Abstract
Low androgen levels are associated with an increased risk of coronary artery disease (CAD), thrombosis and myocardial infarction (MI), suggesting that androgen has a protective role. However, little is known about the underlying molecular mechanism. Our genome-wide association study identified the ADTRP gene encoding the androgen-dependent TFPI regulating protein as a susceptibility gene for CAD and MI. The expression level of ADTRP was regulated by androgen, but the molecular mechanism is unknown. In this study, we identified the molecular mechanism by which androgen regulates ADTRP expression and tested the hypothesis that androgen plays a protective role in cardiovascular disease by activating ADTRP expression. Luciferase assays with an ADTRP promoter luciferase reporter revealed that androgen regulated ADTRP transcription in a dose- and time-dependent manner, and the effect was abolished by three different androgen inhibitors, including pyrvinium pamoate, bicalutamide, and cyproterone acetate. Chromatin-immunoprecipitation showed that the androgen receptor bound to a half androgen response element (ARE, TGTTCT) located at + 324 bp from the ADTRP transcription start site. The ARE is required for concentration-dependent transcriptional activation of ADTRP. HL-60 monocyte adhesion to EAhy926 endothelial cells (ECs) and transmigration across the EC layer, the two processes critical to development of CAD and MI, were inhibited by androgen, but the effect was rescued by ADTRP siRNA and exacerbated by overexpression of ADTRP and its downstream genes PIK3R3 and MIA3. These data suggest that one molecular mechanism by which androgen confers protection against CAD is stimulation of ADTRP expression.
Keywords: Androgen, Androgen receptor (AR), ADTRP, Atherosclerosis, Coronary artery disease (CAD), Myocardial infarction (MI), Monocyte adhesion to endothelial cells, Monocyte transmigration across endothelial, cells, Transendothelial migration of monocytes
1. Introduction
Coronary artery disease (CAD) and its main complication myocardial infarction (MI) are the leading cause of death worldwide [1–5]. CAD and MI are caused by genetics factors, environmental factors and interactions among these factors. Many risk factors have been identified for CAD and MI, including male gender, smoking, hyperlipidemia, diabetes, hypertension and obesity. There has been a long history of interest in the relationship between androgen and cardiovascular disease. Lower serum androgen/testosterone levels were associated with insulin resistance, diabetes and metabolic syndrome, which all increase risk of cardiovascular disease [6]. Moreover, many studies showed that lower serum androgen/testosterone levels were associated with atherosclerosis, cardiovascular risk, stroke and cardiovascular mortality in men [6]. Androgen can bind to its receptor, the androgen receptor (AR), in the cytoplasm, which activates the AR and translocates it to the nucleus. The AR is a DNA-binding transcriptional factor that regulates gene expression by directly binding to the androgen response element (ARE) at the promoter/regulatory region of target genes [7–10]. Knockout of the AR gene increased atherosclerosis in mice under the ApoE−/− knockout background [11,12]. Thus, the atheroprotective effect of androgen was also inferred in animal models. However, the molecular mechanism by which androgen reduces risk of atherosclerotic CAD is unknown.
Genome-wide association studies (GWAS) have been effective in identifying genomic variants that confer risk or protection of CAD and MI [13,14]. Our first GWAS for CAD and MI in a non-European ancestry population identified a susceptibility single nucleotide polymorphism (SNP) for CAD and MI, rs6903956 in the ADTRP gene encoding the androgen-dependent TFPI regulating protein [15]. The ADTRP protein showed homology to AIG1 (androgen-induced protein 1). ADTRP was found to be expressed on the cell surface of endothelial cells (ECs) and regulate the expression of TFPI (Tissue Factor Pathway Inhibitor) [16], which is a key natural inhibitor of coagulation and thrombosis [16]. Recently, we reported that ADTRP regulates cell cycle progression, proliferation, and apoptosis by global regulation of gene expression [17]. Moreover, we also reported that ADTRP regulates the levels of MIA3/TANGO1 (another CAD and MI-associated gene identified by GWAS), collagen VII and ApoB by positively regulating the expression of PIK3R3 and activation of AKT [18]. The ADTRP-MIA3-collagen VII/ApoB network was associated with monocyte adhesion to endothelial cells and transmigration of monocytes across the endothelial cell layer, the two cellular processes directly relevant to atherosclerosis [18]. Chinetti-Gbaguidi et al. recently showed that ADTRP is expressed in human macrophages and atherosclerotic lesions, and its expression is regulated by the peroxisome proliferator-activated receptor (PPAR)γ in human primary macrophages [19].
Similar to AIG1 [20], the expression levels of both ADTRP mRNA and ADTRP protein on the endothelial cell surface were up-regulated by androgen (dihydrotestosterone) [16], but the molecular mechanism remains to be identified. In the present study, we characterized the molecular mechanism for the regulation of ADTRP by androgen. We found that the transcriptional activation of ADTRP is induced by androgen and by the direct binding of the AR to the ADTRP promoter/regulatory region. Importantly, we demonstrated that androgen exerts the atheroprotective effect by decreasing monocyte adhesion to endothelial cells and monocyte transmigration across endothelial cells via up-regulation of ADTRP.
2. Materials and methods
2.1. Cell culture
The EAhy926 endothelial cell line was purchased from Shanghai Institute of Biochemistry and Cell Biology of SIBCB (Shanghai, China) and maintained in the human endothelial basal growth medium supplemented with 15% fetal bovine serum (FBS) from Gibco, Life Technologies, CA, USA. The HeLa cell line was purchased from ATCC (American Type Culture Collection, VA, USA) and maintained in the Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% FBS. The HL-60 cells line was purchased from Pricells (Wuhan, Hubei, China) and maintained in the 1640 medium supplemented with 20% FBS from Gibco, Life Technologies, California, USA. All cells were cultured at 37 °C in a humidified incubator with 5% CO2.
2.2. Plasmids and siRNA
The promoter/regulatory region of the ADTRP gene starting from −789 bp to + 724 bp from the transcriptional start site (TSS) was amplified by PCR analysis using human genomic DNA as the template, the Prime STAR HS DNA polymerase (TaKaRa, Dalian, China) and PCR primers ADTRP-F-KpnI 5′-CGGGGTACCCCTCCTTGTCCAGCCTACAG-3′ and ADTRP-R-XhoI 5′-CCGCTCGAGCCCCTCTTTGA GCTCATCTG-3′. The PCR product was digested with restriction enzymes Kpn I and Xho I (TaKaRa, Dalian, China), and sub-cloned into the multiple cloning site of the pGL3-Basic luciferase vector. This generates a luciferase reporter for the ADTRP promoter, pGL3-Basic-ADTRPp-Luc, in which the ADTRP promoter/regulatory region is inserted upstream of the firefly luciferase coding region.
The half ARE sequence at the position of + 324 bp from the TSS was mutated from TGTTCT to AAAAAT and TAAAAA using site-directed mutagenesis as described elsewhere [21,22], resulting in two mutant reporters pGL3-Basic-ADTRPp-Luc-Mut1 and pGL3-Basic-ADTRPp-Luc-Mut2. The primers for mutagenesis from TGTTCT to AAAAAT included a forward primer 5′-TGCATATACCACTTCCT AAAAATGAGCTGGTATACTTTCC-3′ and a reverse primer 5′-GGAAAGTATACCAGCTCATTT TTAGGAAGTGGTATATGCA-3′ (for pGL3-Basic-ADTRPp-Luc-Mut1). The primers for mutagenesis from TGTTCT to TAAAAA included a forward primer 5′-TGCATATACCACTTCCTTAAAAAGAGCTGGTATACTTTCC-3′ and a reverse primer 5′-GGAAAGTATACCAGTTCTTTTTAAGGAAGTGGTATATGCA-3′ (for pGL3-Basic-ADTRPp-Luc-Mut 2).
A plasmid with the full-length cDNA for the human AR gene, pEZ-M02 AR, was purchased from Gene Copoeia (Guangzhou, China) (human genome build hg19 mRNA: NC_000023.11, Entrez GeneID 367, Ensembl: ENSG00000169083). The full length cDNA for AR was amplified by PCR analysis using pEZ-M02 AR as the template and primers AR (920aa) 2763 bp F-BglII: 5′-GGAAGATCTGGATGGAAGTGCAGTTAGGGCTGGG-3’and R-XhoI:5′-CCGCTCGAGTCACTGGGTGTGGAAATAGATGGGCT-3′. The PCR product was digested using restriction enzymes Bgl II and Xho I (TAKARA, Dalian, China) and sub-cloned into the multiple cloning site of eukaryotic tag expression vector pCMV-Myc, generating a mammalian expression plasmid for AR, pCMV-Myc-AR.
The full-length cDNA for PIK3R3 (NC_000001.11, Entrez GeneID 8503, Ensembl: ENSG00000117461) was obtained by RT-PCR analysis using RNA samples isolated from HeLa cells. The PCR product was digested with EcoR I and Kpn I (TAKARA, Japan) and sub-cloned into the multiple cloning site of pCDNA3.1(−), resulting in an expression plasmid for PIK3R3, pCDNA3.1(−)-PIK3R3.
The expression plasmid for MIA3/TANGO1, pcDNA3.1(+)-TANGO1-HA, was as described previously [23–28] and kindly provided by Dr. Vivek Malhotra and colleagues.
We purchased siRNAs from Genepharma (Suzhou, China). The sequences for siRNAs are 5′-GGAUCCUCUUUCUCUACAATT-3′ (sense) and 5′-UUGUAGAGAAAGAGGAUCCTT-3′ (antisense) (ADTRP), 5′-GGACUUGCUUUAUGGGAAAdTdT-3′ (sense) and 5′-UUUCCCAUAAAGCAAGUCCdTdT-3′(antisense) (PIK3R3), and 5′-GGUGAAGUCUGAAUGCCAUTT-3′ (sense) and 5′-AUGGCAUUCAGACUUCACCTT-3′(antisense) (MIA3/TANGO1).
2.3. Androgen receptor inhibitors
Three different inhibitors for the AR were analyzed for their effects on androgen-induced transcriptional activation of the ADTRP promoter. The first potent AR inhibitor is pyrvinium pamoate salt hydrate from Sigma-Aldrich, MO, USA. Pyrvinium pamoate inhibits the AR activity by inhibiting androgen receptor-dependent gene expression via a distinct signaling mechanism [29]. It does not block DNA occupancy by the AR or binding to the ligand-binding domain of the androgen receptor [30]. Pyrvinium pamoate binds to the DNA binding domain of AR at the interface to the minor groove [31].
The second AR inhibitor is bicalutamide from Sigma-Aldrich, MO, the USA. It is a selective antagonist for the AR and blocks androgen binding to the AR and subsequent AR activation [32].
The third AR inhibitor is cyproterone acetate (Selleck Chemicals, Texas, USA). Cyproterone acetate is a potent competitive AR antagonist, which directly blocks binding of androgen to the AR and subsequent activation of the AR [33].
2.4. Cell transfection
For plasmid DNA, HeLa cells were transfected with 500 ng of wild type (WT) pGL3-Basic-ADTRPp-Luc or mutant reporters together with 25 ng of Renilla luciferase reporter plasmid using Lipofectamine®2000 (Invitrogen, Thermo Fisher Scientific, the Massachusetts, USA) according to the manufacturer's instruction. Transfection of EAhy926 cells was carried out using the Fu GENE® HD Transfection Reagent from Promega (Madison, Wisconsin, USA).
Transfection of siRNA was performed for HeLa and EAhy926 cells using Lipofectamine® RNAi MAX (Invitrogen, Thermo Fisher Scientific, USA) and Fu GENE®HD Transfection Reagent (Promega, Madison, Wisconsin, the USA), respectively.
2.5. Dual luciferase assays for the ADTRP promoter
Cells were cultured and transfected as describe above. Cells were harvested 48 h after transfection and used for dual luciferase assays using the Dual-luciferase reporter assay system (Promega, Madison, Wisconsin, the USA) according to the manufacturer's instructions and as described [34–36]. The firefly luciferase activity was normalized to the corresponding Renilla luciferase activity. Each luciferase assay was performed in triplicate and repeated at least three times.
2.6. Cell proliferation and toxicity assay
HeLa and EAhy926 cells (5 × 103 per well) were plated in 96-well plates, and transfected as described above. The transfected cells were then treated for 12 h with androgen (testosterone, 5 nM) and an AR-antagonist (pyrvinium pamoate, bicalutamide or cyproterone acetate) at the concentration of 0 nM, 0.02 nM, 0.05 nM, 0.1 nM, 0.25 nM and 0.5 nM, respectively. Cell proliferation assays were carried out using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) according to the manufacturer's instruction [17]. Each assay was performed in triplicate and repeated at least three times.
2.7. Chromatin immunoprecipitation (ChIP) assays
ChIP was performed using the chromatin immunoprecipitation linked PCR assay with the EZ-ChIP kit (Merck Millipore, Germany). Cross-linked chromatin was extracted from HeLa cells and EAhy926 endothelial cells transfected with the Myc-tagged AR expression plasmid pCMV-Myc-AR and immunoprecipitated using an anti-Myc antibody (Merck Millipore, Germany). Rabbit IgG was used as a negative control, whereas an anti-RNA polymerase II antibody (Merck Millipore, Germany) was used as a positive control. The precipitated DNA was used for PCR detection of the ARE bound by the AR and the promoter bound the RNA polymerase II. The PCR primers for the ARE include forward primer 5′-GCATATACCACTTCCTTGTTCTGAGCTG-3′ and reverse primer 5′-TCATATATTTCCACCTTGCACCATTTG-3′. The negative control for PCR was performed for a distal, non-ARE site using forward primer 5′-CTTCCCAGTTTACAGTATGTTGTTATAGCA-3′ and reverse primer 5′-GAGTTCTGCATTGTAACCATGTTTTTTT-3′. The PCR primers for the promoter (RNA polymerase II) included the forward primer 5′-GCAACTTCCTTGTTATATGCAAATGAAAAG-3′ and reverse primer 5′-CTGTTTCACTAGGGCCTGGGGCA-3′. Each ChIP assay was replicated at least three times.
2.8. Monocyte adhesion to endothelial cells assay
An assay for monocyte adhesion to ECs was carried out as described [18,37]. In brief, EAhy926 endothelial cells were transfected with siRNA or plasmid DNA as described above. The cells were seeded in 6-well plates at a density of 8 × 104 cells/well, grown to fully confluent and stimulated with oxidized LDL (0.8 mg/ml ox-LDL, Yesen, Shanghai, China) for 6 h. Human HL-60 promyelocytic leukemia cells were cultured and fluorescently labeled with Calcein AM (Fanbo, Beijing, China) for 60 min at 37 °C, and seeded into the wells coated with EAhy926 endothelial cells. After 1 h, the media were removed and fluorescence intensity was measured with a standard fluorometer with an inverse microscope (Nikon, The Tokyo metropolitan, Japan). The fluorescent units reflecting the number of adhered HL-60 cells to EAhy926 cells (exit: 490 nm, emission: 515 nm) was determined and analyzed with Image Pro Plus 6.0 software (Media Cybernetics, Bethesda, MD, the USA). Each experiment was repeated at least three times.
2.9. Transmigration of monocytes across endothelial cells
Transmigration of monocytes across endothelial cells was analyzed by a Transwell assay using the Boyden chamber method as described [17,18,37]. In brief, HL-60cells (3 × 105) were loaded into 0.4 µm pore size Transwell membranes within a Boyden chamber (Corning), which were plated with confluent EAhy926 endothelial cells transfected with siRNA or plasmid DNA. Cells were stimulated for 6 h with 0.25 µg of ox-LDL 24 h after the transfection. The HL-60 cells migrated to the bottom chamber 24 h after their administration were counted under an Olympus 81 FV1000 microscope. Each experiment was repeated at least three times.
2.10. Expression analyses
Real-time RT-PCR analysis was performed as described previously by us [17,18,38]. The primer sequences used are as follows:
ADTRP Forward: 5′-GCCGCATCCTATGGCTCTACTTTG-3′.
ADTRP Reverse: 5′-CAAGTAGGTAGATGCTGGCGATGA-3′.
MIA3 Forward: 5′-TACAAGCGGAGAATTGAAGAAATGG-3′.
MIA3 Reverse: 5′-GCCAGTTTTCATGAGCTTTCTTCT-3′.
GAPDH Forward: 5′-ATGGGGAAGGTGAAGGTCG-3′.
GAPDH Reverse: 5′-GGGGTCATTGATGGCAACAATA-3′.
PIK3R3 Forward: 5′-ATGTACAATACGGTGTGGAGTATG-3′.
PIK3R3 Reverse: 5′-GCTGGAGGATCCATTTCAAT-3′.
Western blot analysis was carried out as described previously by us [17,18]. The primary antibodies used include a rabbit polyclonal antibody against ADTRP (C6orf105) (1:800 dilution; Sigma, MO, USA), a rabbit polyclonal antibody against MIA3/TANGO1 (1:500; Santa Cruz Biotechnology, Dallas, TX, USA), a mouse monoclonal antibody against α-tubulin (1:5000; Merck Millipore, a part of Merck KGaA, Darmstadt, Germany), a polyclonal anti-goat antibody against ApoB (1:1000, Abcam, MA, USA), and a rabbit polyclonal antibody against collagen VII (1:500, Abcam, MA, USA). The secondary antibodies were HRPlabeled rabbit anti-goat antibody, goat anti-rabbit antibody and goat anti-mouse antibody (1:20,000 dilution) (Thermo Fisher Scientific).
2.11. Statistical analysis
The data were presented as mean ± standard error of means (SEM) unless specifically noted and were from at least three independent experiments. Statistical analysis was carried out using a Student's t-test or ANOVA with SPSS version17.0 software (SPSS, Chicago, IL, USA). Statistical significance was considered at a P value of 0.05 or less.
3. Results
3.1. Androgen induces ADTRP transcriptional activation in a dose-dependent and time-dependent manner
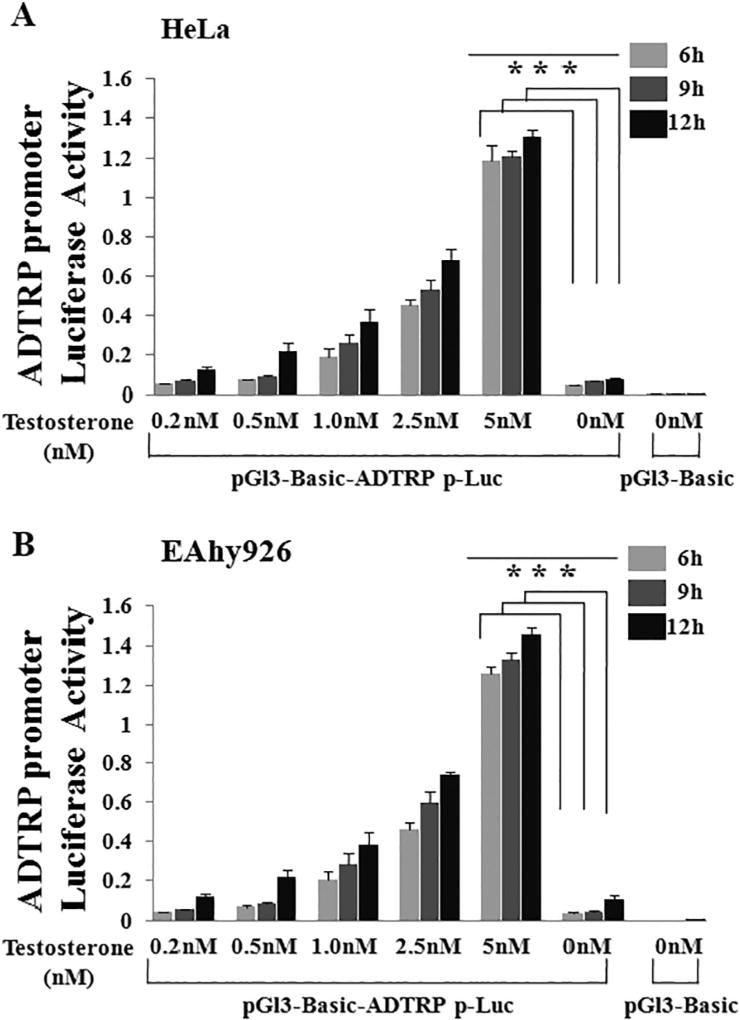
Lupu et al. used immunostaining analysis to show that androgen increased the cell surface expression level of the ADTRP protein by about 2-fold [16]. However, it is unknown whether the regulation of ADTRP expression by androgen is on the transcriptional level. We constructed an ADTRP promoter luciferase reporter to demonstrate that androgen activates ADTRP expression on the transcriptional level. HeLa cells were transfected with 500 ng of the pGL3-Basic-ADTRP-Luc reporter plasmid and 25 ng of Renilla luciferase reporter plasmid, treated with different concentrations of testosterone, and collected at different time points for luciferase assays. Luciferase assays revealed that testosterone induced transcriptional activation of the ADTRP promoter in a dose-dependent manner in HeLa cells (Fig. 1A). Testosterone also induced activation of the ADTRP promoter in a time-dependent manner in HeLa cells, especially at lower concentrations of androgen (Fig. 1A). Similar observations were made in EAhy926 endothelial cells (Fig. 1B).
Fig. 1.
ADTRP expression is induced by testosterone in HeLa and EAhy926 endothelial cells in a dose-and time-dependent manner. An ADTRP promoter luciferase reporter pGl3-Basic-ADTRPp-luc reporter (500 ng) and a Renilla luciferase plasmid (25 ng) were cotransfected into HeLa cells (A) or EAhy926 cells (B), cultured for 48 h, and used for luciferase assays. Treatments with testosterone were made at the time points of 36 h (12 h treatment), 39 h (9 h treatment) and 42 h (6 h treatment). Cells were collected and lysed and used for dual luciferase assays. ***, P < 0.001 (n = 4).
3.2. Androgen receptor inhibitors and antagonists block androgen-induced transcriptional activation of the ADTRP promoter
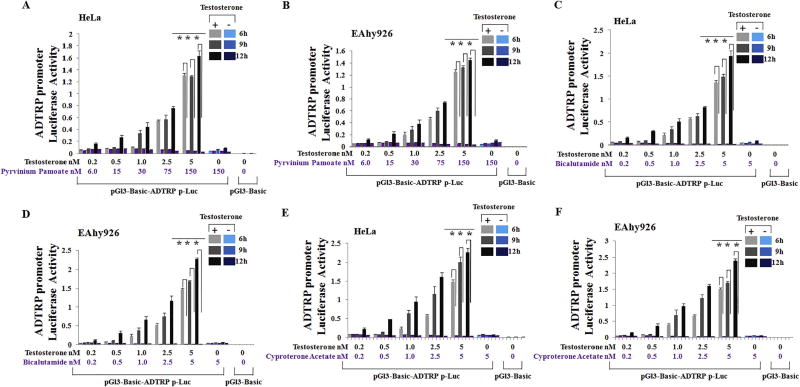
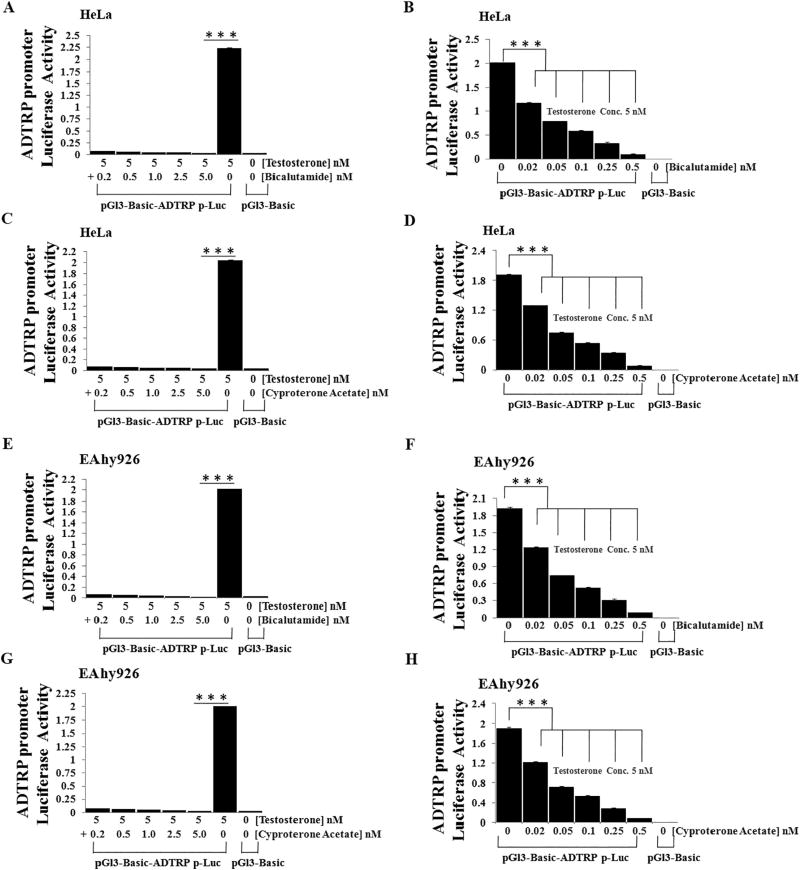
To further verify that androgen regulates ADTRP transcriptional activation, we studied three androgen receptor inhibitors and antagonists for their effects on transcriptional activation of the ADTRP promoter using luciferase assays. Testosterone induced strong transcriptional activation of the ADTRP promoter, but AR inhibitor pyrvinium pamoate abolished androgen-induced transactivation of the ADTRP promoter in HeLa cells (Fig. 2A) and EAhy926 cells (Fig. 2B). Similar results were obtained for AR antagonist bicalutamide (Fig. 2C–D) and cyproterone acetate (Fig. 2E–F). Further luciferase assays showed that bicalutamide and cyproterone acetate blocked androgen-induced ADTRP transcriptional activation in a dose-dependent manner in both HeLa cells (Fig. 3A–D) and EAhy926 cells (Fig. 3E–H). The doses of bicalutamide and cyproterone acetate used in the study were not toxic to cells (Fig. S1), suggesting that the effects of androgen antagonists on activation of ADTRP promoter were not due to toxic effects on cell growth.
Fig. 2.
AR inhibitors and antagonists block transcriptional activation of the ADTRP promoter stimulated by testosterone. HeLa cells and EAhy926 cells were treated and cultured as described in the Fig. 1 legend except for addition of a treatment group with both testosterone and an androgen inhibitor or antagonist. Cells were collected and lysed and used for dual luciferase assays. (A) Effect of AR inhibitor pyrvinium pamoate on androgen-induced ADTRP transactivation in HeLa cells. (B) Effect of AR inhibitor pyrvinium pamoate on androgen-induced ADTRP transactivation in EAhy926 cells. (C) Effect of AR antagonist bicalutamide on androgen-induced ADTRP transactivation in HeLa cells. (D) Effect of AR antagonist bicalutamide on androgen-induced ADTRP transactivation in EAhy926 endothelial cells. (E) Effect of AR antagonist cyproterone acetate on androgen-induced ADTRP transactivation in HeLa cells. (F) Effect of AR antagonist cyproterone acetate on androgen-induced ADTRP transactivation in EAhy926 endothelial cells. ***, P < 0.001 (n =4).
Fig. 3.
AR antagonists block testosterone-induced ADTRP transactivation in a dose-dependent manner. HeLa cells and EAhy926 cells were treated and cultured as described in the Fig. 2 legend except that the concentration of testosterone was fixed at 5 nM and that different concentrations of AR antagonists were used. Cells were collected and lysed and used for dual luciferase assays. (A) Effect of AR antagonist bicalutamide (0–5 nM) on androgen-induced ADTRP transactivation in HeLa cells. (B) Effect of AR antagonist bicalutamide (0–0.5 nM) on androgen-induced ADTRP transactivation in HeLa cells. (C) Effect of AR antagonist cyproterone acetate (0–5 nM) on androgen-induced ADTRP transactivation in HeLa cells. (D) Effect of AR antagonist cyproterone acetate (0–0.5 nM) on androgen-induced ADTRP transactivation in HeLa cells. (E) Effect of AR antagonist bicalutamide (0–5 nM) on androgen-induced ADTRP transactivation in EAhy926 endothelial cells. (F) Effect of AR antagonist bicalutamide (0–0.5 nM) on androgen-induced ADTRP transactivation in EAhy926 endothelial cells. (G) Effect of AR antagonist cyproterone acetate (0–5 nM) on androgen-induced ADTRP transactivation in EAhy926 endothelial cells. (H) Effect of AR antagonist cyproterone acetate (0–0.5 nM) on androgen-induced ADTRP transactivation in EAhy926 endothelial cells. ***, P < 0.001 (n = 4).
3.3. Androgen activates ADTRP transcription through the androgen responsive element (ARE) at the ADTRP promoter/regulatory region
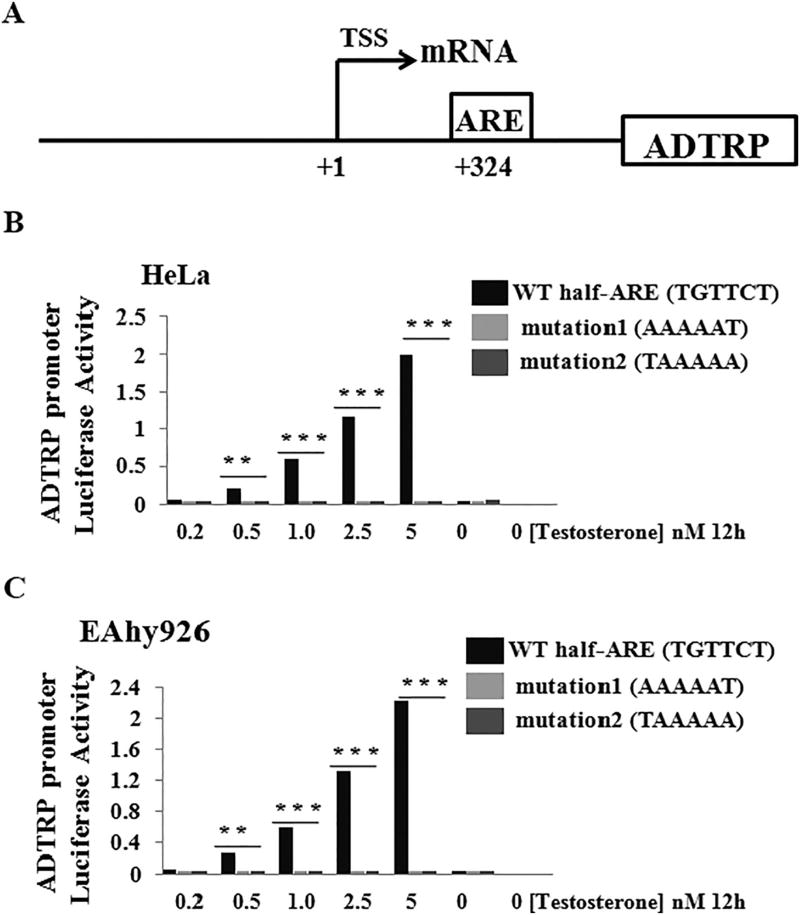
To identify the molecular mechanism by which androgen regulates transcriptional activation of the ADTRP promoter, we searched for potential binding sites for the AR at the ADTRP promoter/regulatory region and found one half androgen-responsive element (ARE) (TGTTCT) at the position of + 324 bp from the transcriptional start site (Fig. 4A). We mutated the half ARE site from TGTTCT to either AAAAAT (mutation 1) or TAAAAA (mutation 2). Compared to the WT ADTRP promoter, both mutant promoters lost responses to testosterone induction in both HeLa cells (Fig. 4B) and EAhy926 endothelial cells (Fig. 4C). These data suggest that the half ARE is responsible for the androgen-induced transcriptional activation of the ADTRP promoter.
Fig. 4.
Androgen-induced ADTRP transactivation is mediated by a half androgen response element in the ADTRP promoter/regulatory region. (A) Schematic diagram showing the structure of the ADTRP promoter/regulatory region. ARE, a half androgen-responsive element (TGTTCT); TSS, transcriptional start site (marked as position + 1). (B) Transcriptional activation activity (luciferase activity) from wild type (WT) and mutant ADTRP promoters with the ARE mutated in HeLa cells. Mutation 1, ARE TGTTCT mutated to AAAAAT; Mutation 2, ARE TGTTCT mutated to TAAAAA. (C) Transcriptional activation activity (luciferase activity) from wild type (WT) and mutant ADTRP promoters with the ARE mutated in EAhy926 endothelial cells. Cells were treated with different concentrations of testosterone varying from 0 to 5 nM for 12 h. **, P < 0.01 (n = 4); ***, P < 0.001 (n = 4).
3.4. Androgen receptor interacts with the half ARE at the ADTRP promoter/regulatory region
We performed chromatin immunoprecipitation (ChIP) analysis to determine whether the AR binds to the half ARE at the ADTRP promoter/regulatory region in vivo. HeLa cells were transfected with an expression plasmid for a Myc-tagged AR. The protein-DNA complex formed between the half ARE and Myc-tagged AR was immunoprecipitated using an anti-Myc antibody. The DNA associated with AR was pulled down by ChIP and detected by PCR analysis using primers spanning the ARE (Fig. 5A). As shown in Fig. 5B, the anti-Myc (AR) antibody successfully pulled the ARE down, but the control IgG failed to pull the ARE down. As negative controls, neither anti-Myc nor IgG pulled DNA at a distal non-specific control region down (Fig. 5A–B). We used an anti-RNA Polymerase II antibody as a positive control for ChIP analysis (Fig. 5C). The anti-RNA Polymerase II antibody, but not the negative control rabbit IgG, was able to pull DNA at the TSS of the ADTRP promoter down (Fig. 5C). Similar data were obtained in EAhy926 endothelial cells (Fig. 5D–E). These data suggest that the AR binds to the half ARE at the ADTRP promoter/regulatory region.
Fig. 5.
Chromatin immunoprecipitation (ChIP) for detection of the interaction between the AR and a half ARE at the ADTRP promoter/regulatory region. (A) Schematic diagram of the promoter/regulatory region of the human ADTRP gene. TSS, the transcription start site (+ 1 position); ARE, androgen-responsive element. (B) ChIP analysis showing that a Myc-tagged AR interacts with the ARE at the proximal ADTRP promoter/regulatory region in HeLa cell. An anti-Myc antibody pulled down chromatin containing the ADTRP ARE, but not a non-specific, control distal genomic DNA fragment. (C) Positive control for ChIP in HeLa cells. An anti-RNA Polymerase II antibody pulled down the chromatin containing the ADTRP TSS, but not a non-specific, control distal genomic DNA fragment. (D) Similar ChIP analysis as in (B), but in EAhy926 endothelial cells. (E) Similar control ChIP analysis as in (C), but in EAhy926 endothelial cells. ***, P < 0.001 (n = 4); NS, not significant.
3.5. Androgen has similar effects on endothelial cell proliferation, cell cycle progression, migration and apoptosis as ADTRP
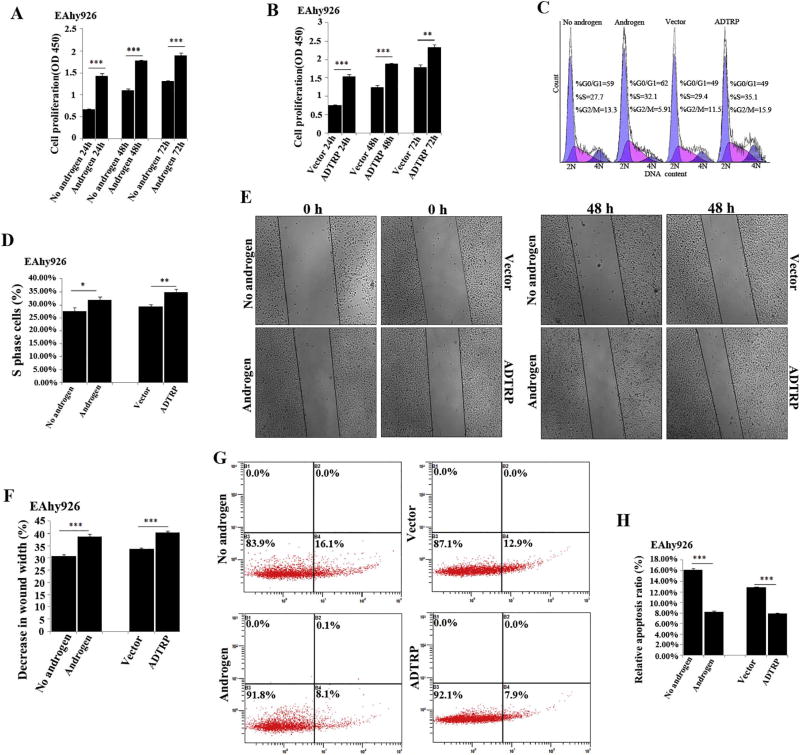
We previously reported that ADTRP regulates the proliferation, cell cycle progression and apoptosis of endothelial cells [17]. Because androgen regulates the expression of ADTRP, we hypothesized that androgen may have similar effects on endothelial cell functions as ADTRP. As shown in Fig. 6A–B, similar to overexpression of ADTRP, EAhy926 cells treated with androgen showed significantly increased proliferation at three time points of 24, 48 and 72 h compared to cells treated with vehicle. For the number of cells at the S phase, similar to overexpression of ADTRP, EAhy926 cells treated with androgen showed a significantly increased number of cells compared to cells treated with vehicle (Fig. 6C–D). Similarly, androgen had similar effect on promoting migration of EAhy926 cells as overexpression of ADTRP (Fig. 6E–F). For apoptosis, similar to overexpression of ADTRP, EAhy926 cells treated with androgen showed a significantly decreased rate of apoptosis compared to cells treated with vehicle (Fig. 6G–H).
Fig. 6.
Androgen has similar effects on EC proliferation, cell cycle progression, migration and apoptosis as overexpression of ADTRP. (A) Cell proliferation analysis for EAhy926 cells treated with androgen vs. vehicle (no androgen) control. ***, P < 0.001 (n = 3). (B) Cell proliferation analysis for EAhy926 cells with overexpression of ADTRP (transfection with an expression plasmid for ADTRP) or without ADTRP overexpression (transfection with the empty vector). **, P < 0.01 (n = 3); ***, P < 0.001 (n =3). (C) Representative flow cytometry data showing the cell number at the S phase of cell cycle for EAhy926 cells with or without overexpression of ADTRP vs. vector, and treated with androgen vs. vehicle. (D) Summary data from (C). *, P < 0.05 (n = 3); **, P < 0.01 (n = 3). (E) Representative images for scratch-based cell migration analysis for EAhy926 cells with or without overexpression of ADTRP vs. vector, and treated with androgen vs. vehicle. (F) Summary data from (E). ***, P < 0.001 (n =3). (G) Representative flow cytometry images for analysis of apoptosis for EAhy926 cells with or without overexpression of ADTRP vs. vector, and treated with androgen vs. vehicle. (H) Summary data from experiments as shown in (G), ***, P < 0.001 (n = 3).
3.6. Androgen regulates the levels of ADTRP-downstream genes/proteins PIK3R3, MIA3/TANGO1, collagen VII and ApoB
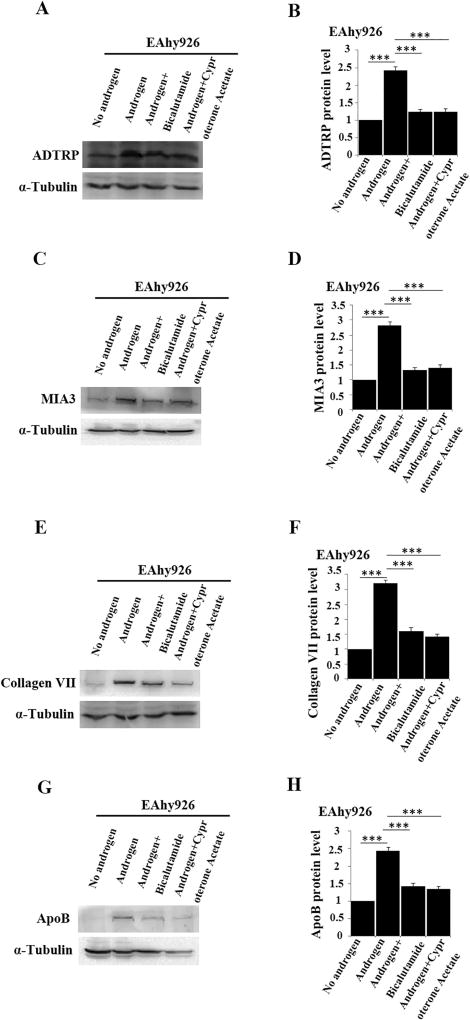
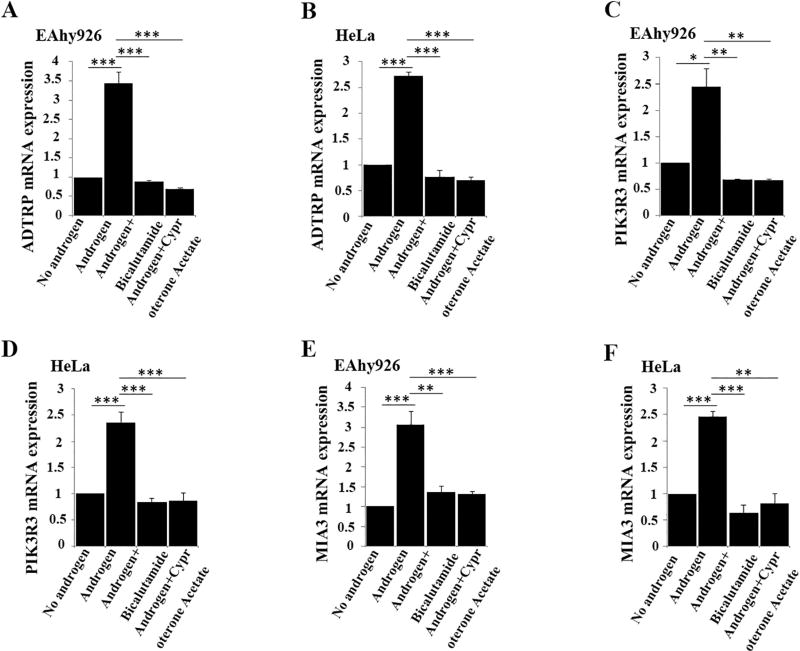
We previously reported that ADTRP regulates the expression of PIK3R3, which activates AKT and increases the expression level of MIA3/TANGO1, resulting in increased levels of collagen VII and ApoB [18]. As androgen regulates the expression of ADTRP, we examined the effect of androgen on the levels of PIK3R3, MIA3/TANGO1, collagen VII and ApoB. Western blot analysis showed that similar to the effect of overexpression of ADTRP (Fig. 7A–B) androgen treatment of EAhy926 cells significantly increased the levels of MIA3/TANGO1 (Fig. 7C–D), collagen VII (Fig. 7E–F), and ApoB (Fig. 7G–H), whereas the effects were almost abolished by androgen inhibitors bicalutamide and cyproterone acetate. Similar effects of androgen were observed on the mRNA levels for PIK3R3 and MIA3 by real-time RT-PCR analysis (Fig. 8). The real-time RT-PCR analysis was not performed for COL7A1 encoding collagen VII and ApoB because their regulation is on the level of protein trafficking [18].
Fig. 7.
Androgen regulates the levels of ADTRP-downstream targets PIK3R3 and MIA3/TANGO1 as well as the levels of collagen VII and ApoB downstream of MIA3/TANGO1. (A) Western blot analysis showing that the expression level of ADTRP in EAhy926 cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (B) Summary data from (A). (C) Western blot analysis showing that the expression level of MIA3/TANGO1 in EAhy926 cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (D) Summary data from (C). (E) Western blot analysis showing that the expression level of collagen VII in EAhy926 cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (F) Summary data from (E). (G) Western blot analysis showing that the expression level of ApoB in EAhy926 cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (H) Summary data from (G). ***, P < 0.001 (n = 3).
Fig. 8.
Androgen regulates the levels of ADTRP-downstream targets PIK3R3 and MIA3/TANGO1. (A) Real-time RT-PCR analysis showing that the expression level of ADTRP in EAhy926 cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (B) Real-time RT-PCR analysis showing that the expression level of ADTRP in HeLa cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (C) Real-time RT-PCR analysis showing that the expression level of PIK3R3 in EAhy926 cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (D) Real-time RT-PCR analysis showing that the expression level of PIK3R3 in HeLa cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (E) Real-time RT-PCR analysis showing that the expression level of MIA3 in EAhy926 cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. (F) Real-time RT-PCR analysis showing that the expression level of MIA3 in HeLa cells was induced by androgen and inhibited by androgen receptor antagonists bicalutamide and cyproterone acetate. *, P < 0.05 (n = 4); **, P < 0.01 (n =4); ***, P < 0.001 (n =4).
3.7. Androgen inhibits monocyte adhesion to endothelial cells, a functional process directly relevant to atherosclerosis, via activating ADTRP expression
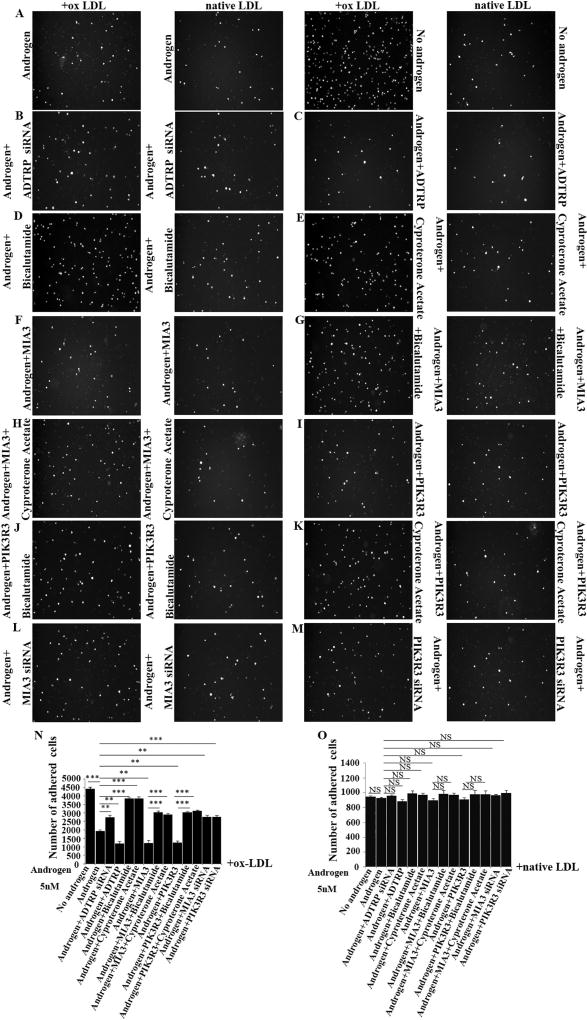
Reduced androgen levels were shown to be associated with risk of cardiovascular disease, therefore, androgen is protective of cardiovascular disease [18,39]. The molecular and cellular mechanisms by which androgen reduces risk of cardiovascular disease are unknown. Because androgen induces transcriptional activation of ADTRP, a susceptibility gene for CAD and MI, we hypothesized that androgen reduces risk of cardiovascular disease by activating expression of ADTRP. To test the hypothesis, we characterized the effects of androgen in combination with knockdown or overexpression of ADTRP on monocyte adhesion to the endothelium (a layer of ECs), the first event involved in the initiation of atherosclerosis [37]. Upon stimulation with oxidized-LDL (ox-LDL), significantly less HL-60 cells showed adhesion to EAhy926 cells plated on the bottom of culture wells when treated with androgen than treatment with vehicle (P < 0.001) (Fig. 9A and N). The data are consistent with the protective effect of androgen on atherosclerosis. The effect of androgen was partially rescued by knockdown of ADTRP expression with siRNA (Fig. 9B, N) and by siRNAs for PIK3R3 and MIA3/TANGO1 (Fig. 9L–M and N). The data suggest that androgen reduces monocyte adhesion to endothelial cells by activating ADTRP expression and expression of ADTRP-downstream genes PIK3R3 and MIA3/TANGO1 (9C, 9F, 9I and 9 N). On the contrary, overexpression of ADTRP by transient transfection of EAhy926 cells with an ADTRP expression further reduced HL-60 cell adhesion to endothelial cells by androgen (Fig. 9C and N). As controls, androgen receptor antagonists bicalutamide and cyproterone acetate blocked the androgen-induced reduction of adhesion of HL-60 to endothelial cells (Fig. 9D–E and N). Similar to ADTRP, overexpression of ADTRP-downstream genes PIK3R3 and MIA3 also further reduced HL-60 cell adhesion to endothelial cells by androgen (Fig. 9F, I and N), but the effects were abolished by bicalutamide or cyproterone acetate (Fig. 9G–H, J–K, and N).
Fig. 9.
Androgen inhibits ox-LDL-mediated monocyte adhesion to endothelial cells by inducing expression of ADTRP and its downstream genes PIK3R3 and MIA3/TANGO1. (A) Representative images for analysis of monocyte adhesion to EAhy926 endothelial cells stimulated by ox-LDL (native LDL as control) in the presence and absence of testosterone treatment (5 nM). (B) Effect of ADTRP siRNA on androgen-blocked HL-60 monocyte adhesion to EAhy926 endothelial cells. (C) Effect of ADTRP overexpression on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (D) Effect of bicalutamide on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (E) Effect of cyproterone acetate on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (F) Effect of MIA3/TANGO1 overexpression on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (G) Effect of both MIA3/TANGO1 overexpression and bicalutamide on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (H) Effect of both MIA3/TANGO1 overexpression and cyproterone acetate on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (I) Effect of PIK3R3 overexpression on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (J) Effect of both PIK3R3 overexpression and bicalutamide on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (K) Effect of both PIK3R3 overexpression and cyproterone acetate on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (L) Effect of MIA3/TANGO1 siRNA on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (M) Effect of PIK3R3 siRNA on androgen-blocked monocyte adhesion to EAhy926 endothelial cells. (N) Summary data from experiments in (A–M) with ox-LDL treatment. (O) Summary data from experiments in (A–M) with native LDL treatment. **, P < 0.01 (n = 4); ***, P < 0.001; NS, not significant.
The effects of androgen and ADTRP as well as ADTRP-downstream genes PIK3R3 and MIA3 on monocyte adhesion to endothelial cells occurred only when cells were treated with ox-LDL, but not with native LDL (Fig. 9A–E, F–M and O).
3.8. Androgen inhibits monocyte transmigration across a layer of endothelial cells, a functional process directly relevant to atherosclerosis, via activating ADTRP expression
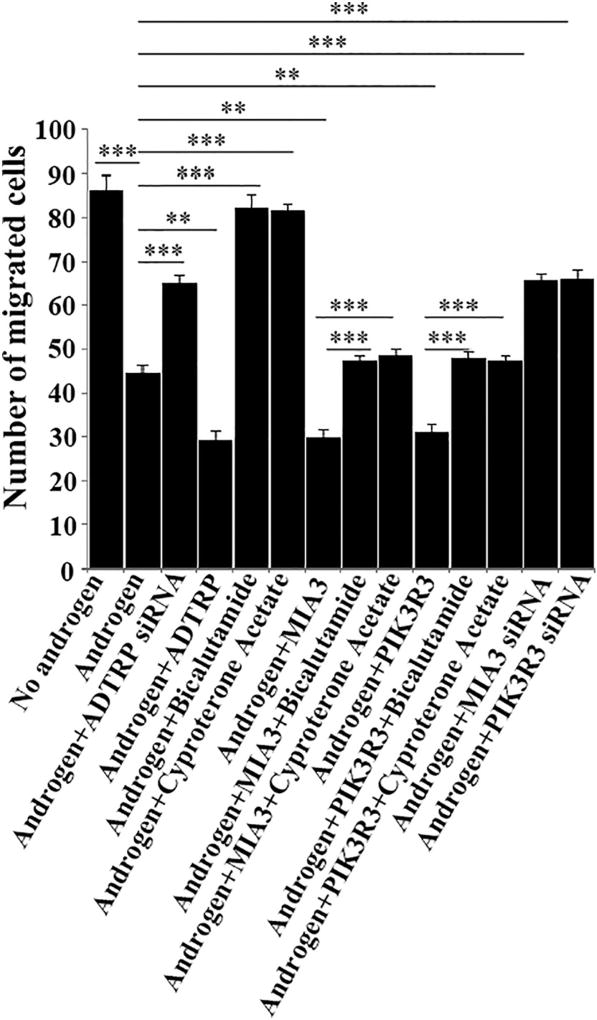
Trans-endothelial migration of monocytes is one of the most important processes in the initiation of atherosclerosis [18,37]. We coated the Transwell membrane with confluent EAhy926 endothelial cells transfected with siRNA or plasmid DNA and inserted it into the Boyden chamber. Cells were treated with ox-LDL or other agents. HL-60 cells were then loaded on the top of the endothelial cell layer. The HL-60 cells migrated across the endothelial layer were counted and analyzed. Androgen treatment significantly inhibited ox-LDL-induced HL-60 migration across the layer of EAhy926 cells (Fig. 10), which is consistent with the protective effect of androgen on atherosclerosis. Knockdown of ADTRP expression by siRNA attenuated the effect of androgen (Fig. 10), suggesting that androgen reduces trans-endothelial migration of monocytes by activating ADTRP expression. Similar to ADTRP siRNA, PIK3R3 and MIA3/TANGO1 siRNAs also attenuated the effect of androgen on monocyte transmigration of endothelial cells (Fig. 10). Consistent with this, overexpression of ADTRP by transient transfection of EAhy926 cells with an ADTRP expression plasmid further reduced HL-60 cell migration across a layer of EAhy926 endothelial cells by androgen (Fig. 10). As controls, androgen receptor antagonists bicalutamide and cyproterone acetate blocked the androgen-induced reduction of HL-60 migration across to a layer of EAhy926 endothelial cells (Fig. 10). Similar to ADTRP, overexpression of ADTRP-downstream genes PIK3R3 and MIA3 in EAhy926 cells further reduced HL-60 cell migration across a layer of EAhy926 endothelial cells by androgen, and the effects were abolished by androgen receptor antagonists bicalutamide and cyproterone acetate (Fig. 10).
Fig. 10.
Androgen inhibits ox-LDL-mediated transmigration of monocytes across a layer of EAhy926 endothelial cells by inducing expression of ADTRP and its downstream genes PIK3R3 and MIA3. HL-60 monocytes (3 × 105) were loaded on the top of confluent EAhy926 endothelial cells transfected with siRNA or plasmid DNA and stimulated with ox-LDL (native LDL as control) in the presence and absence of testosterone treatment (5 nM) cultured on 0.4 µm pore size Transwell membranes within a Boyden chamber. Twenty-four hours later, the HL-60 cells that have migrated to the bottom of the Transwell and into the chamber were manually counted under a microscope. Each experiment was repeated at least three times. Each experiment was repeated at least three times. ***, P < 0.001 (n = 4).
4. Discussion
Previously, Lupu et al. used qRT-PCR to show that ADTRP mRNA expression was up-regulated in endothelial cells by 30 nM dihydrotestosterone treatment for 24 h [16]. In addition, immunostaining showed that the cell surface expression of the ADTRP protein was also increased by dihydrotestosterone treatment [16]. However, it is unknown whether androgen regulates ADTRP expression at the transcriptional level. In this study, we further assessed the regulation of ADTRP expression by androgen using luciferase assays of an ADTRP promoter luciferase reporter gene. Testosterone treatment increased the ADTRP promoter luciferase reporter activity in a dose-and time-dependent manner and three different androgen inhibitors/antagonists, including pyrvinium pamoate, bicalutamide and cyproterone acetate, abrogated the effects of testosterone (Figs. 1–3). These data suggest that androgen regulation of ADTRP expression is on the transcriptional level. ChIP analysis showed that the androgen receptor binds to the half ARE (TGTTCT) at the promoter/regulatory region of ADTRP (Fig. 5). Mutational analysis showed that the half ARE is critical to the transcriptional activation of ADTRP and mutations of the ARE abolished the transcriptional activation of ADTRP (Fig. 4). Our data suggest that androgen induces transcriptional activation of ADTRP through direct binding of the androgen receptor to the ARE at the ADTRP promoter/regulatory region.
Atherosclerosis in coronary arteries is the cause of CAD and MI [40,41]. Atherosclerosis is initiated with the activation of endothelial cells (endothelium) by ox-LDL or other inflammatory molecules, which attracts adhesion of monocytes to the endothelium [40,41]. The monocytes then transmigrate across the endothelium and become macrophages. Macrophages ingest ox-LDL and other lipids, attract other cells, and induce apoptosis, which leads to formation of foam cells and plaques, the hallmark of atherosclerosis [40,41]. The molecular mechanism by which a low level of androgen increases the risk of atherosclerosis is unknown. Our data in this study identified one potential mechanism (the first molecular mechanism to the best of knowledge): androgen regulates expression of CAD/MI susceptibility gene ADTRP and reduces risk of cardiovascular disease by activating ADTRP expression (Figs. 6 and 7). Androgen inhibited monocyte adhesion to EAhy926 endothelial cells and blocked monocyte transmigration across the endothelial cells (Figs. 6 and 7). These effects of androgen were reversed by knockdown of ADTRP expression (Figs. 6 and 7). The data suggest that ADTRP expression is important for androgen-mediated inhibition of monocyte adhesion to endothelial cells and monocyte transmigration across the endothelial cells, the two key processes in atherosclerosis. Together, our data suggest that the molecular mechanism by which androgen confers protection against CAD and MI is stimulation of ADTRP expression.
Although knockdown of ADTRP expression by siRNA reversed the effects of androgen on monocyte adhesion to endothelial cells and migration of monocytes across endothelial cells, the reversal was not 100% (Figs. 6 and 7). The data suggest that in addition to ADTRP, other downstream target genes regulated by androgen may also be involved in the action of androgen on atherosclerosis. As the androgen receptor is a transcriptional factor, it regulates expression of numerous genes. Future studies are needed to identify other downstream target genes of the androgen and AR and to distinguish which other AR-regulated genes are also involved in reducing the risk of atherosclerosis, in addition to ADTRP.
In conclusion, the data in this study showed that androgen induced up-regulation of ADTRP expression on the transcriptional level and the underlying molecular mechanism is the direct binding of the androgen receptor to the half ARE at the promoter/regulatory region, which activate transcription of ADTRP. Most importantly, we showed that androgen blocked two key endothelial functions/processes associated with atherosclerotic CAD: monocyte adhesion to the endothelium and transmigration of monocytes across the endothelium, and the effects were mediated by activating the transcription of the CAD susceptibility gene ADTRP, at least partly.
Supplementary Material
Acknowledgments
Funding
This study was supported by the China National Natural Science Foundation Key Program (31430047), Chinese National Basic Research Programs (973 Programs 2013CB531101 and 2012CB517801), a Key Project in the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2011BAI11B19), Hubei Province's Outstanding Medical Academic Leader Program, Hubei Province Natural Science Key Program (2014CFA074), the China National Natural Science Foundation grants (91439129 and NSFC-J1103514), NIH/NHLBI grants R01 HL121358 and R01 HL126729, Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education, and the “Innovative Development of New Drugs” Key Scientific Project (2011ZX09307-001-09).
We thank Dr. Vivek Malhotra, Dr. Ishier Raote and Dr. Maria Ortega Bellido for providing the mammalian expression plasmids for MIA3/TANGO1. We thank other members of the Wang laboratory for help and assistance. We greatly appreciate all study subjects for their participation in this project.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbadis.2017.06.015.
Conflict of interest
K.L., W.Z., E.P., S.H.C., C.P., M.B., J.H., and V.L. are current or formal employees of Bayer. Other authors are the members of Wang Laboratory, which received funding from Bayer AG.
Disclosures
This project received funding from Bayer AG.
Transparency document
The http://dx.doi.org/10.1016/j.bbadis.2017.06.015 associated with this article can be found, in online version.
References
- 1.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Huang Y, Yin D, Wang D, Xu C, Wang F, Yang Q, Wang X, Li S, Chen S, Xiong X, Huang Y, Zhao Y, Wang L, Zhu X, Su Z, Zhou B, Zhang Y, Wang L, Chang L, Xu C, Li H, Ke T, Ren X, Cheng X, Yang Y, Liao Y, Tu X, Wang QK. Meta-analysis identifies robust association between SNP rs17465637 in MIA3 on chromosome 1q41 and coronary artery disease. Atherosclerosis. 2013;231:136–140. doi: 10.1016/j.atherosclerosis.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Wang X, Wang J, Zhao Y, Wang D, Tan C, Fa J, Zhang R, Wang F, Xu C, Huang Y, Li S, Yin D, Xiong X, Li X, Chen Q, Tu X, Yang Y, Xia Y, Xu C, Wang QK. Genomic variant in CAV1 increases susceptibility to coronary artery disease and myocardial infarction. Atherosclerosis. 2016;246:148–156. doi: 10.1016/j.atherosclerosis.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu C, Yang Q, Xiong H, Wang L, Cai J, Wang F, Li S, Chen J, Wang C, Wang D, Xiong X, Wang P, Zhao Y, Wang X, Huang Y, Chen S, Yin D, Li X, Liu Y, Liu J, Wang J, Li H, Ke T, Ren X, Wu Y, Wu G, Wan J, Zhang R, Wu T, Wang J, Xia Y, Yang Y, Cheng X, Liao Y, Chen Q, Zhou Y, He Q, Tu X, Wang QK. Candidate pathway-based genome-wide association studies identify novel associations of genomic variants in the complement system associated with coronary artery disease. Circ. Cardiovasc. Genet. 2014;7:887–894. doi: 10.1161/CIRCGENETICS.114.000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong X, Xu C, Zhang Y, Li X, Wang B, Wang F, Yang Q, Wang D, Wang X, Li S, Chen S, Zhao Y, Yin D, Huang Y, Zhu X, Wang L, Wang L, Chang L, Xu C, Li H, Ke T, Ren X, Wu Y, Zhang R, Wu T, Xia Y, Yang Y, Ma X, Tu X, Wang QK. BRG1 variant rs1122608 on chromosome 19p13.2 confers protection against stroke and regulates expression of pre-mRNA-splicing factor SFRS3. Hum. Genet. 2014;133:499–508. doi: 10.1007/s00439-013-1389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeap BB. Androgens and cardiovascular disease. Curr. Opin. Endocrinol. Diabetes Obes. 2010;17:269–276. doi: 10.1097/MED.0b013e3283383031. [DOI] [PubMed] [Google Scholar]
- 7.Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015;36:3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marszall MP, Sroka WD, Sikora A, Chelminiak D, Ziegler-Borowska M, Siodmiak T, Moaddel R. Ligand fishing using new chitosan based functionalized androgen receptor magnetic particles. J. Pharm. Biomed. Anal. 2016;127:129–135. doi: 10.1016/j.jpba.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa H, Ueda T, Ito S, Shiraishi T, Taniguchi H, Kayukawa N, Nakanishi H, Ushijima S, Kanazawa M, Nakamura T, Naya Y, Hongo F, Kamoi K, Okihara K, Ukimura O. Androgen suppresses testicular cancer cell growth in vitro and in vivo. Oncotarget. 2016;7:35224–35232. doi: 10.18632/oncotarget.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell-and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagman JB, Wilhelmson AS, Motta BM, Pirazzi C, Alexanderson C, De Gendt K, Verhoeven G, Holmang A, Anesten F, Jansson JO, Levin M, Boren J, Ohlsson C, Krettek A, Romeo S, Tivesten A. The androgen receptor confers protection against diet-induced atherosclerosis, obesity, and dyslipidemia in female mice. FASEB J. 2015;29:1540–1550. doi: 10.1096/fj.14-259234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourghardt J, Wilhelmson AS, Alexanderson C, De Gendt K, Verhoeven G, Krettek A, Ohlsson C, Tivesten A. Androgen receptor-dependent and independent atheroprotection by testosterone in male mice. Endocrinology. 2010;151:5428–5437. doi: 10.1210/en.2010-0663. [DOI] [PubMed] [Google Scholar]
- 13.Bai Y, Nie S, Jiang G, Zhou Y, Zhou M, Zhao Y, Li S, Wang F, Lv Q, Huang Y, Yang Q, Li Q, Li Y, Xia Y, Liu Y, Liu J, Qian J, Li B, Wu G, Wu Y, Wang B, Cheng X, Yang Y, Ke T, Li H, Ren X, Ma X, Liao Y, Xu C, Tu X, Wang QK. Regulation of CARD8 expression by ANRIL and association of CARD8 single nucleotide polymorphism rs2043211 (p.C10X) with ischemic stroke. Stroke. 2014;45:383–388. doi: 10.1161/STROKEAHA.113.003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu C, Wang F, Wang B, Li X, Li C, Wang D, Xiong X, Wang P, Lu Q, Wang X, Yang Q, Yin D, Huang Y, Ji L, Wang N, Chen S, Cheng X, Liao Y, Ma X, Su D, Chen G, Xia H, Shi L, Tu X, Wang QK. Minor allele C of chromosome 1p32 single nucleotide polymorphism rs11206510 confers risk of ischemic stroke in the Chinese Han population. Stroke. 2010;41:1587–1592. doi: 10.1161/STROKEAHA.110.583096. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, Cheng X, Li C, Yang R, Wang CC, Wu G, Lu QL, Bai Y, Huang YF, Yin D, Yang Q, Wang XJ, Dai DP, Zhang RF, Wan J, Ren JH, Li SS, Zhao YY, Fu FF, Huang Y, Li QX, Shi SW, Lin N, Pan ZW, Li Y, Yu B, Wu YX, Ke YH, Lei J, Wang N, Luo CY, Ji LY, Gao LJ, Li L, Liu H, Huang EW, Cui J, Jia N, Ren X, Li H, Ke T, Zhang XQ, Liu JY, Liu MG, Xia H, Yang B, Shi LS, Xia YL, Tu X, Wang QK. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat. Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 16.Lupu C, Zhu H, Popescu NI, Wren JD, Lupu F. Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood. 2011;118:4463–4471. doi: 10.1182/blood-2011-05-355370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo C, Wang F, Qin S, Chen Q, Wang QK. Coronary artery disease susceptibility gene ADTRP regulates cell cycle progression, proliferation, and apoptosis by global gene expression regulation. Physiol. Genomics. 2016;48:554–564. doi: 10.1152/physiolgenomics.00028.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo C, Wang F, Ren X, Ke T, Xu C, Tang B, Qin S, Yao Y, Chen Q, Wang QK. Identification of a molecular signaling gene-gene regulatory network between GWAS susceptibility genes ADTRP and MIA3/TANGO1 for coronary artery disease. Biochim. Biophys. Acta. 2017;1863:1640–1653. doi: 10.1016/j.bbadis.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinetti-Gbaguidi G, Copin C, Derudas B, Vanhoutte J, Zawadzki C, Jude B, Haulon S, Pattou F, Marx N, Staels B. The coronary artery disease-associated gene C6ORF105 is expressed in human macrophages under the transcriptional control of PPARgamma. FEBS Lett. 2015;589:461–466. doi: 10.1016/j.febslet.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Seo J, Kim J, Kim M. Cloning of androgen-inducible gene 1 (AIG1) from human dermal papilla cells. Mol. Cell. 2001;11:35–40. [PubMed] [Google Scholar]
- 21.Tian XL, Kadaba R, You SA, Liu M, Timur AA, Yang L, Chen Q, Szafranski P, Rao S, Wu L, Housman DE, DiCorleto PE, Driscoll DJ, Borrow J, Wang Q. Identification of an angiogenic factor that when mutated causes susceptibility to Klippel-Trenaunay syndrome. Nature. 2004;427:640–645. doi: 10.1038/nature02320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou B, Si W, Su Z, Deng W, Tu X, Wang Q. Transcriptional activation of the Prox1 gene by HIF-1α and HIF-2α in response to hypoxia. FEBS Lett. 2013;587:724–731. doi: 10.1016/j.febslet.2013.01.053. [DOI] [PubMed] [Google Scholar]
- 23.Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 2009;136:891–902. doi: 10.1016/j.cell.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 24.Santos AJM, Nogueira C, Ortega-Bellido M, Malhotra V. TANGO1 and Mia2/cTAGE5 (TALI) cooperate to export bulky pre-chylomicrons/VLDLs from the endoplasmic reticulum. J. Cell Biol. 2016;213:343–354. doi: 10.1083/jcb.201603072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malhotra V, Erlmann P, Nogueira C. Procollagen export from the endoplasmic reticulum. Biochem. Soc. Trans. 2015;43:104–107. doi: 10.1042/BST20140286. [DOI] [PubMed] [Google Scholar]
- 26.Malhotra V, Erlmann P. The pathway of collagen secretion. Annu. Rev. Cell Dev. Biol. 2015;31:109–124. doi: 10.1146/annurev-cellbio-100913-013002. [DOI] [PubMed] [Google Scholar]
- 27.Nogueira C, Erlmann P, Villeneuve J, Santos AJ, Martinez-Alonso E, Martinez-Menarguez JA, Malhotra V. SLY1 and Syntaxin 18 specify a distinct pathway for procollagen VII export from the endoplasmic reticulum. Elife. 2014;3:e02784. doi: 10.7554/eLife.02784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos AJ, Raote I, Scarpa M, Brouwers N, Malhotra V. TANGO1 recruits ERGIC membranes to the endoplasmic reticulum for procollagen export. Elife. 2015;4:e10982. doi: 10.7554/eLife.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii I, Harada Y, Kasahara T. Reprofiling a classical anthelmintic, pyrvinium pamoate, as an anti-cancer drug targeting mitochondrial respiration. Front. Oncol. 2012;2:137. doi: 10.3389/fonc.2012.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiegering A, Uthe FW, Huttenrauch M, Muhling B, Linnebacher M, Krummenast F, Germer CT, Thalheimer A, Otto C. The impact of pyrvinium pamoate on colon cancer cell viability. Int. J. Color. Dis. 2014;29:1189–1198. doi: 10.1007/s00384-014-1975-y. [DOI] [PubMed] [Google Scholar]
- 31.Lim M, Otto-Duessel M, He M, Su L, Nguyen D, Chin E, Alliston T, Jones JO. Ligand-independent and tissue-selective androgen receptor inhibition by pyrvinium. ACS Chem. Biol. 2014;9:692–702. doi: 10.1021/cb400759d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadar MD, Hussain M, Bruchovsky N. Prostate cancer: molecular biology of early progression to androgen independence. Endocr. Relat. Cancer. 1999;6:487–502. doi: 10.1677/erc.0.0060487. [DOI] [PubMed] [Google Scholar]
- 33.Kim JH, Yoo BW, Yang WJ. Hepatic failure induced by cyproterone acetate: a case report and literature review. Can. Urol. Assoc. J. 2014;8:E458–E461. doi: 10.5489/cuaj.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Z, Si W, Li L, Zhou B, Li X, Xu Y, Xu C, Jia H, Wang QK. MiR-144 regulates hematopoiesis and vascular development by targeting meis1 during zebrafish development. Int. J. Biochem. Cell Biol. 2014;49:53–63. doi: 10.1016/j.biocel.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Zhou M, Wang J, Zhao Y, Li S, Zhou B, Su Z, Xu C, Xia Y, Qian H, Tu X, Xiao W, Chen X, Chen Q, Wang QK. Role of microRNA-27a in down-regulation of angiogenic factor AGGF1 under hypoxia associated with high-grade bladder urothelial carcinoma. Biochim. Biophys. Acta. 2014;1842:712–725. doi: 10.1016/j.bbadis.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Zhou B, Ma R, Si W, Li S, Xu Y, Tu X, Wang Q. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–169. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 37.Archacki SR, Angheloiu G, Moravec CS, Liu H, Topol EJ, Wang QK. Comparative gene expression analysis between coronary arteries and internal mammary arteries identifies a role for the TES gene in endothelial cell functions relevant to coronary artery disease. Hum. Mol. Genet. 2012;21:1364–1373. doi: 10.1093/hmg/ddr574. [DOI] [PubMed] [Google Scholar]
- 38.You SA, Archacki SR, Angheloiu G, Moravec CS, Rao S, Kinter M, Topol EJ, Wang Q. Proteomic approach to coronary atherosclerosis shows ferritin light chain as a significant marker: evidence consistent with iron hypothesis in atherosclerosis. Physiol. Genomics. 2003;13:25–30. doi: 10.1152/physiolgenomics.00124.2002. [DOI] [PubMed] [Google Scholar]
- 39.Navarro-Dorado J, Orensanz LM, Recio P, Bustamante S, Benedito S, Martinez AC, Garcia-Sacristan A, Prieto D, Hernandez M. Mechanisms involved in testosterone-induced vasodilatation in pig prostatic small arteries. Life Sci. 2008;83:569–573. doi: 10.1016/j.lfs.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Wang Q. Molecular genetics of coronary artery disease. Curr. Opin. Cardiol. 2005;20:182–188. doi: 10.1097/01.hco.0000160373.77190.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q. Advances in the genetic basis of coronary artery disease. Curr Atheroscler Rep. 2005;7:235–241. doi: 10.1007/s11883-005-0012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.