than the insulin signaling pathway, to control systemic healthspan benefits without detectable developmental consequences. Further analysis revealed a potent role of EGF signaling, acting via downstream phospholipase C-γ plc-3 and inositol-3-phosphate receptor itr-1, to promote healthy aging associated with low lipofuscin levels, enhanced physical performance, and extended lifespan. This study identifies HPA-1 and HPA-2 as novel negative regulators of EGF signaling and constitutes the first report of EGF signaling as a major pathway for healthy aging. Our data raise the possibility that EGF family members should be investigated for similar activities in higher organisms.
Introduction
The increase in human life expectancy has been accompanied by a focused appreciation of the significant need to maintain general health and vigor late into life (Glatt et al., 2007; Kirkland & Peterson, 2009). As such, extending healthspan—the period of maintained function and stress-resistance that precedes debilitating decline—has become a central goal of current aging research. Improving overall robustness as well as maintaining the integrity of individual organ systems are both likely to contribute to healthspan extension and an increased quality of life.
Locomotory decline is a conserved feature of aging animals that is typically accompanied by a progressive loss of muscle mass and muscle strength called sarcopenia (Fisher, 2004; Lang et al., 2009). As an inescapable component of normal human aging, sarcopenia impacts the entire elderly population, and is thought to be a major underlying cause of loss of independence, frailty, and morbidity. Given the seemingly universal association of old age with diminished mobility, genetic analyses in invertebrate models may contribute novel insights into conserved molecular causes of locomotory decline (Augustin & Partridge, 2009; Tatar, 2009). Multiple studies in C. elegans have documented diminished locomotion with age scored as crawling on solid agar plates (Croll et al., 1977; Bolanowski et al., 1981; Johnson, 1987; Duhon & Johnson, 1995; Herndon et al., 2002; Glenn et al., 2004; Huang et al., 2004; Hsu et al., 2009). Another C. elegans locomotory phenotype, more easily quantitated by unaided observers, is the rate of body bends/unit time when animals are placed in liquid (“swimming” (Pierce-Shimomura et al., 2008)). Swimming rate progressively declines with age (Duhon & Johnson, 1995; Restif & Metaxas, 2008) (and data herein). Physical deterioration of muscle (without notable cell death) has been correlated with locomotory decline, with relatively subtle changes such as in actin filament organization (Glenn et al., 2004) preceding a dramatic loss of sarcomere units and fat infiltration of muscle that resembles sarcopenia in higher organisms (Herndon et al., 2002). A proportion of muscle deterioration might be attributed to diminished neuronal signaling--administration of muscarinic agonist arecoline can extend locomotory healthspan (Glenn et al., 2004), and altering serotonin signaling can suppress early phases of locomotory aging (Murakami et al., 2008). Lowering the strength of insulin receptor signaling prolongs locomotory healthspan and physical integrity of muscle (Herndon et al., 2002) as well as extends lifespan. Dietary restriction regimens can also improve locomotory function in aging animals (Huang et al., 2004; Hsu et al., 2009). Overall, however, there has been little systematic evaluation of genetic influences on locomotory decline and much remains to be learned about the molecular systems modulating this process.
With an interest in identifying genes that impact locomotory decline, we screened sets of healthspan candidates by RNAi knockdown, scoring for enhanced swimming prowess in old age. Among genes suggested to encode insulin receptor-related proteins (Dlakic, 2002), we identified hpa-1 and hpa-2 (high performance in advanced age), which proved to modulate multiple aging phenotypes in C. elegans healthspan. Effects of HPA-1 and HPA-2 occur largely independently of insulin signaling—instead, we show that hpa-1 and hpa-2 act through the EGF receptor/phospholipase Cγ/ IP3 receptor pathway. This work identifies novel upstream negative regulators of EGF pathway activities that are also noteworthy because their genetic manipulation exerts a greater proportionate impact on healthspan than on the longevity endpoint. In addition, this work is the first to document the impact of EGF signaling and the downstream PLCγ/IP3R signaling branch in C. elegans healthspan and lifespan. We suggest EGF signaling may be a conserved mechanism for adult maintenance and healthy aging.
Results
Inactivation of hpa-1 and hpa-2 specifically extends locomotory capacity late in life
Although the problem of locomotory decline is a pervasive and universal component of aging biology, genetic influences on this process have not yet been systematically identified. In C. elegans, expression of individual genes can be knocked down by feeding animals the corresponding double stranded RNA expressed in bacterial food (Kamath & Ahringer, 2003). We initiated screening for gene knockdowns that altered adult locomotory healthspan by first testing selected sets of genes implicated in, or associated with, those genes known to influence longevity.
Among signaling pathways that modulate longevity, the insulin/IGF (IIS) pathway activity has emerged as a conserved and potent mechanism for modulating both healthspan and lifespan (Tatar et al., 2003; Broughton & Partridge, 2009). The 40 insulin-like ligands encoded by the C. elegans genome (Malone & Thomas, 1994; Pierce et al., 2001) are all thought to act via the sole DAF-2 insulin receptor homolog. Activation of the DAF-2 insulin receptor initiates a kinase cascade that includes AGE-1 (PI-3 kinase), and pathway activation ultimately phosphorylates FOXO family transcription factor DAF-16 to inhibit its activity (Tatar et al., 2003). Conversely, decreased DAF-2 signaling promotes DAF-16 transcriptional functions and causes daf-16-dependent lifespan extension (Kenyon et al., 1993; Hsu et al., 2003). Additional, less-characterized molecular modulators of insulin pathway activity are likely to also influence signaling. For example, a bioinformatic study identified 54 proteins related to the extracellular ligand binding domain of insulin receptor (Dlakic, 2002). These insulin receptor-related proteins share sequence similarities in the extracellular ligand-binding domain and contain secretion signal sequences, but lack the transmembrane domain and intracellular kinase domains characteristic of classic receptor kinases. Thus, this group of proteins resemble secreted proteins that might bind ligand more than they resemble intact transmembrane receptor kinases. Functional studies on this gene group have not yet been reported.
We constructed or obtained 54 clones for the IGF receptor-related genes and used these for food-delivered RNAi inactivation to test effects on locomotory healthspan (Fig. 1A; Table S1, Supporting Information). As a first-pass screen for changes in locomotory ability in aging adults, we scored body bend frequency (swimming rates) in aging post-reproductive adults (11 days from the hatch, 25°C; day 3 is the first day of reproductive adult life at this temperature and day 11 is middle/late adult life of the ∼ 20 day lifespan). We then verified that RNAi clones conferring statistically significant changes later in life did not impact locomotion rates in young adult life to rule out developmental or general behavioral effects of RNAi knockdown that might confer chronic hypo- or hyper-active swimming. In this way, we sought to identify genetic knockdowns that specifically changed swimming prowess in aging adults.
Fig. 1. hpa-1 and hpa-2 deficiencies promote healthy aging in C. elegans.

(A) Summary of screen strategy for RNAi interventions that confer high performance in advanced age (Hpa phenotype). We fed bacteria expressing dsRNA to synchronized L1 larvae of ts sterile mutant spe-9, 25°C, and counted the number of body bends during 30 second swim trials in M9 buffer at day 11 post-hatching (post-reproductive animals in mid/late adult life, slightly more than halfway through maximum wt adult lifespan at this temperature; ∼ 60% alive).
(B) RNAi inactivation of hpa-1 and hpa-2 extends swimming healthspan in wild type N2. Negative control was fed with empty vector. Each trial count was for 30 animals collected from a population of 150-250 synchronized animals (three trials). Error bars represent the standard error of the mean (s.e.m.). Unpaired two tailed t-test (control versus hpa-1, hpa-2 or positive control age-1 RNAi on each day), ***P< 0.0001.
(C) hpa-1 (tm3256) and hpa-2 (tm3827) deletion mutants exhibit enhanced swimming performance in advanced age. age-1 (hx546) mutants were used as a positive control for Hpa phenotype. Day 3 is the first day of adult life in animals raised at 25°C, as indicated by egg-laying onset (two trials). Error bars represent s.e.m. Unpaired two tailed t-test (N2 versus mutant on each day), ***P< 0.0001. Note that differences in onset of decline between RNAi-treated animals (panel 1B) and actual deletion mutants (panel 1C) may reflect the partial gene inactivation effects of RNAi.
(D) Quantitative measurement of AGE pigment levels for wild type N2, positive control age-1 (hx546), hpa-1 (tm3256), and hpa-2 (tm3827), 25°C. The AGE pigment score is normalized as the ratio of fluorescence units for AGE pigments / tryptophan fluorescence (three independent experiments, 50 animals per strain). Error bars, s.e.m., P values, unpaired two-tailed t-test, *P< 0.05 and **P< 0.01. Although age pigment scores are low relative to wt on the first day of adult life in both hpa-1 and hpa-2, only hpa-1 maintains low age pigment levels later into adulthood.
(E) The hpa-1 mutant has low age pigment levels in old age. Images are taken under fluorescent light with identical exposures, 25°C. The gut houses autofluorescent lipofuscin, sequestered into lysosomes (Clokey & Jacobson, 1986). Top, N2; bottom, hpa-1 (tm3256).
(F) The age-associated decline in pumping rate is delayed in aging hpa-1 and hpa-2 deletion mutants. hpa-1 (tm3256) and hpa-2 (tm3827) deletion mutants exhibit enhanced pumping performance in advanced age. At least fifteen animals were recorded for each trial (two trials). Day 3 is first day of adult life in animals raised at 25°C, as indicated by egg-laying onset. Error bars represent s.e.m. Unpaired two tailed t-test (N2 versus mutant on each day), ***P< 0.0001.
(G) Comparative survival curves for wild type N2, age-1 (hx546), hpa-1 (tm3256), and hpa-2 (tm3827). Details of data in Table S3, Supporting information.
In the screen of 54 insulin receptor-related genes, we identified nine genes for which RNAi knockdown conferred enhanced swimming vigor in aging adults (Table S1, Supporting Information). Of these, we elected to focus on detailed analysis of H25K10.5 and T11F1.8 because RNAi directed against these consistently conferred strongest effects and because deletion alleles became available during the course of our study. We designate these receptor-related genes as hpa-1 and hpa-2, respectively, for the phenotype of high performance in advanced age (hpa). For hpa-1 (RNAi) and hpa-2 (RNAi), animals swim at control rates in young day5 adults, but swim faster than wild type as older day11 adults (Fig. 1B). In young hpa-1 (RNAi) and hpa-2 (RNAi) adults pharyngeal pumping rates, defecation rates, dauer formation, vulval development, and brood size are all similar to controls (Fig. S2A-C and G; Table S2A, Supporting Information), supporting that hpa-1 and hpa-2 (RNAi) do not exert major impact on C. elegans development or basic behaviors up to young adulthood. Instead, phenotypes are apparent in aging adults.
We verified that 6× outcrossed deletion mutants of hpa-1 (tm3256) and hpa-2 (tm3827) exhibited increased swimming prowess at old age (Fig. 1C). For deletion mutants, swimming was indistinguishable from wild type on the first day of adult life (day3), with differences in swim vigor becoming already apparent at day5, and maintained later into adulthood (day11). (The different onsets for phenotype expression of deletion mutants vs. hpa (RNAi) might be attributed to the variable/partial knockdown capacities of RNAi). Consistent with hpa-1 and hpa-2 (RNAi) phenotypes, hpa-1 (tm3256) and hpa-2 (tm3827) deletions, as well as their double mutant combination, did not affect C. elegans development or basic behaviors up to young adulthood (Fig. S2D-F, H and Fig. S3; Table S2B, Supporting Information). We conclude that diminishing hpa-1 and hpa-2 activities extends swimming locomotory healthspan without a major impact on basic swimming or other functions during development.
We previously documented that WT C. elegans reared on agar plates progress through successive stages of crawling impairment, classifying animals that move vigorously in response to an eyelash hair touch as class A, those that are uncoordinated in response to touch as class B, and those that are virtually paralyzed except for the head as class C (Herndon et al., 2002). To evaluate an independent measure of locomotory capacity when hpa-1 and hpa-2 are deficient, we scored the relative prevalence of ABC classes during adult life consequent to RNAi knockdown. We found that hpa-1 (RNAi) and hpa-2 (RNAi) animals exhibit diminished rates of decline in this comparative plate locomotion assay (Fig. S4, Supporting Information), in further support that locomotory aging is delayed when either hpa-1 or hpa-2 is lacking.
hpa-1 and hpa-2 deficiencies improve multiple measures of favorable healthspan
hpa-1 and hpa-2 might specifically impact locomotory aging or, alternatively, might systemically affect the overall quality of C. elegans aging. To address whether hpa-1 and hpa-2 affect expression of other age-associated phenotypes, we monitored additional indicators that can reflect the quality of aging: age pigment accumulation (Gerstbrein et al., 2005), pharyngeal pumping (Huang et al., 2004; Chow et al., 2006), and survival curve properties. Lipofuscin and advanced glycation end products (referred to here together as age pigments) accumulate in aging organisms across species (Perriere & Gouy, 1996; Ulrich & Cerami, 2001). We previously demonstrated via in vivo age pigment quantitation that age pigments accumulate at accelerated rates in aging C. elegans and that long-lived strains tend to have low levels of age pigments in early and middle adult life (Gerstbrein et al., 2005). Moreover, same-age adults with low age pigment levels have longer life expectancy than age-matched siblings reared in the same environment that stochastically have high age pigment levels, suggesting that age pigment scores reflect a “physiological”, rather than chronological, age. Using in vivo fluorometric analysis, we precisely quantitated age pigment values in young and aging hpa-1 and hpa-2 adults (Fig. 1D,E). We find that hpa-1 and hpa-2 have low age pigment scores on the first day of adult life. Only hpa-1, however, maintains differences later into adulthood. Thus, hpa-1 exhibits low age pigments though most of adult life, suggesting a systemic impact on healthy aging. The consequences of transiently low age pigments as found in hpa-2 are not known, and thus we are unable to comment as to whether a transient low age pigment period early in life might be expected to influence the quality of aging in later adulthood.
As another measure of maintained tissue function/integrity over time, we analyzed pharyngeal pumping frequency in mid-life. Previous work documented that pumping frequency decline is dramatic in early adult life (Huang et al., 2004). We find that hpa-1 and hpa-2 deficiencies slow this decline significantly without affecting pumping activity on the first day of adult life (Fig. 1F). Our data indicate improved maintenance of pharyngeal function as hpa-1 and hpa-2 mutant adults age.
We also compared survival curves of hpa-1 and hpa-2 deficient animals to those of wild type and long-lived age-1 mutants. Analysis of deletion mutants supported that hpa-1 and hpa-2 deficiencies extend median and mean lifespan to improve mid-life survival in aging cultures (Fig. 1G). For example, in the lifespan trial in Fig. 1G, median lifespan of WT was 13 days, of age-1 and hpa-2 was 15 days, and of hpa-1 was 17 days (15% increase in age-1, 31% increase in hpa-1, and 15% increase in hpa-2). Although the exact magnitude of mid-life survival showed some variation between repeat trials, we found that the median and mean lifespans for hpa-1 and hpa-2 were increased with statistical significance in all trials (Table S3, Supporting Information). Interestingly, for hpa mutant strains, the maximum lifespan changes (scored as the mean of the top 10% survivors) were more modest or lacking as compared to the median lifespan phenotypes (Max: N2 21.3 +/-0.1; age-1 32.3 +/-0.7 (51%); hpa-1 23.5+/-0.7 (10%); hpa-2 22.0+/- 0.6 (NS)). Thus, unlike age-1 reduction-of-function, hpa-1 and hpa-2 deletions exert a proportionally greater impact on median lifespan as compared to maximum lifespan.
We also calculated mortality rates over adult life for hpa-1 and hpa-2 deletion mutants (Johnson, 1987) (Fig. S5, Supporting Information). Like WT, mortality rates rise exponentially with age in hpa backgrounds, but hpa mutants show the same rate of change as WT (Fig. S5 A-C, Supporting Information). Slopes of plots of log mortality vs. age (Fig. S5 D-F, Supporting Information) suggest better function throughout life rather than a change in the rate of decline, per se.
In sum, on the basis of multiple assessments of organ and animal vigor in mid/late adult life, we conclude that HPA-1 and HPA-2 are newly identified modulators of healthy aging in C. elegans.
HPA-1 and HPA-2 can promote locomotory healthspan through a pathway distinct from insulin signaling and dietary restriction
HPA-1 and HPA-2 were initially identified by homology to extracellular ligand-binding regions of the insulin receptor (InsR) (Dlakic, 2002). To address whether hpa-1 and hpa-2 extend locomotory healthspan via the insulin/IGF signaling pathway, we first tested for a requirement for critical downstream IIS transcription factors in the execution of the hpa-1 and hpa-2 age-associated RNAi swimming phenotypes. Downstream transcription factor FOXO/DAF-16 is essential for the longevity phenotype of InsR/daf-2 (rf) mutants (Kenyon et al., 1993) and daf-16 null mutants have early-onset age pigment elevation (Gerstbrein et al., 2005), consistent with a progeric condition when daf-16 is lacking. We find that hpa-1 and hpa-2 RNAi partially restored swimming prowess in young daf-16 null mutants (∼29 bends/30min for daf-16 vs. ∼35 bends/30min for hpa-1 (RNAi) daf-16 and ∼33 bends/30min for hpa-2 (RNAi) daf-16, day5), suggesting that hpa-1 and hpa-2 knockdown effects are, at least in part, daf-16-independent.
Transcription factor HSF-1 is also required for daf-2 (rf) to extend lifespan (Hsu et al., 2003) and to protect against proteotoxic stress (Cohen et al., 2006). We find that a shift of the temperature-sensitive hsf-1 mutant to the non-permissive temperature accelerates swimming decline in young adults (Fig. 2A). hpa-1 and hpa-2 RNAi can extend swimming healthspan in the hsf-1 mutant (∼7 bends/30min day8 for hsf-1 vs. ∼13 bends/30min day8 for hpa-1 and hpa-2 (RNAi) hsf-1, day8). We conclude that hsf-1 is needed for normal locomotory healthspan, and that knockdown of hpa-1 and hpa-2 extends locomotory healthspan via a mechanism that is at least in part hsf-1-independent.
Fig. 2. hpa-1 and hpa-2 (RNAi) can extend locomotory healthspan via a pathway distinct from insulin/IGF-1 signaling (IIS) and some dietary restriction (DR) indicators.
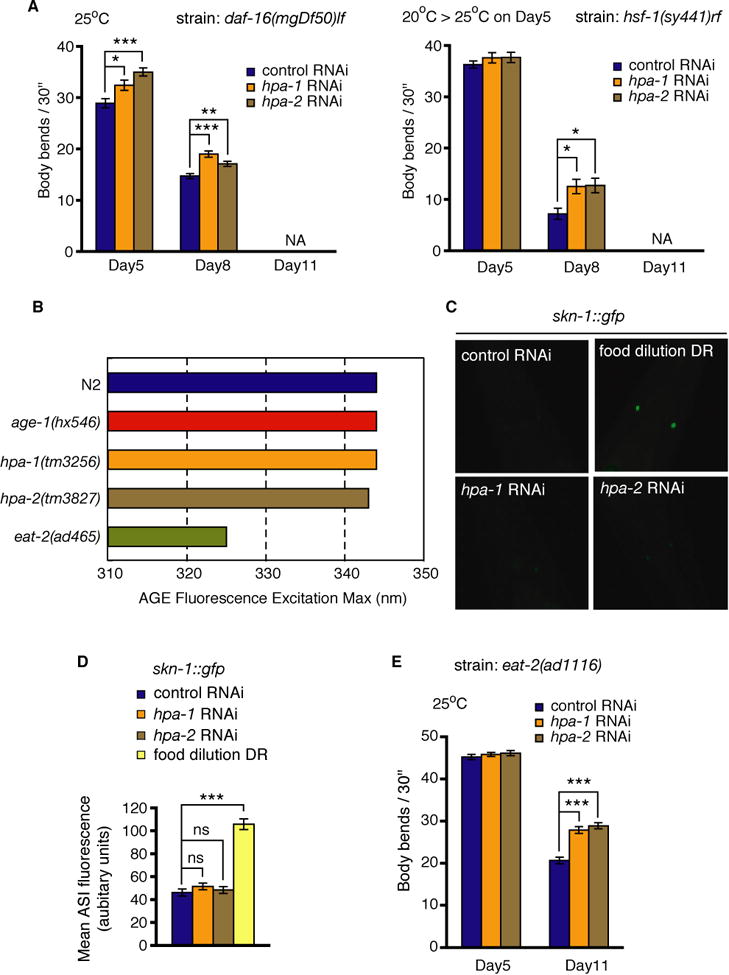
(A) Assays of hpa-1 and hpa-2 (RNAi) effects on representative mutants of the insulin signaling pathway. daf-16 (mgDf50) is a progeric null mutant; hsf-1 (sy441) is a progeric temperature-sensitive mutant. Mutants were fed with bacteria expressing indicated dsRNA; control is empty vector. Error bars represent s.e.m., unpaired two tailed t-test (control versus hpa-1 or hpa-2 RNAi on each day, *P< 0.01, **P< 0.001, ***P< 0.0001, NA; not available since population is largely dead). Note that to avoid developmental consequences of genetic disruption, temperature-sensitive hsf-1 (sy441) mutants were grown at permissive temperature of 15-20°C until day 5 after hatching (first day adults) and were then transferred to restrictive temperature 25°C at the beginning of adult life. Thus hsf-1 activities are disrupted only during adulthood. Since hpa-1 (RNAi) and hpa-2 (RNAi) can still extend locomotory healthspan when key donwstream transcription factors in the IIS pathway are deficient, hpa-1 and hpa-2 appear able to act, at least in part, independently of the IIS pathway. Note that temperature shifts required for some studies can influence lifespan both by temperature and genetic changes; direct comparison between the same experimental days in different temperature regimens thus cannot be made.
(B) The excitation maximum for the age pigment fluorescence shifts to ∼320 nm for impaired feeding DR mutant eat-2 (ad465), while it remains constant at 340 nm for wild-type, age-1 (hx546), hpa-1 (tm3256) and hpa-2 (tm3827) animals. The shift is seen for other DR eat mutants, unc-26, and growth in liquid culture, which induces DR, but not for other longevity mutants.
(C) Representative images of SKN-1∷GFP in the ASI neurons after RNAi treatments or DR. Note significant increase under food dilution DR.
(D) Activation of SKN-1 in the ASI neurons is induced by DR but not by hpa-1 and hpa-2 (RNAi). We quantitated fluorescence at 25°C in the ASI neurons in adults harboring a skn-1∷gfp reporter, which increases GFP expression under the DR conditions with UV-killed bacteria, consistent with a previous report (Bishop & Guarente, 2007). hpa-1 and hpa-2 RNAi treatments do not change the level of SKN-1∷GFP in the ASI neurons, supporting that their knockdown is not associated this type of DR activation, or that if they do, SKN-1 activation occurs upstream of HPA function. Error bars, s.e.m.; p value in unpaired two-tailed t-test, ***P < 0.0001.
(E) Effects of hpa-1 and hpa-2 RNAi on DR constitutive mutant eat-2. eat-2 (ad1116) mutants were fed with bacteria expressing dsRNA (control is empty vector) at 25°C. Error bars represent s.e.m., unpaired two tailed t-test (control versus hpa-1 or hpa-2 (RNAi) on each day, ***P< 0.0001).
Long-lived insulin receptor daf-2 (rf) and PI3 kinase age-1 (rf) (Herndon et al., 2002) exhibit extended locomotory healthspan. We knocked down hpa-1 and hpa-2 in daf-2 (rf) and age-1 (rf) mutants to show that this intervention further extends swimming healthspan (Fig. S6, Supporting Information). Although epistasis analysis on reduction-of-function mutations rather than deletion mutations is not definitive (Huang & Sternberg, 2006), the additive effects of low insulin pathway signaling and hpa knockdown are consistent with the interpretation that the two HPA proteins identified by similarity to insulin receptor ligand-binding sequences can modulate locomotory healthspan at least in part via an IIS-independent pathway. The identity of this alternative pathway thus became a question of interest.
A second major pathway for longevity and healthspan benefit conserved across species is dietary restriction (DR), and thus we considered whether hpa-1 and hpa-2 RNAi might extend locomotory healthspan by activating DR. However, several lines of evidence argue against this possibility. First, as noted in Fig. S2, pharyngeal pumping is normal in young hpa-1 and hpa-2 RNAi and deletion mutants and enhanced in aging animals (Fig. 1F), so hpa-1 or hpa-2 deficiency does not physically limit feeding (Fig. S2A, D Supporting Information). Second, hpa-1 or hpa-2 deficiency exhibits the characteristic fluorometric shift in age pigment excitation maximum that exclusively characterizes DR mutants and animals treated with every DR regimen we have tested to date (Fig. 2B) (Gerstbrein et al., 2005). Third, hpa-1 and hpa-2 (RNAi) do not cause the induction of a SKN-1∷GFP reporter in the ASI neurons that occurs in WT under a modified food limitation DR protocol (Fig. 2D, E) (Bishop & Guarente, 2007). Finally, hpa-1 and hpa-2 RNAi further enhances the old-age swimming prowess of the eat-2 feeding limited DR mutant (Lakowski & Hekimi, 1998), which suggests that hpa-1 and hpa-2 act via a mechanism distinct from, but additive with, feeding-limited DR (Fig. 2B). Thus, although we have not tested all DR regimens (Greer & Brunet, 2009), and genetic interactions with eat-2 must be interpreted with attention to experimental concerns that DR might not be optimized in the eat-2 background in this experiment (Huang & Sternberg, 2006; Mair & Dillin, 2008), our compiled data fail to implicate hpa-1 and hpa-2 in any of several of probed DR mechanisms.
Activation of the conserved EGF signaling pathway confers healthspan benefits in C. elegans
Our genetic observations suggesting that HPA-1 and HPA-2 can act, at least in part, via a pathway distinct from the IIS pathway to influence locomotory healthspan prompted us to revisit bioinformatic analysis of HPA-1 and HPA-2. Alignments of HPA-1 and HPA-2 revealed primary sequence homologies to EGF receptor ligand-binding domains that appeared potentially more significant than the relationship to insulin receptor ligand-binding domains (Fig. S7A, Supporting Information). More specifically, HPA-1 and HPA-2, which are related in sequence to each other, are similar in ligand-binding Leucine Rich domains (L domains) to mammalian EGF Receptor-Related Protein (ERRP), a secreted negative regulator of EGF receptor (EGFR), which itself exhibits homology to the extracellular domain of mammalian EGFR (Yu et al., 2001) (Fig. S7B, Supporting Information). This sequence relationship prompted us to address whether HPA-1 and HPA-2 might act via the EGF signaling pathway.
We first examined how the EGF signaling pathway itself impacts healthspan and lifespan—a question that, surprisingly, had not yet been addressed in the facile nematode model. The genetics of the C. elegans EGF pathway have been characterized in exquisite detail by Sternberg and colleagues, with a focus on EGF signaling roles in development (Moghal & Sternberg, 2003), and a more recent observation of a role in behavioral quiescence (Van Buskirk & Sternberg, 2007). We found that a gain-of-function mutation affecting the EGF receptor, let-23 (sa62), increases swimming vigor later in life (Fig. 3A). Conversely, temperature-sensitive reduction-of-function of the EGF receptor mutant let-23 (n1045), which is impaired at 20°C, exhibits the opposite effect on swimming healthspan—an accelerated decline (Fig. 3B). let-23 (gf) delays the decline in muscle nuclear GFP signal dimunition that characterizes sarcopenia in elderly MYO-3∷GFP-NLS transgenic animals (Herndon et al., 2002) (Fig. 3C, D), suggesting that both muscle function and integrity are maintained longer in EGFR/let-23 (gf) mutants. The let-23 (gf) mutant also exhibits low age pigment levels later in life, consistent with a general healthy aging trajectory (Fig. 3E, F). Finally, analysis of survival curves indicates that EGFR/let-23 (gf) cultures survive more robustly in middle adulthood (i.e., 29% increase of median lifespan, 9% increase of maximum lifespan, and EGFR/let-23 (rf) cultures survive less robustly in middle adulthood than matched wild type controls (i.e., 19% decrease of median lifespan, 8% decrease of maximum lifespan) (Fig. 3G, H). We conclude that EGF receptor activity modulates the quality of aging, with EGFR activation promoting extended healthspan, and EGFR inactivation associated with age-related declines. Consistent with this conclusion, we find that the reduction-of-function EGF mutant lin-3 (n1058) exhibits reduced swimming vigor in mid-adulthood and a shortened median lifespan (Fig. S8A, B, Supporting Information).
Fig. 3. Activation of the EGF receptor LET-23 promotes healthy aging.

(A) Activation of the EGFR let-23 extends swimming healthspan. The gain-of-function let-23 (sa62) mutant strongly increases swimming performance in advanced age. Data are for 25°C, though similar effects are observed also at 20°C. Error bars represent s.e.m. Unpaired two tailed t-test (N2 versus let-23 (sa62) mutant on each day), ***P< 0.0001.
(B) Disruption of EGFR activity accelerates swimming decline in aging animals. For swimming assay, the reduction-of-function let-23 (n1045) rf mutant and N2 wild type were grown continuously at restrictive temperature of 20°C. Error bars represent s.e.m. Unpaired two tailed t-test (N2 versus let-23 (n1045) mutant on each day), ***P< 0.0001.
(C) Age-related deterioration of C. elegans body wall muscle as indicated by GFP fluorescence decline of a pmyo-3NLS∷GFP fusion. The GFP reporter is localized to muscle nuclei, and becomes progressively sequestered and dims with age (Herndon et al., 2002). Representative whole-animal view of the wild type and the let-23 (sa62) gf mutant expressing GFP in the nuclei of body wall muscle at ages indicated (25°C). When the EGFR is activated, signal dimming occurs more slowly, consistent with muscle healthspan extension.
(D) Quantitation of the number of fluorescent nuclei in the wild type and the let-23 (sa62) gf mutant. 15 animals of each strain were scored in 2 independent trials. Error bars, s.e.m. P value for unpaired two-tailed t-test, ***P < 0.0001.
(E) Age pigments accumulate at a reduced rate when EGFR is activated. Representative same-exposure photographs of fluorescent species in the intestine of wild type N2 versus let-23 (sa62) mutants at day 11 (25°C).
(F) Measurement of AGE pigment accumulation for wild type N2 and let-23 (sa62) gf. AGE pigment scores were normalized as the relative fluorescence units for AGE pigments divided by tryptophan fluorescence. Data are from three independent experiments, 50 animals total for each strain; au arbitrary units, error bars, s.e.m.; P value, unpaired two-tailed t-test, *P < 0.01.
(G) The EGFR(gf) mutant exhibits enhanced lifespan, 20°C. Details of data in Table S3, Supporting information.
(H) The EGFR(rf) mutant is short-lived. For lifespan assay, strains were grown at permissive temperature of 25°C until day 5 after hatching, and then transferred to the non-permissive temperature of 15°C. Details of data in Table S3, Supporting information.
The downstream branch of the EGF pathway involving phospholipase Cγ PLC-3 and IP3 receptor ITR-1 promotes healthy aging outcomes of EGFR activation
In C. elegans, EGF signaling activates distinct downstream signaling pathways (Fig. 4A) for specific functional outcomes (Moghal & Sternberg, 2003; Van Buskirk & Sternberg, 2007): EGFR signaling through the RAS-MAPK pathway determines cell fates that affect viability and development of the hermaphrodite vulva (Beitel et al., 1990; Han & Sternberg, 1990; Lackner et al., 1994; Wu & Han, 1994); EGF signaling through diacylglycerol (DAG) regulates behavioral quiescence in larvae at the developmental molts (Van Buskirk & Sternberg, 2007), and EGF signaling acts through phospholipase C-γ/plc-3 and inositol-1,4,5-triphosphate (IP3) to regulate ovulation (Clandinin et al., 1998; Merris et al., 2004). To identify the downstream signaling branch of the EGF pathway that influences adult healthspan, we asked whether RAS/let-60, MAPK/mpk-1, DAG-binding protein/unc-13, PLC-γ/plc-3, or IP3 receptor/itr-1 are required for the old-age swimming prowess observed in the EGFR/let-23 (gf) mutant. We conducted these assays by performing feeding RNAi to knockdown gene activities in the EGFR/let-23 (gf) background and measuring body bend frequency in middle/late adulthood. We found that plc-3 (RNAi) and itr-1 (RNAi) suppress the youthful swimming phenotype of the let-23 (gf) mutant, but RNAi interventions affecting genes in other downstream branches of EGFR signaling do not (Fig. 4B). These data implicate the downstream PLC-γ/IP3 receptor pathway, rather than the RAS pathway used in vulval fate specification or the DAG pathway used in larval quiescence at the molts, in mediating the positive EGFR effects on swimming healthspan.
Fig. 4. The PLC-γ/IP3 receptor signaling branch mediates EGFR healthy locomotory aging benefits.

(A) Epidermal growth factor (EGF) signaling activates multiple pathways in C. elegans. EGF ligands are encoded by the lin-3 gene(Hill & Sternberg, 1992). EGF receptor LET-23 can activate at least three distinct downstream pathways. The LET-60/RAS MAP kinase pathway influences cell fate specification (Beitel et al., 1990; Han & Sternberg, 1990; Lackner et al., 1994; Wu & Han, 1994). Phospholipase Cγ can act via diacyl glycerol binding protein UNC-13 to induce quiescent behavior during larval molts (Van Buskirk & Sternberg, 2007). The ER calcium release channel IP3 receptor acts in another pathway branch to influence ovulation (Clandinin et al., 1998; Merris et al., 2004).
(B) RNAi knockdown of key genes in pathways downstream of EGFR implicates the PLC-γ/IP3 receptor branch in healthy locomotory aging. EGFR/let-23 (sa62) gf mutant swimming in mid/late age was assayed in feeding RNAi experiments for the indicated genes (25°C). Note that itr-1 RNAi was initiated on day 4 to avoid larval arrest caused by itr-1 (RNAi). The fact that this RNAi intervention blocks benefits of EGFR(gf) both supports that itr-1 is a required downstream effector of LET-23 (gf) and indicates that IP3 receptor activity is required during adult life for extended locomotory healthspan. Error bars represent s.e.m. Unpaired two tailed t-test (control versus let-60, mpk-1, plc-3, unc-13 or itr-1 RNAi) ***P< 0.0001. Note that to cross-verify results from MAPK and DAG pathways, we performed RNAi in the “reverse” direction, testing hpa-1 and hpa-2 (RNAi) on let-60 (rf), let-60 (gf), unc-13 and dgk-1 (gf) mutants, see Fig. S9, Supporting information).
(C-H) Genetic activation of IP3 receptor elicits healthy aging and longevity. (C) Activation of the IP3R itr-1 elicits youthful swimming in old age. The itr-1 (sy290) gf mutant exhibits markedly enhanced swimming performance in adulthood at 25°C. Error bars represent s.e.m. Unpaired two tailed t-test (N2 versus itr-1 (sy290) mutant on each day): ***P< 0.0001.
(D) Reduction of IP3 receptor/ITR-1 activity from early adulthood accelerates swimming decline in advanced age. For assay, itr-1 (sa73) rf strains and N2 wild type were grown at permissive temperature of 15°C until day 5 after hatching (first day of sexual maturity), and were then transferred to the non-permissive temperature (25°C). Error bars represent s.e.m. Unpaired two tailed t-test (N2 versus itr-1 (sa73) mutant on each day): ***P< 0.0001. Data indicate that limiting IP3R signaling in only the adult stage has deleterious consequences for locomotory aging.
(E-F) Age pigments are low in aging itr-1 (gf) mutants. (E) Representative photographs of fluorescent species in the intestine of wild type N2 versus itr-1 (sy290) gf worms at day 11. (F) Quantitative measurement of AGE pigment accumulation for wild type N2 and itr-1 (sy290) gf. Indicated are AGE/TRP ratios; unpaired two tailed t-test, **P< 0.001, ***P< 0.0001.
(G) Survival curve for itr-1 (sy290) gf indicates extension of both mean and maximum lifespan, 20°C. Details of data in Table S3, Supporting information. (H) Survival curve for itr-1 (sa73) rf reveals shortening of both mean and maximum lifespan. For lifespan assay, strains were grown at permissive temperature of 15° C until day 5 after hatching, and then transferred to the non-permissive temperature of 20° C. Details of data in Table S3, Supporting information.
To independently confirm that the IP3R pathway can influence multiple healthspan indicators, we examined itr-1 alleles that decrease or increase IP3 receptor signaling for impact on late adult swimming prowess, age pigment accumulation rates and adult survival. We found that gain-of-function IP3 receptor itr-1 (sy290) mutation conferred relatively youthful swimming in 11 day old animals (Fig. 4C), whereas reduction-of-function itr-1 (sa73) mutants exhibited accelerated swimming decline (Fig. 4D). Gain-of-function mutation in IP3 receptor also reduced age pigment accumulation (Fig. 4E, F). Increased ITR-1 activity extends median and maximum lifespan (53% increase of median lifespan, 29% increase of maximum lifespan), whereas reduced ITR-1 activity shortens culture survival (i.e., -11% increase of median lifespan, -31% increase of maximum lifespan) (Fig. 4G, H). We find that the double mutant of progeric lin-3 (rf) with healthy aging itr-1 (gf) exhibits healthspan extension (Fig. S8B, Supporting Information) consistent with positioning of IP3R activity downstream of (or parallel to) EGF/LIN-3 action. Thus, both RNAi studies and analysis of mutant strains indicate that elevated signaling through the IP3 receptor extends C. elegans healthspan and lifespan.
Taken together, our perturbations of EGF pathway components support that beneficial effects of EGFR/LET-23 signaling occur via the IP3 receptor pathway to promote healthy adult aging and longevity in C. elegans. This is a previously undescribed role for the EGF/IP3R pathway in aging biology.
HPA-1 and HPA-2 influence locomotory healthspan through the EGF pathway
Having established the positive impact of EGF signaling on healthspan and defined the downstream pathway operative, we returned to address the hypothesis that HPA-1 and HPA-2 act via the EGF pathway. We confirmed that the swimming healthspan phenotypes induced by hpa-1 (RNAi) and hpa-2 (RNAi) depend on the activity of the EGF signaling pathway by conducting RNAi inactivation in mutants defective for specific components of EGF signaling. In wild-type, hpa-1 (RNAi) and hpa-2 (RNAi) significantly increase late-life swimming vigor (Fig. 1B). In contrast, neither hpa-1 (RNAi) nor hpa-2 (RNAi) extends swimming healthspan in mutants bearing reduction-of-function alleles in EGF pathway genes EGF/lin-3 (Fig. S8D, Supporting Information), EGFR/let-23 (Fig. 5A) or IP3R/itr-1 (Fig. 5B). Furthermore, neither hpa-1 (RNAi) nor hpa-2 (RNAi) can further extend the swimming prowess of gain-of-function mutations in EGFR/let-23 or IP3R/itr-1 (Fig. 5C, D). However, hpa-1 (RNAi) and hpa-2 (RNAi) significantly increase late-life swimming vigor in mutants of either the RAS pathway (let-60 (n1021) rf and let-60 (n1046) gf) or the DAG pathway (unc-13 (e51) rf and dgk-1 (nu62) rf, which increases UNC-13 activity) (Fig. S9, Supporting Information), consistent with roles for HPA-1 and HPA-2 in only the downstream ITR-1 signaling branch. We conclude that the EGF pathway must be operative for beneficial HPA-1 and HPA-2 knockdown effects, and that HPA-1 and HPA-2 exert their most significant effects on healthspan via the EGFR/ITR1 pathway.
Fig. 5. HPA-1 and HPA-2 influence locomotory aging through the EGF pathway.

Effect of EGFR and IP3R mutations on swimming vigor under conditions of hpa-1 and hpa-2 RNAi. (A) let-23 (n1045) rf mutants. (B) itr-1 (sa73) rf mutants. (C) let-23 (sa62) gf mutants. (D) itr-1 (sy290)(gf) mutant were introduced to indicated dsRNAs by bacterial feeding; control is empty vector. Error bars represent s.e.m., unpaired two tailed t-test (control versus hpa-1 or hpa-2 RNAi on each day: unlike wt strains, P values support no significant differences). Note: let-23 (n1045) rf mutants were continuously grown at restrictive temperature of 20°C. itr-1 (sa73) rf strains were grown at permissive temperature of 15°C until day 5 after hatching (first day of egg laying), and then transferred to the non-permissive temperature (25°C). Other experiments were conducted at 25°C. Note that it is unlikely that let-23 (rf) and itr-1 (rf) mutations themselves disrupt RNAi efficacy because positive control sca-1 (RNAi) induced larval arrest and sterility to a similar extent in these backgrounds as in WT, and because we found that age-1 (RNAi) further extended swimming healthspan in let-23 (rf) and itr-1 (rf) mutants (data not shown).
Adult EGF/IP3R signaling can modulate locomotory healthspan
Our studies of itr-1 (rf) used a temperature-sensitive mutation with shifts to non-permissive temperature performed just prior to the reproductive adult stage to avoid developmental defects. Despite the fact that animals were disrupted for itr-1 activity only during adulthood, these interventions still prevent the HPA-1 and HPA-2 RNAi–dependent healthspan effects (Fig. 5B). These experiments suggest that EGFR/IP3R signaling can be activated in the adult to maintain adult robustness in swimming. Our RT/PCR experiments do find EGF isoforms (Dutt et al., 2004; Van Buskirk & Sternberg, 2007) expressed even in mid/late adult in synchronized populations (data not shown) and HPA-1 and HPA-2 translational GFP fusions are co-expressed strongly in adult in posterior intestine, and glial amphid and phasmid socket cells, and a few neurons (Fig. S11, Supporting Information). Thus, HPA-1, HPA-2, and EGF isoforms are expressed during adulthood, when they can act to influence healthspan.
In sum, we document a previously unreported pathway for the regulation of healthy aging and longevity in C. elegans (modeled in Fig. 6), revealing unexpected roles for proteins well known to promote cell specification and cell function, and suggesting unique regulatory mechanisms that control EGF/EGFR/PLC-γ/IP3R signaling relevant to adult maintenance. One possible model for healthspan modulation by HPA-1 and HPA-2 deficiency, suggested by homologies to ligand binding domains of EGRR and EGFR, is that HPA-1 and/or HPA-2 normally bind EGF to limit EGFR signaling. If this negative regulation is relieved, the EGFR pathway involving the downstream EGFR/PLC-γ/IP3R branch is activated to promote healthy aging.
Fig. 6. Model for HPA-1 and HPA-2 modulation of EGF signaling in aging C. elegans.

HPA-1 and HPA-2 are secreted proteins that might bind LIN-3/EGF via domains related to EGF binding regions of EGF receptor to limit signaling. Alternatively, HPA-1 and HPA-2 might interact with EGFR/LET-23 to prevent EGF/LIN-3 binding. In either case, the activities of HPA-1 and HPA-2 normally down-regulate EGF signaling. When HPA-1 or HPA-2 are disrupted, EGF signaling is increased, with the EGFR activating the downstream signaling pathway that includes phospholipase-Cγ (PLC-3) and the IP3 receptor (ITR-1) to promote healthspan as evidenced by low age pigment accumulation, extended locomotory function, and increased median lifespans. Note that activation of ITR-1 signaling in the adult appears necessary to confer a healthspan benefit for locomotory aging. Activation of the EGF or IP3R pathways later in life could be a therapeutic consideration for combating sarcopenia and other aspects of age-related decline.
Discussion
Pursuing an initial interest in hypothesized aging-associated functions of proteins related to extracellular ligand binding domains of DAF-2 insulin receptor, we identified two receptor-related genes, hpa-1 and hpa-2, for which RNAi conferred a high performance in advanced age (Hpa) phenotype for swimming behavior. Further analysis of these novel genes revealed an unexpected but potent role of EGF signaling in promoting system-wide healthy aging and longevity in the adult that appears largely distinct from insulin signaling mechanisms. To the best of our knowledge, this is the first report that EGF signaling constitutes a major healthspan and longevity pathway. Given conservation of EGF signaling pathways and data on mouse knockouts suggestive of roles for EGF in adult maintenance, we speculate that EGF family members may play a conserved role in maintaining adult health that might be exploited for anti-aging therapies.
Newly identified molecular modulators of EGF signaling are candidate secreted EGF binding proteins
We report that EGF signaling is limited by HPA-1 and HPA-2, and suggest this can occur during adult life. Eliminating the negative regulation mediated by HPA-1 and HPA-2 activates EGFR and promotes changes that are associated with improved healthy aging. HPA-1 and HPA-2 encode proteins of 516 and 472 amino acids, respectively, predicted to be secreted due to canonical signal sequences at their N-termini. HPA-1 and HPA-2 exhibit some sequence similarity to ligand binding domains of the EGF-binding EGF receptor-related protein ERRP and to the EGF receptor itself. Rat ERRP is primarily expressed in intestine and liver (Yu et al., 2001) (interestingly, hpa-1 and hpa-2 are expressed in the worm intestine) and can negatively regulate EGFR activities in culture models (Yu et al., 2001; Marciniak et al., 2004) as well as in vivo (Schmelz et al., 2007). Although the mechanism of ERRP action has not yet been clearly defined, some evidence suggests ERRP sequesters EGFR ligand TGF-α to limit signaling. The similarity of HPA-1 and HPA-2 to proteins that bind EGF suggests that one mechanism of their action could be to bind and sequester EGF to negatively regulate EGFR activity. Such an EGF-sequestering regulatory mechanism has been documented for the secreted, but structurally distinct (Klein et al., 2008), Argos protein that binds to Drosophila EGF/Spitz to downregulate EGF signaling (Klein et al., 2004). Our study constitutes the first implication of putative secreted EGF binding proteins in any EGF signaling regulation in C. elegans. Whether HPA-1 and HPA-2 bind C. elegans EGF awaits biochemical confirmation. Regardless of the precise molecular mechanism by which HPA-1 and HPA-2 normally limit EGF signaling, it is noteworthy that these novel regulators of the EGF pathway exhibit a strong biological impact on age-associated phenotypes and without dramatic phenotypes on development.
We note that our RNAi screen identified 7 additional knockdowns of receptor-related proteins that also conferred statistically significant effects on swimming healthspan (Table S1, Supporting Information). Some of these additional hpa genes might also participate in EGF regulation that modulates locomotory aging. Alternatively, some might function as insulin binding proteins as originally proposed (Dlakic, 2002) to influence locomotory aging via IIS.
hpa-1 and hpa-2 function similarly, but not identically, to impact aging
It is somewhat striking that GFP reporters for hpa-1 and hpa-2 are expressed with the same timing and cell expression pattern (Fig. S11, Supporting Information), yet do not appear functionally redundant for basic phenotypes or for aging phenotypes (Fig. S3, Supporting Information). Moreover, we have noted that hpa-1 and hpa-2 RNAi knockdowns and mutants share many phenotypes and depend upon the same downstream signal transduction pathway, suggesting that they enact similar functions. Still, it should be underscored that age pigment accumulation patterns and maximum lifespan phenotypes differ between hpa-1 and hpa-2 (Fig 1D, G), and thus a subset of their activities, or the levels of activity required for specific functions, appear different.
hpa-1 and hpa-2 exert proportionately greater impact on the quality of mid-life than on overall longevity, and might thus be identified as “healthspan” genes
hpa-1 and hpa-2 genes have not been identified in previous genetic or RNAi screens for longevity. Indeed, maximum lifespan increase for hpa-1 (tm3256) is only on the order of 10% and is essentially undetectable for hpa-2 (tm3827). For both hpa mutants, however, the general increases in mid-life vigor as measured by swimming locomotory prowess and mid-life survival are more substantial than longevity phenotypes. Thus the hpa gene activities affect mid-life outcomes proportionately more than lifespan endpoints. The implications of this observation are worth underscoring: there may exist many genes with substantial effect on healthspan but relatively little effect on maximum lifespan. Such a gene class would most likely have been missed in previous screens focused on lifespan extension. Like the study we report here, future genetic screens that focus on healthspan phenotypes may thus uncover novel molecular strategies for healthy aging.
Downstream signaling that alters calcium balance can confer healthspan benefits
After EGFR activation, the downstream signal transduction pathway that promotes healthy aging involves PLC-γ/PLC-3 and IP3 receptor/ITR-1, which regulates ER calcium release and impacts cellular calcium homeostasis. This is the first implication of Ca2+ action through IP3 signaling in promoting nematode healthspan and lifespan. Calcium homeostasis undoubtedly plays an important role in adult maintenance (Imura et al., 2007), and has been suggested to be modulated in the long-lived Klotho mouse (Kurosu et al., 2005). The dramatic benefits of the itr-1 (gf) mutation on healthspan and lifespan, more substantial than in individual hpa mutants, may reflect an optimal level of pathway activity in the itr-1 (gf) mutant background. Such activity levels might be attained by further manipulation of EGF signaling levels in other hpa backgrounds or by adding inputs from other signaling pathways. Regardless of how optimal signaling is attained, our data implicate IP3R as a plausible therapeutic target for healthspan extension.
EGF exerts mechanistically distinct effects on behavioral quiescence and healthy aging
EGF signaling has elegantly been shown to induce a state of behavioral quiescence that precedes the four larval molts that occur as C. elegans grows to adulthood (Van Buskirk & Sternberg, 2007). Over-expression of EGF under control of a heat shock promoter (hspEGF) causes rapid cessation of pumping and slowing of locomotion, even in young adults. This role of EGF signaling is distinct from the HPA-regulated EGF signaling we characterized that systemically affects healthy aging in that: 1) hspEGF is associated with locomotory impairment and cessation of pumping (Van Buskirk & Sternberg, 2007), whereas HPA-1/2 (RNAi)-induced EGFR activation does not impact pumping rates in young adults (Fig. S2, Supporting Information) and is associated with maintained pumping and locomotory activity late into life; and, 2) molecular requirements for downstream EGF signaling are different for quiescence vs. healthspan outcomes, with hspEGF acting via DAG receptor UNC-13 (Van Buskirk & Sternberg, 2007) rather than through the ITR-1/IP3 receptor that we document influences healthspan prowess (Fig. 4B; Fig. S8C and Fig. S9, Supporting Information). Another significant difference in the experimental paradigms of EGF signaling modulation in these two studies is that overexpression studies transiently express specific EGF isoforms under heat shock stress conditions, so that time of expression, EGF concentration and particular isoforms overexpressed are likely different from the EGF signaling that occurs (or is prevented from occurring) as a component of normal aging. Despite the different pathways for EGF signaling in C. elegans aging and quiescence biology, the findings that EGF can promote healthy aging, that EGF can promote quiescence via a mechanism that also intersects in part with activities that influence sleep-like behavior (such as cGMP-dependent kinase EGL-4 (Raizen et al., 2006; Van Buskirk & Sternberg, 2007; Raizen et al., 2008)), and that sleep is known to have a significant affect on aging quality in humans (Neikrug & Ancoli-Israel, 2009), raise the question as to whether precisely modulated EGF signaling via multiple pathways might be a component of a fundamentally conserved rest/rejuvenation mechanism that promotes effective repair processes required for adult maintenance.
EGFs as conserved promoters of healthspan
In mammals, activated EGF signaling has been associated with epithelial and other cancers and secreted negative regulator ERRP has been used as a candidate anti-cancer therapeutic (Majumdar, 2003; Majumdar, 2005). Our data introduce a new way of thinking about EGF signaling in adults—effects on non-proliferating cells can clearly be beneficial. These effects may be conserved in mammals, a hypothesis that has not yet been directly tested. Interestingly, it has been reported that triple knock-out (TKO) of EGF ligands in mice causes accelerated hair and weight loss, dermatitis and skin ulceration with aging (Luetteke et al., 1999), suggesting the possibility of EGF signaling for promoting healthy aging in mammals. The implication of EGFR and IP3R activities in a pathway for healthspan and lifespan suggests antagonistic (i.e., targeted to mammalian ERRP) and agonistic (i.e., targeted to IP3 receptor) perturbations that might be considered in anti-aging strategies directed against functional disability in advanced age.
Experimental procedures
Strains and nematode growth
C. elegans were grown on nematode growth media (NGM) plates streaked with Escherichia coli OP50-1 (a streptomycin-resistant derivative of OP50) at 20°C as described (Brenner, 1974), except that temperature sensitive strains containing itr-1 (sa73) and daf-2 (e1368) were routinely kept at the permissive temperature of 15°C. Ts strains hsf-1 (sy441), let-23 (n1045), and spe-9 (hc88) were maintained at 20°C. Strains used in this study include: BA708 spe-9 (hc88) ts; CB138 unc-24 (e138); DA465 eat-2 (ad465); DA1116 eat-2 (ad1116); DR1572 daf-2 (e1368) ts; GP555 daf-16 (mgDf50); JT73 itr-1 (sa73); KP1097 dgk-1 (nu62); LD001 Is007[skn-1∷GFP, rol-6 (su1006) ]; MT2123 let-23 (n1045); MT2124 let-60 (n1046); MT2136 lin-3 (n1058)/unc-8 (e49) dpy-20; MT7929 unc-13 (e51); PS1524 let-23 (sa62); PS1631 itr-1 (sy290) dpy-20 (e1282); PS3551 hsf-1 (sy441); PD4251 dpy-20 (e1282) ccIs4251 [Pmyo-3NLS/GFP, dpy-20 (+) ];PS1378 itr-1 (sy290) lin-3 (n1058); TJ1052 age-1 (hx546); TM3256 hpa-1 (tm3256); TM3827 hpa-2 (tm3827) and N2 wild type. We outcrossed hpa-1 (tm3256) and hpa-2 (tm3827) six times with N2 to generate strains ZB2844 and ZB2845. The hpa-1 (tm3256) and hpa-2 (tm3827) alleles were tracked and/or sequenced by PCR amplification of genomic sequence encompassing the deletions with specific primers for hpa-1 (tm3256) (5′CGGTTATCTAGGTGTGGCCT3′ and 5′CCATGAGCAATATTACCCGA3′) and for hpa-2 (tm3827) (5′GTAGGTGGTAATTACGCCGA3′ and 5′ ACTCAAACAGCCGACATCGT3′), respectively.
Age synchronization
Egg-bearing animals were collected from NGM plates with OP50. Eggs were extracted in the cleaning solution (0.7 M NaOH with 2% Na-hypochlorite (household bleach)) and then washed with M9 buffer at least three times. Eggs were transferred to 7 ml M9 buffer in a 1 liter flask and incubated overnight at 20°C with fairly vigorous shaking to obtain synchronous L1 animals. The day of egg preparation was scored as day 0. For strains containing either daf-2 (e1368) or itr-1 (sa73), the temperature of incubation was kept at 15°C.
RNAi feeding
RNAi feeding was similar to as described in (Timmons et al., 2001; Kemp et al., 2009). Escherichia coli (HT115) producing dsRNA for individual genes was seeded onto RNAi plates containing 25 mg/ml carbenicillin with 0.2 % lactose to induce the expression of the dsRNA for the gene of interest. The negative control was conducted by seeding the plates with HT115 containing empty vector pL4440. Synchronous L1-stage animals were placed onto each plate. After growing to the young adult stage, animals were transferred away from their progeny to fresh HT115-seeded plates every 1-2 days until the end of the reproductive period (except when animals were sterile, in which case they were transferred to fresh plates at the young adult stage and then were kept on the same plate). For targeting itr-1, animals were initially incubated with HT115 containing pL4440 because itr-1 RNAi induces larval arrest. After growing to young adult stage, animals were transferred to fresh plates with HT115-expressing itr-1 dsRNA every 1-2 day.
Dietary restriction (DR)
For DR protocol, bacterial food deprivation treatments were carried out as described in reference (Gems & Riddle, 2000). Agar plates were spread with a suspension of HT115 bacteria and incubated overnight at room temperature. Plates were then irradiated in a UV Stratalinker (Stratagene, La Jolla, CA), with UV-killing verified by failure to form colonies upon streaking to Luria broth (LB) plate. For the assay, synchronous L1 animals were placed onto the NGM plate with UV-killed bacteria, and were incubated at 25°C. At the L4 stage, animals were transferred to fresh NGM with UV-killed bacteria. RNAi treatments were conducted at 25°C by using RNAi feeding protocol described above.
Swimming analysis
For RNAi screening of 54 insulin-related genes, we used the adult sterile strain, spe-9 (hc88), as the wild type control to avoid progeny overgrowth in age-synchronized population. Synchronous L1 animals containing spe-9 (hc88) were incubated at 25°C, the non-permissive temperature for adult sterility, and then were treated with RNAi feeding protocol as described above. Swimming assays for RNAi-screening were performed on day 11, a mid-to-late adult stage (∼ 60 % alive).
For the swimming assay, single worms were picked off agar plates spread with a lawn of bacteria. If animals were buried into a thick bacterial lawn, they were gently mined with a platinum wire and allowed to crawl on an unseeded agar plate for about 30 seconds to remove adherent bacteria, a protocol that did not change the frequency of body bends per minute (See Note1 in Supporting Information). We then transferred individual animals to 1 ml M9 buffer in a 24-well plate. After a 10-30 second recovery period, we counted the number of body bends during a 30 second trial using a stereomicroscope for observation. A body bend was defined as a change in the reciprocating motion of bending at the mid-body. Only animals that could move away after a touch and could thrash were used for the swimming assay (See Note 2 in Supporting Information).
Lifespan analysis
Synchronous L1 animals were placed onto an NGM plate with OP50, and were incubated at 20-25°C. Lifespan assays were initiated at young adult stage as counted from the hatch. After growing to the young adult stage, animals were transferred away from their progeny to fresh OP50-seeded plates every 1-2 day until the end of the reproductive period. For the temperature-sensitive mutant let-23 (n1045) as well as the control (N2 wild type), animals were grown at 25°C (permissive temperature) until day 5, and then were maintained at 15°C (non-permissive temperature). For temperature-sensitive mutant itr-1 (sa73), the temperature of incubation was kept at 15°C (permissive temperature) during growth up to 5 days in both mutants and N2 wild type, and then shifted to 20°C (non-permissive temperature). For RNAi lifespan experiments, age-synchronous animals were grown at 25°C and were treated with the RNAi feeding protocol as described above. Animals that were lost, or exploded, or died from internal hatching of progeny were censored at the time of the event. Survival analyses were performed using the Kaplan Meier method on censored data, and the significance of differences between survival curves calculated using the log rank test. The statistical software used was GraphPad Prism v.5.02 (GraphPad Software, Inc., La Jolla, CA 92037 USA), which also computed the median lifespan. Maximum lifespan was determined by taking the mean age at death of the longest-lived 10% of a given test population (Sutphin & Kaeberlein, 2008). The unpaired t-test was used to determine statistical significance and calculate P-values for the mean and maximum lifespan.
Pharyngeal pumping decline assays
Age-synchronous animals were grown at 25°C on E. coli strains at 25°C as described above. The number of contractions in the terminal bulb of pharynx was measured on Days 3, 5 and 7. For each strain, 10-15 different animals were scored during a 60 second trial.
In vivo spectrofluorimetric quantitation of age pigments
Age-synchronized animals were grown and collected as described for swimming assays. Autofluorescent peaks corresponding to age pigments (which change with age, excitation/emission pair 340 nm/430 nm) and tryptophan (which remain constant with age, excitation/emission pair 290 nm/330 nm) were measured from 50 worms by using an in vivo spectrofluorimetory (SkinSkan, JY Horiba, Edison, NJ, USA), as described previously (Gerstbrein et al., 2005). Often for analysis the ratio of AGE/TRP is compared to normalize. A Zeiss Axioplan 2 Microscope with a UV cube (excitation bondpass filter centered at 360 nm and 420 nm emission longpass filter) was used to image animals and to verify fluorescence intensity with the results from the spectrofluorimeter.
Fluorescence microscopy
Animals were observed and photographed using a Zeiss Axioplan 2 Microscope with a Real-14 Precision Digital camera. Observations of GFP expression were recorded and color images were taken from the documentation of results with Magnafire software. For counting the number of GFP-labeled nuclei in the muscle, the transgenic strain PD4251 and its double-mutant strain, which carries a gain-of-function let-23 (sa62) allele, were continuously grown at 25°C after age-synchronized preparation, as described above. Animals were then observed using the 40× objective of a fluorescence microscope (Herndon et al., 2002; Cao et al., 2007).
Quantitation of fluorescence intensity in the ASI neuron
The strain LD001, which expresses skn-1∷GFP in the ASI neurons, was observed as described in ref (Bishop & Guarente, 2007). Fluorescence images were collected from worms subjected to RNAi bacteria feeding or dietary restriction (DR) on day 4 at 25°C. Fluorescence intensity in the ASI was quantitated by Image J software [available from part of National Institute of Health (NIH) website: http//rsb.info.nih.gov/ij/], as described previously.
Supplementary Material
Acknowledgments
We thank the following people and institutes for providing the strains used in this study: A. Fire for PS4251, P. Sternberg for PS1524, the National BioResource Project at Tokyo Women's Medical University School of Medicine for tm3256 and the Caenorhabditis Genetic Center supported by NIH NCRR for many strains. We also thank A. Fire for plasmid vectors; J. Ahringer for RNAi library; B. Grant for helpful discussion; M. Barr and C. Rongo for critical reading of this manuscript; and H. Suda and T. Johnson for advice on mortality rate analysis. This work was supported by grants from the National Institute on Aging (AG024882), The Ellison Medical Foundation, and The Paul Glenn Foundation.
References
- Augustin H, Partridge L. Invertebrate models of age-related muscle degeneration. Biochim Biophys Acta. 2009;1790:1084–1094. doi: 10.1016/j.bbagen.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Beitel GJ, Clark SG, Horvitz HR. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348:503–509. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bolanowski MA, Russell RL, Jacobson LA. Quantitative measures of aging in the nematode Caenorhabditis elegans. I. Population and longitudinal studies of two behavioral parameters. Mech Ageing Dev. 1981;15:279–295. doi: 10.1016/0047-6374(81)90136-6. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton S, Partridge L. Insulin/IGF-like signalling, the central nervous system and aging. Biochem J. 2009;418:1–12. doi: 10.1042/BJ20082102. [DOI] [PubMed] [Google Scholar]
- Cao Z, Wu Y, Curry K, Wu Z, Christen Y, Luo Y. Ginkgo biloba extract EGb 761 and Wisconsin Ginseng delay sarcopenia in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2007;62:1337–1345. doi: 10.1093/gerona/62.12.1337. [DOI] [PubMed] [Google Scholar]
- Chow DK, Glenn CF, Johnston JL, Goldberg IG, Wolkow CA. Sarcopenia in the Caenorhabditis elegans pharynx correlates with muscle contraction rate over lifespan. Exp Gerontol. 2006;41:252–260. doi: 10.1016/j.exger.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW. Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell. 1998;92:523–533. doi: 10.1016/s0092-8674(00)80945-9. [DOI] [PubMed] [Google Scholar]
- Clokey GV, Jacobson LA. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Croll NA, Smith JM, Zuckerman BM. The aging process of the nematode Caenorhabditis elegans in bacterial and axenic culture. Exp Aging Res. 1977;3:175–189. doi: 10.1080/03610737708257101. [DOI] [PubMed] [Google Scholar]
- Dlakic M. A new family of putative insulin receptor-like proteins in C. elegans. Curr Biol. 2002;12:R155–157. doi: 10.1016/s0960-9822(02)00729-7. [DOI] [PubMed] [Google Scholar]
- Duhon SA, Johnson TE. Movement as an index of vitality: comparing wild type and the age-1 mutant of Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 1995;50:B254–261. doi: 10.1093/gerona/50a.5.b254. [DOI] [PubMed] [Google Scholar]
- Dutt A, Canevascini S, Froehli-Hoier E, Hajnal A. EGF signal propagation during C. elegans vulval development mediated by ROM-1 rhomboid. PLoS Biol. 2004;2:e334. doi: 10.1371/journal.pbio.0020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AL. Of worms and women: sarcopenia and its role in disability and mortality. J Am Geriatr Soc. 2004;52:1185–1190. doi: 10.1111/j.1532-5415.2004.52320.x. [DOI] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstbrein B, Stamatas G, Kollias N, Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell. 2005;4:127–137. doi: 10.1111/j.1474-9726.2005.00153.x. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Chayavichitsilp P, Depp C, Schork NJ, Jeste DV. Successful aging: from phenotype to genotype. Biol Psychiatry. 2007;62:282–293. doi: 10.1016/j.biopsych.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Glenn CF, Chow DK, David L, Cooke CA, Gami MS, Iser WB, Hanselman KB, Goldberg IG, Wolkow CA. Behavioral deficits during early stages of aging in Caenorhabditis elegans result from locomotory deficits possibly linked to muscle frailty. J Gerontol A Biol Sci Med Sci. 2004;59:1251–1260. doi: 10.1093/gerona/59.12.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Sternberg PW. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell. 1990;63:921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Hill RJ, Sternberg PW. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature. 1992;358:470–476. doi: 10.1038/358470a0. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Feng Z, Hsieh MY, Xu XZ. Identification by machine vision of the rate of motor activity decline as a lifespan predictor in C. elegans. Neurobiol Aging. 2009;30:1498–1503. doi: 10.1016/j.neurobiolaging.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LS, Sternberg PW. Genetic dissection of developmental pathways. WormBook. 2006:1–19. doi: 10.1895/wormbook.1.88.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura A, Tsuji Y, Murata M, Maeda R, Kubota K, Iwano A, Obuse C, Togashi K, Tominaga M, Kita N, Tomiyama K, Iijima J, Nabeshima Y, Fujioka M, Asato R, Tanaka S, Kojima K, Ito J, Nozaki K, Hashimoto N, Ito T, Nishio T, Uchiyama T, Fujimori T, Nabeshima Y. alpha-Klotho as a regulator of calcium homeostasis. Science. 2007;316:1615–1618. doi: 10.1126/science.1135901. [DOI] [PubMed] [Google Scholar]
- Johnson TE. Aging can be genetically dissected into component processes using long-lived lines of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1987;84:3777–3781. doi: 10.1073/pnas.84.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kemp BJ, Church DL, Hatzold J, Conradt B, Lambie EJ. Gem-1 encodes an SLC16 monocarboxylate transporter-related protein that functions in parallel to the gon-2 TRPM channel during gonad development in Caenorhabditis elegans. Genetics. 2009;181:581–591. doi: 10.1534/genetics.108.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kirkland JL, Peterson C. Healthspan, translation, and new outcomes for animal studies of aging. J Gerontol A Biol Sci Med Sci. 2009;64:209–212. doi: 10.1093/gerona/gln063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DE, Nappi VM, Reeves GT, Shvartsman SY, Lemmon MA. Argos inhibits epidermal growth factor receptor signalling by ligand sequestration. Nature. 2004;430:1040–1044. doi: 10.1038/nature02840. [DOI] [PubMed] [Google Scholar]
- Klein DE, Stayrook SE, Shi F, Narayan K, Lemmon MA. Structural basis for EGFR ligand sequestration by Argos. Nature. 2008;453:1271–1275. doi: 10.1038/nature06978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomura I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro-o M. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner MR, Kornfeld K, Miller LM, Horvitz HR, Kim SK. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev. 1994;8:160–173. doi: 10.1101/gad.8.2.160. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Streeper T, Cawthon P, Baldwin K, Taaffe DR, Harris TB. Sarcopenia: etiology, clinical consequences, intervention, and assessment. Osteoporos Int. 2009 doi: 10.1007/s00198-009-1059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Majumdar AP. Regulation of gastrointestinal mucosal growth during aging. J Physiol Pharmacol. 2003;4(54 Suppl):143–154. [PubMed] [Google Scholar]
- Majumdar AP. Therapeutic potential of EGFR-related protein, a universal EGFR family antagonist. Future Oncol. 2005;1:235–245. doi: 10.1517/14796694.1.2.235. [DOI] [PubMed] [Google Scholar]
- Malone EA, Thomas JH. A screen for nonconditional dauer-constitutive mutations in Caenorhabditis elegans. Genetics. 1994;136:879–886. doi: 10.1093/genetics/136.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak DJ, Rishi AK, Sarkar FH, Majumdar AP. Epidermal growth factor receptor-related peptide inhibits growth of PC-3 prostate cancer cells. Mol Cancer Ther. 2004;3:1615–1621. [PubMed] [Google Scholar]
- Merris M, Kraeft J, Tint GS, Lenard J. Long-term effects of sterol depletion in C. elegans: sterol content of synchronized wild-type and mutant populations. J Lipid Res. 2004;45:2044–2051. doi: 10.1194/jlr.M400100-JLR200. [DOI] [PubMed] [Google Scholar]
- Moghal N, Sternberg PW. The epidermal growth factor system in Caenorhabditis elegans. Exp Cell Res. 2003;284:150–159. doi: 10.1016/s0014-4827(02)00097-6. [DOI] [PubMed] [Google Scholar]
- Murakami H, Bessinger K, Hellmann J, Murakami S. Manipulation of serotonin signal suppresses early phase of behavioral aging in Caenorhabditis elegans. Neurobiol Aging. 2008;29:1093–1100. doi: 10.1016/j.neurobiolaging.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Neikrug AB, Ancoli-Israel S. Sleep Disorders in the Older Adult: A Mini-Review. Gerontology. 2009 doi: 10.1159/000236900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, McIntire SL. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc Natl Acad Sci U S A. 2008;105:20982–20987. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, Liu LX, Doberstein SK, Ruvkun G. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Cullison KM, Pack AI, Sundaram MV. A novel gain-of-function mutant of the cyclic GMP-dependent protein kinase egl-4 affects multiple physiological processes in Caenorhabditis elegans. Genetics. 2006;173:177–187. doi: 10.1534/genetics.106.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- Restif C, Metaxas D. Tracking the swimming motions of C. elegans worms with applications in aging studies. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2008;11:35–42. doi: 10.1007/978-3-540-85988-8_5. [DOI] [PubMed] [Google Scholar]
- Schmelz EM, Xu H, Sengupta R, Du J, Banerjee S, Sarkar FH, Rishi AK, Majumdar AP. Regression of early and intermediate stages of colon cancer by targeting multiple members of the EGFR family with EGFR-related protein. Cancer Res. 2007;67:5389–5396. doi: 10.1158/0008-5472.CAN-07-0536. [DOI] [PubMed] [Google Scholar]
- Sutphin GL, Kaeberlein M. Dietary restriction by bacterial deprivation increases life span in wild-derived nematodes. Exp Gerontol. 2008;43:130–135. doi: 10.1016/j.exger.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M. Can we develop genetically tractable models to assess healthspan (rather than life span) in animal models? J Gerontol A Biol Sci Med Sci. 2009;64:161–163. doi: 10.1093/gerona/gln067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Ulrich P, Cerami A. Protein glycation, diabetes, and aging. Recent Prog Horm Res. 2001;56:1–21. doi: 10.1210/rp.56.1.1. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- Wu Y, Han M. Suppression of activated Let-60 ras protein defines a role of Caenorhabditis elegans Sur-1 MAP kinase in vulval differentiation. Genes Dev. 1994;8:147–159. doi: 10.1101/gad.8.2.147. [DOI] [PubMed] [Google Scholar]
- Yu Y, Rishi AK, Turner JR, Liu D, Black ED, Moshier JA, Majumdar AP. Cloning of a novel EGFR-related peptide: a putative negative regulator of EGFR. Am J Physiol Cell Physiol. 2001;280:C1083–1089. doi: 10.1152/ajpcell.2001.280.5.C1083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


