Abstract
We have studied the localization of synaptogyrin family members in vivo. Both native and green fluorescent protein (GFP)-tagged Caenorhabditis elegans synaptogyrin (SNG-1) are expressed in neurons and synaptically localized. Deletion and mutational analysis with the use of GFP-tagged SNG-1 has defined a 38 amino acid sequence within the C terminus of SNG-1 and a single arginine in the cytoplasmic loop between transmembrane domain 2 and 3 that are required for SNG-1 localization. These domains may represent components of signals that target synaptogyrin for endocytosis from the plasma membrane and direct synaptogyrin to synaptic vesicles, respectively. In chimeric studies, these regions were sufficient to relocalize cellugyrin, a nonneuronal form of synaptogyrin, from nonsynaptic regions such as the sensory dendrites and the cell body to synaptic vesicles. Furthermore, GFP-tagged rat synaptogyrin is synaptically localized in neurons of C. elegans and in cultured hippocampal neurons. Similarly, the C-terminal domain of rat synaptogyrin is necessary for localization in hippocampal neurons. Our study suggests that the mechanisms for synaptogyrin localization are likely to be conserved from C. elegans to vertebrates.
INTRODUCTION
Synaptic vesicles (SVs) contain a restricted set of membrane proteins important for neuronal function (Südhof, 1995; Calakos and Scheller, 1996; Fernandez-Chacon and Südhof, 1999). The mechanisms responsible for targeting these proteins specifically to the SV membrane are poorly understood. Integral membrane proteins are synthesized on the rough endoplasmic reticulum and traffic through the Golgi network. Proteins exiting the trans-Golgi network (TGN) are sorted to different types of transport vesicles. SV proteins are sorted to synaptic vesicle precursors (preSVs), which differ from mature SVs both in morphology and protein composition (Tsukita and Ishikawa, 1980; Okada et al., 1995). Evidence supports both direct routing of SV proteins from TGN to preSVs and indirect routing via the plasma membrane (reviewed by Hannah et al., 1999). preSVs are subsequently transported along axonal processes to the nerve terminal by motor proteins of the kinesin superfamily (reviewed by Hirokawa, 1998). How preSVs mature after delivery to the nerve terminal is unknown. preSVs might fuse with an endosomal compartment and SVs generated by budding from the endosome. A second, but not mutually exclusive possibility, is that preSVs are delivered to the presynaptic plasma membrane at the nerve terminal and then retrieved by the same mechanism(s) used to recycle SVs after regulated exocytosis (Hannah et al., 1999; reviewed by Buckley et al., 2000). Regardless of the exact route of trafficking to the mature organelle, SV proteins must be sorted away from other membrane proteins at several stages during their life cycle.
Signal sequences resident in proteins are thought to mediate the sorting of many proteins to cellular compartments. Several studies have identified domains and motifs necessary for correct localization of SV proteins. Targeting and sorting signals have been identified in synaptophysin (Linstedt and Kelly, 1991), synaptobrevin (Grote et al., 1995; Grote and Kelly, 1996), synaptotagmin (Blagoveshchenskaya et al., 1999; Krasnov and Enikolopov, 2000), and vesicular neurotransmitter transporters (Tan et al., 1998; Varoqui and Erickson, 1998). However, no common motif that could serve as a universal targeting signal has been found by experimental studies or by direct comparison of the primary sequences of SV proteins. Although tyrosine-based signals are one of the major sorting signals used by clathrin pit-mediated endocytosis (Davis et al., 1986, 1987; Collawn et al., 1990; Peters et al., 1990; Thies et al., 1990; Letourneur and Klausner, 1992), only one protein, P-selectin, has been shown to use a tyrosine-based signal to be targeted to synaptic-like microvesicles (SLMVs) in PC12 cells (Blagoveshchenskaya et al., 1999).
The absence of a common sorting signal element in SV proteins contrasts to targeting to other organelles where many distinct proteins use the same sorting signal (Stanley, 1996). At least two different mechanisms of SV protein sorting could account for the lack of a universal SV targeting signal. First, distinct components of SVs may be sorted by independent sorting mechanisms. Such an idea is also supported by other findings. For example, the SV proteins synaptotagmin, synaptophysin, and SV2 are sorted into different classes of organelles when expressed in nonneuronal Chinese hamster ovary cells (Feany et al., 1993). In vivo, SV2 and synaptophysin associate with different kinesin motor proteins and consequently probably reside in distinct vesicle populations (Okada et al., 1995). In addition, in the Caenorhabditis elegans unc-11 (AP180 homolog) mutant, synaptobrevin alone is mislocalized, whereas other SV proteins are unaffected (Nonet et al., 1999). Furthermore, the protein stoned is required selectively for the retrieval of synaptotagmin I from the plasma membrane in Drosophila (Fergestad et al., 1999; Fergestad and Broadie, 2001), perhaps via its interaction with the C2B domain of synaptotagmin I (Littleton et al., 2001). Another possibility is that sorting information is only present in a few proteins, and other components form complexes with these proteins and are secondarily targeted to SVs (Bennett et al., 1992). More likely, a combination of these two mechanisms is used to target SV proteins. Defining distinct trafficking signals for all SV proteins will be complex and ultimately may require defining multiple protein–protein interactions that act coordinately.
Caenorhabditis elegans has several features that make it a valuable system for studying both synaptic function and SV protein localization in vivo. Both forward and reverse genetics have been used to generate mutations in multiple synaptic components, many of which are evolutionarily conserved (Fernandez-Chacon and Südhof, 1999; Nonet, 1999). The unique life cycle of C. elegans allows many mutants (and double and triple mutants) that are severely compromised in neuronal function to survive simplifying detailed analysis of the role of these genes (Nonet et al., 1993, 1997, 1999; Harris et al., 2000). The compact organization and completed sequence description of the genome greatly facilitates the identification and manipulation of existing and novel members of protein families known to have important functions in other organisms (Bargmann, 1998; The C. elegans Sequencing Consortium, 1998). Qualitative and quantitative analysis of the distribution of GFP-tagged protein in live animals under fluorescent microscopy is simple owing to the transparent nature of the organism (Labrousse et al., 1998; Nonet, 1999). Finally, genes that are involved in protein localization can be isolated in classical genetic screens with the use of GFP-tagged transgenes (reviewed by Koushika and Nonet, 2000).
In this study we have characterized signals required for localization of members of the synaptogyrin family, which currently consists of three human genes, three mouse genes, two rat genes and one C. elegans gene (Stenius et al., 1995; Janz and Südhof, 1998; Kedra et al., 1998; Nonet et al., 1999). The first cloned member of this family, rat synaptogyrin (p29), is highly expressed in neurons and neuroendocrine cells where it colocalizes with synaptophysin on SVs and SLMVs (Baumert et al., 1990; Stenius et al., 1995). Synaptogyrin together with synaptophysin, a family distantly related to synaptogyrin, accounts for >10% of the total SV protein content (Jahn et al., 1985; Wiedenmann and Franke, 1985; Baumert et al., 1990; Jahn and Südhof, 1993; Stenius et al., 1995). Moreover, both synaptogyrin and synaptophysin families contain nonneuronal isoforms that are ubiquitously expressed (Haass et al., 1996; Janz and Südhof, 1998; Kedra et al., 1998; Takeshima et al., 1998). Synaptophysin and synaptogyrin share similar membrane topologies with four transmembrane domains where their N and C termini face the cytoplasm (Johnston et al., 1989; Stenius et al., 1995). In PC12 cells, overexpressing synaptogyrin and synaptophysin inhibits exocytosis (Sugita et al., 1999). Double mutant mice lacking both synaptogyrin and synaptophysin show severe reductions in both short-term and long-term synaptic plasticity, suggesting that they are regulators of exocytosis (Janz et al., 1999). sng-1, the C. elegans member, has 30% identity to the rat synaptogyrin and a similar hydrophobicity profile (Nonet, 1999). Analysis of animals expressing green fluorescent protein (GFP)-tagged synaptogyrin (SNG-1) suggests that SNG-1 is localized to synaptic regions (Nonet, 1999). Thus far, no other C. elegans synaptogyrin family members have been identified.
We have identified two primary sequences in SNG-1 that are necessary for its synaptic localization, a C-terminal region containing 38 amino acids and an arginine in the loop facing the cytoplasm. An SNG-1 sequence containing these elements is sufficient to localize rat cellugyrin that is normally not restricted to synaptic regions in C. elegans. Furthermore, rat synaptogyrin is localized to synaptic regions in both C. elegans and hippocampal neurons in culture and the C terminus is necessary for synaptic localization in each system. Our results suggest that the mechanisms used for synaptic localization among synaptogyrin family members are evolutionarily conserved.
MATERIALS AND METHODS
Nematode Strains and Culture
Bristol strain N2, mutants, and transgenic animals were grown at 22.5°C on solid medium as described by Sulston and Hodgkin (1988).
Microscopy and Image Analysis
Live animals were anesthetized with 10 mM sodium azide, mounted on 2% agarose pads, and examined under epifluorescence with the use of an Olympus BX60. Time-averaged images were collected with the use of a DAGE SIT68 camera and a CG-7 frame grabber (Scion Image). Confocal images were taken in a series of 0.5 μM optical sections with the use of an Olympus Fluoview microscope and Fluoview software. A three-dimensional representation of a 50-μm segment of sublateral ventral nerve cord just anterior to the posterior end point of the cord near the tail (boxed area in Figure 4B) was made for quantitative analysis. Total intensity in the boxed area was measured with the use of Fluoview software. Background fluorescence was subtracted as the average intensity of areas adjacent to the boxed region. Total intensity in puncta (closed circles in Figure 4B), which were defined as discrete bright fluorescent patches, was calculated by summing the intensity in each punctum. The percentage of fluorescence in puncta was calculated by dividing the total intensity in the puncta by the total intensity in the boxed area after background subtraction.
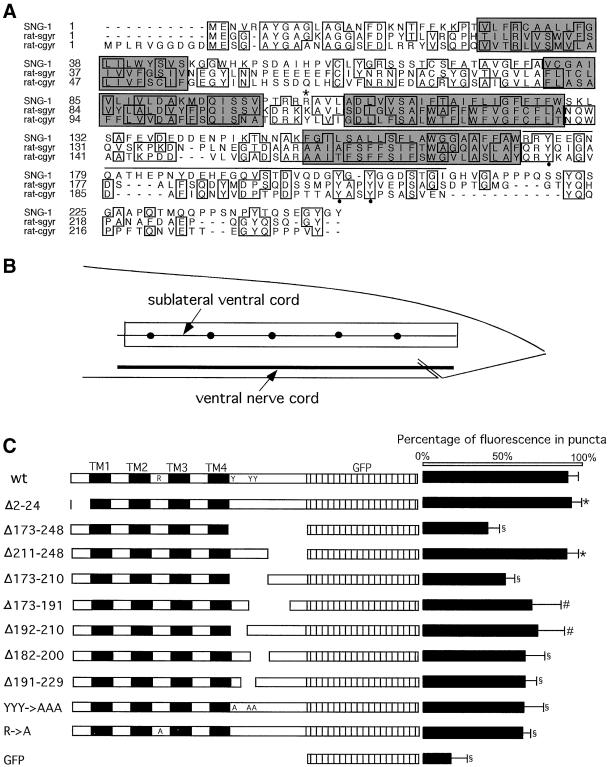
Figure 4.
Fine mapping of SNG-1 localization signals. (A) Sequence alignment of SNG-1, rat synaptogyrin (rat-sgyr), and cellugyrin (rat-cgyr). Amino acids are represented by the standard one-letter code and identical amino acids are boxed. Shaded regions are transmembrane domains. Amino acids 173–210 are overlined. R104 is marked by asterisk. Y174, Y201, and Y203 are marked by closed circles. (B) Diagram of the tail of C. elegans highlighting the ventral nerve cord and two ventral sublateral nerve cords. A box outlines the region chosen for quantitative analysis of the distribution of SNG-1::GFP proteins. Closed circles represent puncta along the sublateral nerve cord. (C) Schematic diagram of the mutated region of SNG-1 is shown on the left. The percentage of fluorescence in the puncta is plotted on the right. Values are means ± SD. ∗, not significantly different from jsIs219 (p > 0.50). §, significantly smaller than jsIs219 (p < 0.001). #, significantly smaller than jsIs219 (p < 0.002). Genotypes of the lines used (with plasmid construct name in parentheses): wt jsIs219 (pSY3), Δ2–24 jsEx353 (pSY17), Δ173–248 jsEx355 (pSY18), Δ211–248 jsEx369 (pSY36), Δ173–210 jsEx373 (pSY39), Δ173–191 jsEx38 (pSY42), Δ192–210 jsEx386 (pSY43), Δ182–200 jsEx388 (pSY44),Δ 191–229 jsEx390 (pSY45), YYY >AAA jsEx497 (pSY60), R>A jsEx547 (pSY62), GFP jsEx489 (pSY1).
Immunocytochemistry
Immunocytochemistry with antibodies against synaptotagmin (SNT-1) (Nonet et al., 1993) and GFP (mouse monoclonal; CLONTECH, Palo Alto, CA) was performed as described previously (Saifee et al., 1998). The polyclonal antibody against SNG-1 was generated in chicken with a purified bacterial product representing amino acids 164–248 fused to a his6 tag. The fusion product was purified under denaturing conditions (8 M urea) with the use of Ni-NTA agarose (Qiagen, Chatsworth, CA), and dialyzed into phosphate-buffered saline. The resulting serum was then immunoabsorbed against the SNG-1 fusion protein bound to nitrocellulose as described by Smith and Fisher (1984). Immunocytochemistry with an antibody against rat synaptophysin (gift of P. De Camilli, Yale University, New Haven, CT) was performed as previously described (Rao et al., 1998).
Plasmid Construction
All deletions were made with the use of Pfu polymerase and DpnI digestion as previously described (Fisher and Pei, 1997). All oligonucleotides listed below are 5′ to 3′.
pSY1: Oligonucleotides TGAGTTGTATTGCATTCCAGATCTAG and GTCCGG ATCCATAAGCACGCACGTTC were used in a polymerase chain reaction (PCR) to amplify the sng-1 promoter, a 1.8-kb sequence upstream of the start codon. The PCR product was digested with BglII and BamHI, and was ligated with pPD95.69 digested with BamHI. A. Fire, J. Ahnn, G. Seydoux, and S. Xu kindly provided pPD95.69. Detailed description of pPD95.69 available at ftp.ciwemb.edu/PNF:byName:/FireLabWeb/FireLabInfo/FireLabVectors/1995_Vector_Kit/
pSY12: Oligonucleotides CATTCTCAGATCTATGGAAGGGGG-TGCGTACGGAGC and CATTACCGGTCCGTAGCCCTGCGACTGGTAGCCC corresponding to the beginning and the end of the coding region of rat synaptogyrin were used in a PCR to amplify the gene from pCMV-Sgyr, a clone containing rat synaptogyrin cDNA (Stenius et al., 1995). The PCR product was digested with BglII and Age I, and was ligated with pSY1 digested with BamHI and Age I.
pSY14: Oligonucleotides CATTCTCAGATCTATGCCCTTGAGG-GTCGGCGGCG and CATTACCGGTCCGTACACTGGGGGAGGCT-GGTAG corresponding to the beginning and the end of the coding region of rat cellugyrin were used in a PCR to amplify the gene from pCMV-Cgyr, a clone containing the rat cellugyrin cDNA (Janz and Südhof, 1998). The PCR product was digested with BglII and Age I, and was ligated with pSY1 digested with BamHI and Age I.
pSY17: Oligonucleotides CTTTTATTTTAGCATGCCCGCGGTT-TTGTTCAGG and CCTGAACAAAACCGCGGGCATGCTAAAAT-AAAAG were designed to delete the region encoding amino acids 2–24 in pSY3, a clone containing the sng-1 genomic DNA, including the coding region and the promoter (Nonet, 1999).
pSY18: Oligonucleotides GCATTTTTCGCATGGCGGCCGCTAGAAAAAATGAG and CTCATTTTTTCTAGCGGCCGCCATGCGA-AAAATGC were designed to delete the region encoding amino acids 173–248 in pSY3.
pSY26: Oligonucleotides GAAGATCTCGCCACCATGGAAGGG-GGTGCGTAC and CATTACCGGTCCGTAGCCCTGCGACTGGTAGCCC corresponding to the beginning and the end of the coding region of rat synaptogyrin were used in a PCR to amplify the gene from pCMV-Sgyr. The PCR product was digested with BglII and Age I, and was ligated with pEGFP (CLONTECH), a clone containing human CMV promoter and the EGFP gene, digested with the same enzymes.
pSY31: Oligonucleotides CGG GAT CCC CGT CGT TAC GAA GAA GG and CGA CCG GTC CAT AAC CAT ATC CTT CCG corresponding to the beginning and the end of the coding region of the C-terminal domain of SNG-1 were used to PCR from pSCG110a, a clone contain partial SNG-1 cDNA. The PCR product was digested with BamHI and Age I and was ligated with pSY14 digested with the same enzymes.
pSY36: Oligonucleotides GGAGGAGACTCAACCCGGGTACCGGTAG and CTACCGGTACCCGGGTTGAGTCTCCTCC were designed to delete the region encoding amino acids 211–248 in pSY3.
pSY39: Oligonucleotides CATTTTTCGCATGGATCGGACATGTTGG and CCAACATGTCCGATCCATGCGAAAAATG were designed to delete the region encoding amino acids 173–210 in pSY3.
pSY42: Oligonucleotides GCATTTTTCGCATGGCAAGTGTCGACAGAC and GTCTGTCGACACTTGCCATGCGAAAAATGC were designed to delete the region encoding amino acids 173–191 in pSY3.
pSY43: Oligonucleotides GATGAACATTTTGGACATGTTGGCGCACCT and AGGTGCGCCAACATGTCCAAAATGTTCATC were designed to delete the region encoding amino acids 192–210 in pSY3.
pSY44: Oligonucleotides GGAAATCAAGCAACTGGATACGGAGGAGAC and GTCTCCTCCGTATCCAGTTGCTTGATTTCC were designed to delete the region encoding amino acids 182–200 in pSY3.
pSY45: Oligonucleotides GATGAACATTTTGGACAAACCATGCAACAAC and GTTGTTGCATGGTTTGTCCAAAATGTTCATC were designed to delete the region encoding amino acids 191–229 in pSY3.
pSY46: Oligonucleotides CAGCGGTATCAGATTGGACCGGTC-G-CCACC and GGTGGCGACCGGTCCAATCTGATACCGCTG were designed to delete the region encoding amino acids 176–235 in pSY26.
pSY47:Oligonucleotides GCCGTGCTAGCCTTGGGACCGGTAG-TAAAAA and TTTTTCTACCGGTCCGAAGGCTAGCACGGC were designed to delete the region encoding amino acids 179–243 in pSY12.
pSY57: Oligonucleotides CTTTTATTTTAGCATGCCGCGGACAAGAAGGAGAGCTG and CAGCTCTCCTTCTTGTCCGCGGC-ATGCTAAAATAAAAG were designed to delete the region encoding amino acids 1–99 of SNG-1 in pSY3 and a SacII restriction site was engineered into the deletion site. The resulting plasmid is designated pSY54. Oligonucleotides CAGATAAGCAATGCCCCGCGGATGAGTAAAGGAGAAG and CTTCTCCTTTACTCATCCGCG-GGGCATTGCTTATCT were used to delete the region encoding amino acids 109–234 of cellugyrin in pSY14 and a SacII restriction site was engineered into the deletion site. The resulting plasmid is designated pSY53. A HindIII and SacII fragment from pSY54 was inserted into pSY53 digested with the same enzymes.
pSY59: Oligonucleotides GATCTCGCCACCATGACCATCCTGCGCGTC and GACGCGCAGGATGGTCATGGTGGCGAGATC were designed to delete the sequences encoding amino acids 2–26 in pSY26.
pSY60: Oligonucleotides GACGTGCAAGATGGTGCAGGAGCG-GGAGGAGACTCAACC and GGTTGAGTCTCCTCCCGCTCCTGCACCATCTTGCACGTC were designed to engineer the changes Y201A and Y203A in pSY3. Oligonucleotides CGCATGGCGTCGTGCAGAAGAAGGAAATC and GATTTCCTTCTTCTGCACGACG-CCATGCG were designed to engineer the change Y174A into the resulting plasmid.
pSY62: Oligonucleotides CCAACAAGAAGGGCAGCTGTCCTA-GCAGAT and ATCTGCTAGGACAGCTGCCCTTCTTGTTGG were designed to engineer the R104A change in pSY3.
Germline Transformation
Nematodes were transformed with the use of the method described by Mello et al. (1991). Plasmids were injected at 5 ng/μl in conjunction with the dominant rol-6(su1003) transformation marker (Kramer et al., 1990) plasmid pRF4 at 140 ng/μl. Several independent transformed lines were obtained for each construct examined.
Neuron Culture and Transfection
Hippocampal neuronal cultures were prepared as previously described (Banker and Cowan, 1977; Goslin and Banker, 1989). After dissociation and before plating, neurons were transfected by a lipid-mediated gene transfer method with the use of the Effectene kit (Qiagen). Approximately 5 × 105 cells were incubated with 1 μg of DNA for 2h at 37°C in the presence of the transfection reagents and were then plated onto coverslips in fresh medium at a density of 18,000 cells/cm2. This procedure resulted in a 0.01–0.05% transfection efficiency. Neurons 5–7 d old were fixed and immunostained with an antibody against synaptophysin.
RESULTS
Endogenous SNG-1 and GFP-tagged SNG-1 Are Localized to SVs in C. elegans
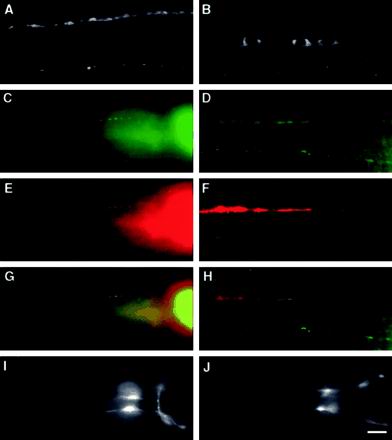
To assess the localization of endogenous SNG-1, an antibody was raised against the C terminus of the protein. Immunostaining wild-type animals with the use of this antibody revealed a typical localization pattern observed for other SV proteins, including SNT-1, SNB-1 (synaptobrevin), and RAB-3 (Nonet et al., 1993, 1997, 1998). Immunoreactivity was concentrated in nervous system regions rich in synapses, including the nerve ring (Figure 1A), the dorsal nerve cord (Figure 1B) and the ventral nerve cord (not shown). Discrete puncta were found in the ventral and dorsal sublateral process bundles (Figure 1B; our unpublished results). The SNG-1 protein also accumulated in the presynaptic varicosities of SAB motor neurons innervating the head muscle (Figure 1A). Finally, the endogenous protein was undetectable in neuronal cell bodies, commissures that run circumferentially along the body, and the dendrites of sensory neurons in the nose. Wild-type animals double immunolabeled with antibodies against SNG-1 and SNT-1 showed that native SNG-1 colocalized with SNT-1 (our unpublished results). Taken together, these results suggest that endogenous SNG-1 is localized to SVs in C. elegans, although localization to another membrane such as synaptic endosomes cannot be excluded.
Figure 1.
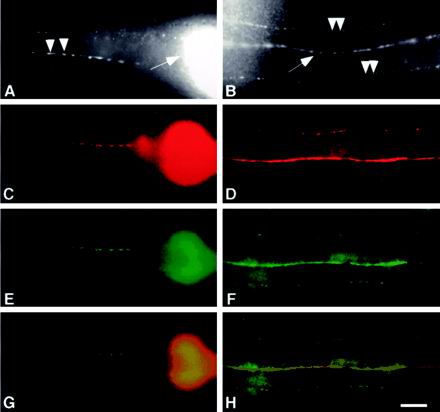
SNG-1::GFP is localized to synaptic regions in C. elegans. (A and B) Whole mount immunocytochemistry of adult wild-type animals stained with a primary antibody directed against SNG-1 and a fluorescein isothiocyanate (FITC)-conjugated secondary antibody. (A) Lateral view of the head illustrates the synaptic varicosities along an SAB motor neuron axon (arrowheads). The nerve ring (arrow) of the animal is also visible. (B) Dorsal view shows the dorsal nerve cord (arrow) and small fluorescence puncta (arrowheads) in dorsal sublateral nerve cords. (C–F) Whole mount immunocytochemistry of adult jsIs219 animals visualized by incubation with primary antibodies directed against SNT-1 (C and D) and GFP (E and F). Cy3- and FITC-conjugated secondary antibodies are used respectively. (G and H) Merged images. (C, E, and G) Lateral views of the nerve ring and the presynaptic varicosities of SAB neurons. (D, F, and H) Dorsal views of the dorsal and dorsal sublateral nerve cords ([C–H] genotype: jsIs219). Bar, 10 μm.
We next asked whether GFP-tagged SNG-1 fusion protein is localized like the endogenous protein. jsIs219 animals harbor a GFP fusion to the C terminus of sng-1 under the control of the sng-1 promoter (SNG-1::GFP) stably integrated into the genome (Nonet, 1999). No behavioral defects have been observed in jsIs219 animals. In jsIs219, the pattern of GFP fluorescence observed was similar to the typical localization pattern seen in immunolocalization experiments with endogenous SNG-1, except that faint GFP fluorescence was detected in the neuronal cell bodies in the head ganglia and the ventral nerve cord (Figure 2B). Double staining jsIs219 with antibodies against GFP (Figure 1, E and F) and SNT-1 (Figure 1, C and D) revealed similar localization pattern in the nerve ring, nerve cords, and the SAB axons. The merged images indicate that SNG-1::GFP colocalizes with SNT-1 (Figure 1, G and H). These results strongly suggest that SNG-1::GFP is localized to SVs, similar to endogenous SNG-1.
Figure 2.
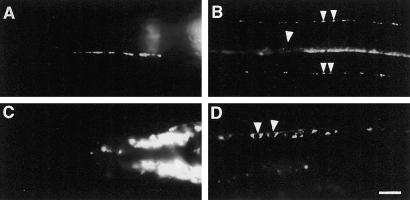
SNG-1::GFP localization depends on unc-104. (A–D) Images of live anesthetized animals expressing GFP-tagged SNG-1. (A) Lateral view of the head of a jsIs219 animal showing an SAB axon with presynaptic varicosities. (B) Ventral view of the ventral nerve cord and ventral sublateral nerve cords of a jsIs219 animal. A punctate pattern of fluorescence is seen along all three nerve cords. Individual puncta are seen in the sublateral cords (small arrowheads). Neuronal cell bodies are barely detectable in the ventral nerve cord (large arrowhead). (C) Lateral view of the head of the axonal transport mutant unc-104(e1265) showing fluorescence accumulations in the head ganglia. (D) Ventral view of the ventral nerve cord of an unc-104(e1265) hermaphrodite. Fluorescence accumulates in the neuronal cell bodies (arrowheads) along the nerve cord [genotype of A and B, jsIs219; C and D, unc-104(e1265) jsIs219]. Bar, 10 μm.
An additional line of evidence supporting this conclusion is the dependence of the synaptic localization of SNG-1::GFP on the unc-104 gene. The unc-104 gene encodes a kinesin-like molecule that is required for the axonal transport of SVs (Hall and Hedgecock, 1991; Otsuka et al., 1991). Mutations in this gene result in the accumulation of SVs in neuronal cell bodies (Hall and Hedgecock, 1991). SV proteins such as RAB-3, SNB-1, and SNT-1 all require unc-104 for synaptic localization (Nonet et al., 1993, 1997, 1998). In contrast, localization of nonvesicular localized proteins such as SYD-2, RIM, UNC-11, and UNC-13 does not require unc-104 (Nonet et al., 1999; Zhen and Jin, 1999; Kohn et al., 2000; Hadwiger and Nonet, unpublished observations). In unc-104(e1265) mutants, SNG-1::GFP was concentrated in neuronal cell bodies in both the head ganglia and the ventral nerve cord (Figure 2, C and D). GFP fluorescence was greatly diminished in ventral axonal regions (Figure 2D) and absent from sublateral nerve cords, SAB axons and the dorsal nerve cord (our unpublished results). This result demonstrates that SNG-1::GFP localization requires the UNC-104 kinesin like other SV proteins, suggesting that it is localized to SVs.
For the purposes of this study, any GFP fusion protein that is considered to be localized to SVs has the following features: 1) strong punctate signal along the nerve cords; 2) accumulation of GFP fluorescence in the nerve ring and presynaptic varicosities of SAB neurons with little process staining; 3) no signal in sensory dendrites in the nose and commissures along the body, and neuronal cell bodies undetectable or barely visible; and 4) a dependency of the localization pattern on the unc-104 gene.
SNG-1::GFP Localization Does Not Require Several Synaptic Proteins We Examined
Biochemical studies have revealed many protein–protein interactions between different SV proteins (Bennett et al., 1992; Calakos and Scheller, 1994; Galli et al., 1996; Schivell et al., 1996). To assess whether localization of SNG-1 is mediated by protein–protein interactions with other SV protein(s), we examined SNG-1::GFP or native SNG-1 localization in mutants lacking distinct SV-associated proteins, including RAB-3 and RBF-1 (rabphilin), SNB-1, and SNT-1 (Table 1; Nonet et al., 1993, 1997, 1998; Staunton, Ganetzky, and Nonet, unpublished observations). In these single mutant backgrounds, the localization of SNG-1::GFP or native SNG-1 was indistinguishable from that in jsIs219 or wild-type animals (our unpublished results). We also determined whether SNG-1::GFP localization depends on unc-11 and aex-3 genes, which are required for the localization of SNB-1 and RAB-3 in C. elegans, respectively (Table 1; Iwasaki et al., 1997; Nonet et al., 1999). The localization of SNG-1::GFP was not affected by mutations in either gene. In addition, the gross localization pattern of SNG-1::GFP was not affected by genes that have been implicated in SV endocytosis such as unc-26 and dpy-23 (Table 1; Harris et al., 2000; Baum and Garriga, personal communication). For all mutants analyzed, except aex-3, null alleles were used to examine the localization of SNG-1::GFP or native SNG-1. Thus, SNG-1::GFP/SNG-1 localization does not depend on the individual SV proteins considered, suggesting that SNG-1 localization to synaptic regions might be mediated by information in its own sequence.
Table 1.
Mutants in which SNG-1 localization is examined
| Gene | Protein homolog | Function | Allele used |
|---|---|---|---|
| snt-1 | Synaptotagmin | Regulating Ca2+-triggered fusion | md290, null |
| snb-1a | Synaptobrevin | v-Snare, essential for fusion | js124, null |
| rab-3a | RAB-3 | Regulating synaptic vesicle docking | js49, null |
| rbf-1a | Rabphilin | RAB-3 binding protein, potentiate SNARE complex formation independent of RAB-3 | js232, null |
| unc-11 | AP180 | Required for SNB-1 localization | e47, null |
| aex-3 | DENN | GDP/GTP exchange factor, required for RAB-3 localization | y255, severe |
| unc-26 | Synaptojanin | Phosphotidyl inositol-5-phosphatase, required for endocytosis | s1710, null |
| dpy-23 | AP-2 subunit μ2 | Required for endocytosis | e840, null |
SNG-1 localization is examined by incubation with primary antibody against SNG-1 and FITC-conjugated secondary antibody. Others were examined by fluorescence of GFP-tagged SNG-1.
A Synaptic Localization Signal Resides in C-Terminal Domain of SNG-1
The topology of rat synaptogyrin suggests that the N terminus, C terminus, and the small loop between the second and the third transmembrane domain of SNG-1 reside in the cytoplasm (Stenius et al., 1995; Nonet, 1999). Reasoning that the localization of SNG-1 might be mediated by interactions with cytoplasmic factors, we deleted cytoplasmic domains of SNG-1 and assessed the ability of the lesioned proteins to localize in vivo (Figure 4C). As shown in Figure 3, A and B, when the N terminus was deleted, the localization of the mutated protein was similar to the full-length protein. However, when the C terminus was deleted, the pattern of GFP fluorescence was altered. SNG-1 (Δ173–248)::GFP decorated the entire cell in a distribution likely to reflect localization to the plasma membrane. Fluorescence was observed in the sensory dendrites in the nose (Figure 3E) and in many commissures along the body (Figure 3F). Moreover, fluorescence was more defuse and less punctate along neuronal processes (Figure 3F). The fraction of GFP in the puncta of the ventral sublateral cord was dramatically decreased and the process regions were more clearly visible (Figures 3F and 4C). The process regions of SAB neurons were also much brighter compared with those in jsIs219 (our unpublished results). Finally, bright fluorescence was seen in neuronal cell bodies along the ventral nerve cord and in the head ganglia (Figure 3, E and F). These results suggest that the C-terminal domain of SNG-1 is required for the correct localization of SNG-1, whereas the N-terminal domain is dispensable.
Figure 3.
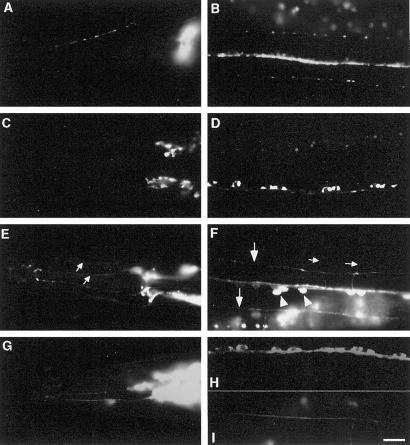
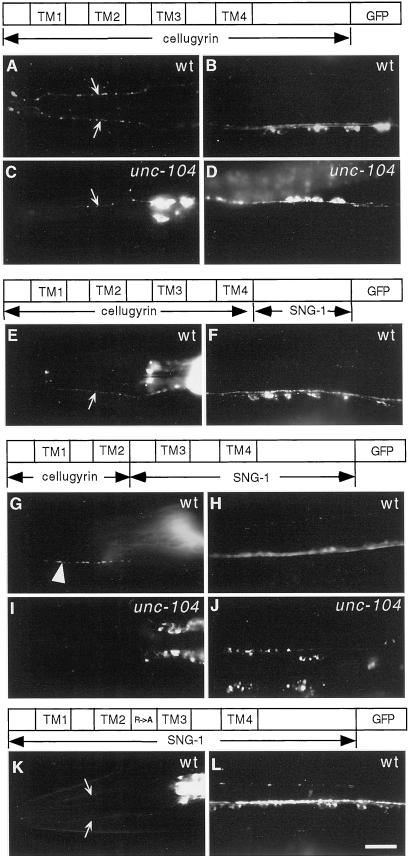
Deletion of C-, but not N-terminal domain causes mislocalization of SNG-1::GFP. (A–I) Images of live anesthetized transgenic animals expressing mutated GFP-tagged SNG-1. (A, C, E, and G) Lateral views of the head. (B, D, F, and H) Ventral views of the ventral nerve cords. (I) Dorsal view of the dorsal nerve cords. (A and B) SNG-1(Δ2–24)::GFP accumulates in presynaptic varicosities along an SAB axon (A) and shows a punctate pattern along all three nerve cords in a wild-type background (genotype: jsEx353, plasmid: pSY17). (C and D) SNG-1(Δ2–24)::GFP accumulates in neural cell bodies in head ganglia (C) and along the ventral nerve cord (D) in an unc-104(e1265) background [genotype: unc-104(e1265); jsEx353, plasmid: pSY17]. (E and F) SNG-1(Δ173–248)::GFP is observed in sensory dendrites (small arrows in E) and commissures (small arrows in F) in a wild-type background. Fluorescence is distributed diffusely along ventral sublateral cords (large arrows in F). GFP also accumulates in the neuronal cell bodies along the ventral nerve cord (arrowheads in F) (genotype: jsEx355, plasmid: pSY18). (G–I) SNG-1(Δ173–248)::GFP shows a similar pattern in an unc-104(e1265) background as in a wild-type background. Fluorescence decorates sensory dendrites (G), commissures, and accumulates in neuronal cell bodies along the ventral nerve cord (H). The dorsal nerve cords are also visible (I) [genotype: unc-104(e1265); jsEx355, plasmid pSY18]. Bar, 10 μm.
To provide additional support that the C-terminal deletion localizes to a cellular compartment distinct from SVs, we examined the localization of the mutant proteins in a unc-104 background. The GFP fluorescence pattern of the fusion protein lacking the N terminus in unc-104(e1265) was similar to that of full-length SNG-1::GFP, namely, GFP accumulated in the neuronal cell bodies of the ventral nerve cord and the head ganglia (Figure 3, C and D) and was absent from the dorsal nerve cord and the varicosities of SAB neurons. In contrast, the C-terminal truncated protein showed a similar distribution pattern in both an unc-104(e1265) and a wild-type background (Figure 3, G–I). To eliminate the possibility that GFP is detached from the SNG-1(Δ173–248)::GFP protein, an eight amino acid FLAG tag was placed at the N terminus of the mutant protein [FLAG::SNG-1(Δ173–248)::GFP]. An identical distribution pattern of FLAG::SNG-1(Δ173–248)::GFP was revealed by antibodies directed against each of the FLAG and GFP tags (our unpublished results), demonstrating that the fusion protein is intact and the localization pattern of the epitopes reflects the distribution pattern of the fusion protein. Immunodetection of FLAG::SNG-1::GFP with the use of both anti-FLAG and anti-GFP showed that the FLAG tag had no effect on localization (our unpublished results). Taken together, these data demonstrate that the C-terminal region of SNG-1 is necessary for its localization to SVs.
Fine Mapping of Localization Signal within C Terminus of SNG-1
To further map the localization signal within the C-terminal domain, we performed a systematic deletion analysis within the C terminus. Figure 4C shows a schematic diagram of the mutated regions. A quantitative approach was used to determine the degree of in vivo synaptic localization of each mutated protein (see MATERIALS AND METHODS). The percentage of the fluorescence intensity in the puncta within a segment of the sublateral ventral cord just anterior of the tail was used as an index for synaptic localization. The localization index of GFP alone was determined as a control.
As shown in Figure 4C, when the whole C-terminal domain was deleted, the localization index was dramatically decreased compared with the full-length fusion protein. However, when amino acids 211–248 were deleted, the localization index was unchanged. In contrast, when amino acids 173–210 were deleted, the localization index was decreased almost to the same extent as when the whole C-terminal domain was deleted. Further division of this region led to partial localization. These results were consistent with the distribution patterns observed by fluorescence microscopy, indicating that the localization signal is broadly distributed within amino acids 173–210 in the C terminus. There are three tyrosines conserved between rat synaptogyrin and SNG-1 within this defined region (Figure 4A). We investigated the role of these tyrosine residues in the localization of SNG-1 because such tyrosine-based signals have been shown to mediate endocytosis (Davis et al., 1986, 1987; Collawn et al., 1990; Peters et al., 1990; Thies et al., 1990; Letourneur and Klausner, 1992; Haucke and De Camilli, 1999). The Y174A Y201A Y203A triple mutant protein was partially mislocalized (Figure 4C). This suggests that the three tyrosines in the C terminus play a role in SNG-1 localization although they are not solely responsible for it. In summary, our data are consistent with a role for the C terminus in targeting SNG-1 for endocytosis from the plasma membrane. However, they do not exclude a role of the C terminus in the sorting of SNG-1 to SV precursors at the level of the trans-Golgi.
A Single Arginine in Small Cytoplasmic Loop Is Involved in Proper Localization of SNG-1
The cytoplasmic loop between transmembrane domain 2 and 3 of SNG-1 is well conserved among members of the synaptogyrin family (Figure 4A). Because rat synaptogyrin and SNG-1 are localized in C. elegans (Figure 6), but rat cellugyrin is not (Figure 5), we focused our attention on differences in this domain because they might identify a signal regulating the selective targeting of synaptogyrin to SVs, rather than to other transport vesicles. Specifically, we noted that a tyrosine in cellugyrin that is substituted by a positively charged amino acid in both synaptogyrin and SNG-1. Hence, we mutated the corresponding amino acid, arginine, in SNG-1 to an alanine by site-directed mutagenesis. In transgenic animals, R104A mutant SNG-1::GFP was partially mislocalized. The fraction of GFP signal in synaptic puncta was decreased (Figure 4C). In addition, GFP signal was retained in the neuronal cell bodies and was present in the commissures and sensory dendrites (Figure 5, K and L). Although this finding does not directly address which sorting step is disrupted by this domain, our data suggest that it plays a role in sorting synaptogyrin to SVs.
Figure 6.
Rat synaptogyrin is localized to synaptic regions in C. elegans. (A and B) Images of live anesthetized animals expressing GFP-tagged rat synaptogyrin. (A) Ventral view of a wild-type animal showing a punctate GFP pattern in the ventral nerve cords. (B) Ventral view of an unc-104(e1265) animal showing GFP accumulations in neuronal cell bodies along the ventral nerve cord. (C–H) Images of wild-type animals expressing GFP-tagged synaptogyrin double stained with antibodies against GFP (C and D) and SNG-1 (E and F). FITC- and Cy-3–conjugated secondary antibody are used respectively. (G and H) Merged images. (C, E, and G) Lateral views of the head. (D, F, and H) Ventral views of the ventral nerve cords. (I and J) Lateral views of the head of wild-type animals expressing GFP-tagged rat synaptogyrin lacking the C-terminal domain. Fluorescence accumulates in selective processes in the nerve ring (I) and neuronal cell bodies of the head ganglia (I and J). GFP is also present in sensory dendrites in the nose (I and J). Genotypes of strains used (plasmid construct in parentheses): A, C–H, jsEx403 (pSY12); B, unc-104(e1265); jsEx403 (pSY12); I and J, jsEx421 (pSY47). Bar, 10 μm.
Figure 5.
Localization signals of SNG-1 target rat cellugyrin to synaptic regions in C. elegans. Images of live anesthetized animals expressing diagrammed constructs (A, C, E, G, I, and K). Lateral views of the head (B, D, F, H, J, and L). Ventral views of the ventral nerve cords. (A) Cellugyrin is seen in sensory dendrites (labeled with small arrows), commissures, and neural cell bodies (B) in a wild-type background and an unc-104 background (C and D). A chimera containing cellugyrin sequences to the end of the transmembrane segments and the SNG-1 C-terminal domain shows a similar distribution pattern as cellugyrin (F and G). In a chimera with additional SNG-1 sequences, the protein now behaves like SNG-1 and accumulates in the presynaptic varicosities along an SAB axon (large arrowhead in H) and shows a punctate pattern along the ventral nerve cords (I) in a wild-type background. In an unc-104(e1265) background, the chimera accumulates in neural cell bodies in the head ganglia (J) and the ventral nerve cord (K). SNG-1(R104A) is partially mislocalized because fluorescence is present in sensory dendrites (K) and commissures as well as cell bodies (L). Genotype of strains used (with plasmid construct in parentheses): A and B, jsEx375 (pSY14); C and D, unc-104(e1265); jsEx375 (pSY14); E and F, jsEx357 (pSY31); J and H, jsEx498 (pSY57); I and J, unc-104(e1265); jxEx498 (pSY57); K and L, jsEx547 (pSY62). Bar, 10 μm.
Cellugyrin-SNG-1 Chimera Is Localized to Synaptic Regions
We next assessed whether a region of SNG-1 containing the two identified primary sequence motifs necessary for SNG-1 localization are sufficient to localize rat cellugyrin to synaptic regions. GFP-tagged full-length cellugyrin expressed in C. elegans under the control of the sng-1 promoter showed a very different localization pattern from SNG-1::GFP. Cellugyrin::GFP accumulated in neuronal cell bodies of the head ganglia and the ventral nerve cord (Figure 5B). GFP was also distributed in the sensory dendrites in the nose and commissures running circumferentially along the body (Figure 5, A and B). Along the nerve cords and SAB axons, GFP was much less punctate (our unpublished results). This distribution pattern was not affected by mutation in unc-104 gene (Figure 5, C and D). These results demonstrate that rat cellugyrin is not restricted to synaptic regions in C. elegans, which is consistent with its endogenous localization in rat where it is excluded from SVs (Janz and Südhof, 1998).
When the C-terminal domain of cellugyrin was replaced by the C-terminal domain of SNG-1, the localization pattern of the chimera was indistinguishable from cellugyrin alone (Figure 5, E and F). However, when a region of SNG-1 containing both the C terminus and the cytoplasmic loop was used to replace the corresponding region of cellugyrin, the chimera was localized to synaptic regions in a manner similar to SNG-1::GFP (Figure 5, G and H). The only difference between the two localization patterns was the slightly greater fluorescence intensity in neuronal cell bodies in animals expressing the cellugyrin::GFP chimera compared with animals expressing SNG-1::GFP (Figure 5H). Furthermore, in unc-104(e1265) background, the chimeric protein was accumulated in neuronal cell bodies (Figure 5, I and J). The GFP-tagged cellugyrin portion or the SNG-1 portion of the chimeric protein was not localized to synaptic regions by itself, instead, the fusion protein was accumulated in the neuronal cell bodies (not shown). These results suggest that the region of SNG-1 that contains the identified localization motifs is sufficient to localize cellugyrin to synaptic regions. This does not exclude the possibility that the third and fourth transmembrane domain and the second intravesicular loop of SNG-1 may aid in the localization of the chimera.
Mechanisms for Synaptogyrin Localization Are Evolutionarily Conserved
To determine whether mechanisms of synaptogyrin localization are evolutionarily conserved, we assessed whether the localization signal(s) of rat synaptogyrin can be recognized in C. elegans. A GFP fusion to the C terminus of rat synaptogyrin was expressed in a wild-type genetic background under the control of the sng-1 promoter. In transgenic animals, GFP-tagged rat synaptogyrin showed a punctate pattern along the major nerve cords and the sublateral nerve cords (Figure 6A). GFP was concentrated in the nerve ring and in the presynaptic varicosities of SAB neurons (our unpublished results). In contrast, in an unc-104(e1265) mutant background, fluorescence accumulated in the cell bodies along the ventral nerve cord (Figure 6B) and in the head ganglia (our unpublished results). Furthermore, GFP-tagged rat synaptogyrin colocalized with native SNG-1 shown by double immunostaining with the use of antibodies against GFP and SNG-1 (SNG-1 antibody did not cross-react with rat synaptogyrin) (Figure 6, C–H). These results suggest that rat synaptogyrin is localized to synaptic regions in C. elegans, indicating that the localization signal(s) of rat synaptogyrin are recognized by the C. elegans SNG-1 localization machinery. We next asked whether the C terminus of rat synaptogyrin is necessary for its localization in C. elegans. GFP-tagged synaptogyrin lacking the C terminus was expressed in wild-type animals. Fluorescence was detected in neuronal cell bodies in the head ganglia and the sensory dendrites in the nose (Figure 6, I and J), but not synaptic regions, suggesting the C terminus of synaptogyrin is necessary for its localization in C. elegans.
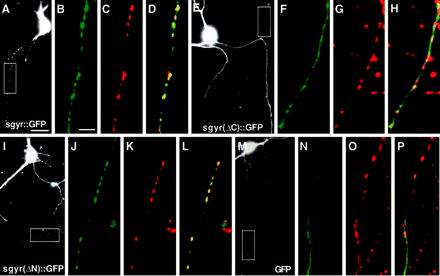
To determine whether rat synaptogyrin was localized similarly to other SV proteins in mammalian neurons, we examined the localization of synaptogyrin in hippocampal neurons. First, GFP was fused to the C terminus of full-length synaptogyrin and the fusion gene was transfected into cultured hippocampal primary neurons. Then 5- to 7-day-old neurons were fixed to preserve GFP fluorescence and stained with an antibody against synaptophysin. Figure 7B shows that the full-length GFP-tagged synaptogyrin exhibited a punctate pattern along the axonal process of the hippocampal neuron, similar to the pattern observed by anti-synaptophysin immunostaining (Figure 7, A and C). GFP was also seen in neuronal cell bodies and dendritic processes, where it was evenly distributed with no obvious puncta (our unpublished results). This aberrant localization may be the result of overexpression of the fusion protein. These results suggest that axonal GFP-tagged synaptogyrin is largely localized to synaptic regions in hippocampal neurons.
Figure 7.
Deletion of C-terminal domain of rat synaptogyrin causes mislocalization in hippocampal neurons. (A, E, I, and M) Images of hippocampal neurons expressing GFP or GFP-tagged synaptogyrin visualized by GFP. Boxed areas are regions shown in after high magnification images. (B–D, F–H, J–L, N–P) High-magnification images of axonal processes visualized by GFP (B, F, J, and N) and incubation with a primary antibody against synaptophysin and an LRSC-conjugated secondary antibody (C, G, K, and L). (D, H, L, and P) are merged images. (A–D) GFP-tagged full-length synaptogyrin expressed with the use of plasmid pSY26 colocalizes with synaptophysin in a punctate pattern along the axonal process. (E–H) GFP-tagged synaptogyrin lacking the C terminus expressed with the use of plasmid pSY46 shows a smooth distribution along the axon (F), whereas synaptophysin is localized to synaptic puncta (G). They do not colocalize in the merged image (H). (I–L) GFP-tagged synaptogyrin lacking N terminus expressed with the use of plasmid pSY59 shows a similar distribution pattern as the full-length protein. (M–P) GFP alone is distributed evenly alone the axonal process. Bar in A is for A, E, I, and M, 10 μM. Bar in B is for B–D, F–H, J–L, and N–P, 5 μm.
To identify whether the localization signals used by C. elegans SNG-1 and rat synaptogyrin were similar we examined the localization of GFP-tagged rat synaptogyrin lacking either the N- or C-terminal domain in cultured hippocampal neurons. Synaptogyrin lacking its N terminus (Figure 7, I–L) showed a distribution pattern similar to the full-length rat synaptogyrin (Figure 7, A–D), suggesting that the N-terminal domain is not required for its correct localization. However, when the C-terminal domain was deleted from rat synaptogyrin, GFP fluorescence was no longer punctate, but showed a diffuse pattern along the axonal processes of cultured hippocampal neurons (Figure 7, E and F). Furthermore, this mutant protein no longer colocalized with synaptophysin immunostaining (Figure 7G) because GFP was present in regions of processes where synaptophysin was absent (Figure 7H). This distribution pattern was similar as that of GFP alone expressed in hippocampal neurons (Figure 7, M–P). These results suggest that the C-terminal domain of synaptogyrin contains a targeting signal that is required for its synaptic localization in mammalian neurons.
DISCUSSION
We have identified the primary sequences that are required for the subcellular localization of both C. elegans and rat synaptogyrin. Synaptic localization of SNG-1 depends upon two sequence motifs located in the cytoplasmic loop separating the second and third transmembrane domains and in the C-terminal domain. These two domains may define signals required for targeting SNG-1 selectively to SVs and for selective endocytosis from the plasma membrane. These regions define bona fide localization signals because they are also sufficient, in the context of other SNG-1 sequences, to localize rat cellugyrin to synaptic regions in C. elegans. The C-terminal domain of rat synaptogyrin contains a similar targeting signal required for localization in hippocampal neurons. Indeed, the rat ortholog is appropriately localized in C. elegans and the analogous C-terminal region of rat synaptogyrin is required in this assay. These data suggest that the rat localization signal is recognized by the C. elegans trafficking machinery. Thus, the mechanisms for synaptogyrin localization appear to be evolutionarily conserved. These results represent the first set of experiments that define the localization signal of a SV integral membrane protein in a model organism with the use of an in vivo assay.
Caenorhabditis elegans as a Model System to Study Protein Targeting
The criteria we have used to examine localization of a SV protein in C. elegans differ considerably from criteria used for most prior studies in PC12 cells to define SV targeting signals. Our studies use primarily visual criteria to assess localization, whereas studies with the use of PC12 cells rely on biochemical fractionation. Due to the small size of C. elegans it is essentially impossible to separate the 2% of neuronal tissue in the C. elegans adult before biochemical fractionation. Our attempts to develop biochemical assays for localization have been severely hampered by this limitation. Despite the limitations, the availability of mutants permit us to examine the dependence of localization on specific trafficking components. For example, examination of the targeting behavior in unc-104 kinesin mutants acts as an indirect assay for sorting to preSVs. As the study of trafficking identifies genes essential for other trafficking steps, these assays will become a powerful tool in our analysis. Our extensive reliance on visual assays does limit the interpretation of some of our in vivo findings. Specifically, although N-terminal–deleted SNG-1 still targets to regions rich in synapses, it is possible that the protein is still absent from mature SVs, and instead accumulates in another vesicular structure in synaptic neuropil (such as synaptic endosomes). It is also difficult to assess in which cellular compartments our mistargeted SNG-1 mutants are accumulating.
Although our assays have limitations, they examine trafficking in functioning neurons in an intact animal. In other studies, we have observed significant differences between the role of targeting signals in PC12 cells and in our in vivo assays. Specifically, we examined the role of previously defined SV targeting signals of synaptobrevin (Grote et al., 1995). We introduced into the C. elegans synaptobrevin gene two specific lesions (M46A and N49A), which altered targeting of synaptobrevin in PC12 cells. Genomic clones containing each lesion completely rescued the lethal phenotype of C. elegans synaptobrevin mutants and the mutant protein localized indistinguishably from wild-type (Wei and Nonet, unpublished observations). These observations imply that the identified sequences do not play as critical roles in synaptobrevin sorting in neurons in vivo. This dichotomy could be explained either by differences between how PC12 cells and neurons sort synaptic proteins, or in differences between synaptobrevin sorting mechanisms in vertebrates and invertebrates. However, our data demonstrating the conservation of synaptogyrin sorting signals through the metazoan lineage suggest that differences between PC12 cells and neurons many be more significant than species differences.
Our in vivo observations also provide a framework within which to interpret functional studies in PC12 cells. Previous experiments reported that C-terminal sequences adjacent to the transmembrane region in rat synaptogyrin are required for its inhibitory effect on Ca2+ dependent exocytosis in PC12 cells (Sugita et al., 1999). Our data suggest that deleting this region probably disrupts protein localization; this in turn could indirectly lead to the reported effects on exocytosis. Our system provides a relatively simple method of examining the role of identified targeting signals in vivo that complement biochemical studies with the use of PC12 cells. Furthermore, in cases where mutants disrupting the gene encoding the protein of interest have been identified, the role of these signals can also be assessed functionally.
The potential to saturate or alter sorting pathways as in response to changes in expression levels of introduced genes is a difficult issue to control for in both this system and in PC12 cells. However, we believe it is unlikely that expression levels are significantly affecting our assays. In C. elegans, expression levels in transgenic animals are often manipulated by varying the ratio of input experimental and marker DNAs (Mello and Fire, 1995). The mutated SNG-1 constructs were injected at a DNA concentration several fold below levels where wild-type SNG-1 protein remains localized indistinguishably from the native protein (our unpublished results). In addition, multiple transgenic lines were created for each construct we examined, and several with similar expression levels behaved identically. Finally, the fluorescent intensity did not vary dramatically in animals expressing different mutant SNG-1 proteins. Taken together, our data suggest that expression levels of the various mutant SNG-1 proteins do not contribute significantly to their mislocalization.
Synaptic Localization Signals in Other Systems
Most of the studies of trafficking pathways of SV proteins have examined the targeting of proteins to SLMVs in PC12 cells. Experimental evidence suggests that SV protein sorting occurs by at least three pathways at a number of distinct stages during the SV life cycle (Calakos and Scheller, 1996; Hannah et al., 1999). SVs are probably not formed directly by budding from the TGN; rather mature SVs probably bud off from another membrane compartment. Experimental evidence supports both the plasma membrane and endosomal intermediates as the donor compartment (Regnier-Vigouroux et al., 1991; Bauerfeind and Huttner, 1993). Pulse chase studies of synaptophysin trafficking in PC12 cells suggest that synaptophysin is routed through the plasma membrane before inclusion into SLMVs. This route is apparently adaptor protein complex 2- (AP-2), clathrin-, and dynamin-dependent and brefeldin A-insensitive (Takei et al., 1996; Shupliakov et al., 1997; Schmidt and Huttner, 1998; Shi et al., 1998). In vitro studies have demonstrated that SVs can be budded from the endosomal compartment with the use of AP-3 and the small GTPase ADP ribosylation factor 1 (Faundez et al., 1997, 1998) as well as being brefeldin A-sensitive (Shi et al., 1998). However, AP-3 δ subunit mutants in mouse are viable and contain an abundance of normal SVs, suggesting this is not an obligate pathway (Kantheti et al., 1998). In addition, recent data have documented that a third route of SLMV formation, from the late endosome, also exists in PC12 cells. In this pathway, P-selectin passes through early and late endosomal intermediates sequentially en route to both the lysosome and SLMVs (Blagoveshchenskaya and Cutler, 2000).
At least in neuroendocrine PC12 cells, different proteins use different pathways to differing extents to traffic to the same destination vesicle, the SLMV (Shi et al., 1998; Blagoveshchenskaya et al., 1999a,b). This suggests that each SV protein contains multiple signals that control the flux of the protein through more than one pathway. These signals may mediate different sorting steps at the TGN, plasma membrane, and early or late endosome. In keeping with this idea multiple signal sequences have been found within the cytoplasmic domain of synaptobrevin and synaptotagmin. In neuroendocrine PC12 cells, a sorting signal of synaptobrevin consisting of residues 41–50 probably functions at the plasma membrane because its deletion prevented endocytosis and SLMV targeting (Grote et al., 1995; Grote and Kelly, 1996). In contrast, residues 31–38 of synaptobrevin are required at a sorting stage other than endocytosis at the plasma membrane because a deletion mutant lacking these amino acids was endocytosed normally but excluded from SLMV (Grote et al., 1995). Although these regions are within the amphipathic helix required for biochemically defined synaptobrevin functions (SNARE complex formation), studies by Hao et al. (1997) revealed little correlation between either endocytosis or SLMV targeting activity and complex formation activity. Furthermore, West et al. (1997) showed that the cytoplasmic sequences of synaptobrevin were sufficient to target synaptobrevin to synaptic sites in hippocampal cultures, but not into SVs, suggesting that the transmembrane sequences may also contain targeting information. Although multiple sorting motifs have been found in the C-terminal domain of synaptotagmin, a detailed biochemical study is lacking that would identify the precise steps in which they play an important role (Blagoveshchenskaya et al., 1999; Krasnov and Enikolopov, 2000).
SNG-1 Sorting Signals in C. elegans
In the C. elegans SNG-1 protein we have also found multiple elements that are required for its proper targeting, namely, a 38 amino acid element in the C-terminal domain and a single amino acid in the cytoplasmic loop. Mutations in these sequences could affect SNG-1 sorting at the TGN, plasma membrane, or endosomal compartments. We observe an accumulation of SNG-1 at the periphery of neuronal cell bodies, suggesting that these elements may be required for endocytosis at the plasma membrane. Lesioning the three tyrosines in the C-terminal targeting domain also resulted in accumulation of mutant protein in the periphery of the soma. Thus, one component of the signal may be a tyrosine-based endocytosis signal. In addition, the distribution of the mutant proteins is not significantly altered by a reduction in SV axonal transport caused by the unc-104 mutant, suggesting that SNG-1 lacking these elements is distributed to vesicles other than preSVs at the TGN. One simple model explaining the behavior of our mutants implicates two signals. First, a TGN signal targeting proteins to preSVs. In absence of this signal synaptogyrin traffics to the plasma membrane in the cell body. The protein accumulates in the plasma membrane rather than maturing via an endocytic pathway in the soma because of the absence of a second signal, perhaps tyrosine-based, which mediates an AP-2–dependent endocytosis. Although our data are consistent with this model, in absence of a clearer understanding of the cellular compartments where targeting mutants accumulate, it is difficult to eliminate many other consistent models.
What Can Diverse Sorting Signals Achieve?
The signal sequences identified in SNG-1 are unique and do not share any obvious homology to other SV protein-sorting sequences. Other sequences identified in other systems also do not show any similarity to each other. This strongly supports the idea that no common primary sequence motif serves as a universal SV targeting signal. What would be the advantage of having each SV protein sorted with the use of distinct targeting signals, instead of with the use of one common signal? First, multiple signals may aid in regulating the stoichiometry of individual proteins in the SV, a small organelle containing only a few molecules of certain critical protein constituents. Therefore, achieving vesicular homogeneity may require that SV formation be tightly controlled at the single protein level. During the sorting process, “adaptor” proteins that interact with multiple vesicle proteins with the use of distinct targeting sequences could ensure a specific protein stoichiometry in each vesicle. In contrast, if different proteins compete with the use of the same targeting signal, the relative stoichiometry may be more stochastic. Second, several different types of related secretory vesicles are present in neurons and secretory cells with unique, but related membrane protein composition (Winkler, 1997). These include peptide-containing dense core secretory vesicles as well as zinc-containing vesicles (Perez-Clausell and Danscher, 1985). Multiple targeting signals may be needed to target the same protein to different classes of related vesicles. Finally, it may be beneficial for certain classes of vesicles to be nonfunctional with respect to secretion until they reach their final destination. Trafficking of distinct secretory vesicle components to the synapse with the use of different pathways is one mechanism of ensuring that preSVs are largely fusion incompetent.
How Is Sorting Achieved?
Identifying trans-acting factors that interact with identified sorting signals is critical to understanding complex sorting processes that are likely to be dependent on both protein–protein and protein–lipid interactions. We have used two approaches to attempt to identify trans-acting factors. First, we have examined the localization of SNG-1 in other mutants lacking specific SV proteins. In particular, we examined the localization of SNG-1 in a synaptobrevin mutant background. In vertebrates, synaptophysin (related to synaptogyrin) interacts with synaptobrevin (Calakos and Scheller, 1994; Galli et al., 1996), and formation of this complex correlates with maturation of synapses (Becher et al., 1999). In C. elegans, synaptogyrin localization was not dependent upon synaptobrevin, nor on the presence of any other SV protein we examined (Table 1). Although our experiments would not detect interactions that are mediated by the synergistic action of several SV proteins, they argue against a piggyback targeting mechanism for synaptogyrin. In a second attempt to identify other proteins that aid in SNG-1 localization we carried out a yeast two-hybrid analysis with the C termini of both SNG-1 and rat synaptogyrin, without success. The focus of our current and future work is identifying mutants that fail to efficiently target SNG-1::GFP with the use of classical genetic mutant screens.
ACKNOWLEDGMENTS
We thank Anne Marie Craig, Huai-Yang Wu, and Kimberly Harms for providing hippocampal cultures and expertise regarding their manipulation; Dr. T. Südhof for providing cellugyrin and synaptogyrin cDNA clones; Dr. P. De Camilli for providing rat synaptogyrin antibody; and the Nonet and Salkoff labs for critical comments on the manuscript. The Caenorhabditis Genetic Center provided some strains used in this work. This work was funded by a grant from the U.S. Public Health Service.
REFERENCES
- Banker GA, Cowan WM. Rat hippocampal neurons in dispersed cell culture. Brain Res. 1977;126:397–442. doi: 10.1016/0006-8993(77)90594-7. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- Bauerfeind R, Huttner WB. Biogenesis of constitutive secretory vesicles, secretory granules and synaptic vesicles. Curr Opin Cell Biol. 1993;5:628–635. doi: 10.1016/0955-0674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- Baumert M, Takei K, Hartinger J, Burger PM, Fischer von Mollard G, Maycox PR, De Camilli P, Jahn R. P29: a novel tyrosine-phosphorylated membrane protein present in small clear vesicles of neurons and endocrine cells. J Cell Biol. 1990;110:1285–1294. doi: 10.1083/jcb.110.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher A, Drenckhahn A, Pahner I, Margittai M, Jahn R, Ahnert-Hilger G. The synaptophysin-synaptobrevin complex: a hallmark of synaptic vesicle maturation. J Neurosci. 1999;19:1922–1931. doi: 10.1523/JNEUROSCI.19-06-01922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Calakos N, Kreiner T, Scheller RH. Synaptic vesicle membrane proteins interact to form a multimeric complex. J Cell Biol. 1992;116:761–775. doi: 10.1083/jcb.116.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya AD, Hewitt EW, Cutler DF. A complex web of signal-dependent trafficking underlies the triorganellar distribution of P-selectin in neuroendocrine PC12 cells. J Cell Biol. 1999a;145:1419–1433. doi: 10.1083/jcb.145.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya AD, Hewitt EW, Cutler DF. Di-leucine signals mediate targeting of tyrosinase and synaptotagmin to synaptic-like microvesicles within PC12 cells. Mol Biol Cell. 1999b;10:3979–3990. doi: 10.1091/mbc.10.11.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya AD, Cutler DF. Sorting to synaptic-like microvesicles from early and late endosomes requires overlapping but not identical targeting signals. Mol Biol Cell. 2000;11:1801–1814. doi: 10.1091/mbc.11.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley KM, Melikian HE, Provoda CJ, Waring MT. Regulation of neuronal function by protein trafficking: a role for the endosomal pathway. J Physiol. 2000;525:11–19. doi: 10.1111/j.1469-7793.2000.t01-2-00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calakos N, Scheller RH. Vesicle-associated membrane protein and synaptophysin are associated on the synaptic vesicle. J Biol Chem. 1994;269:24534–24537. [PubMed] [Google Scholar]
- Calakos N, Scheller RH. Synaptic vesicle biogenesis, docking, and fusion: a molecular description. Physiol Rev. 1996;76:1–29. doi: 10.1152/physrev.1996.76.1.1. [DOI] [PubMed] [Google Scholar]
- Collawn JF, Stangel M, Kuhn LA, Esekogwu V, Jing SQ, Trowbridge IS, Tainer JA. Transferrin receptor internalization sequence YXRF implicates a tight turn as the structural recognition motif for endocytosis. Cell. 1990;63:1061–1072. doi: 10.1016/0092-8674(90)90509-d. [DOI] [PubMed] [Google Scholar]
- Davis CG, Lehrman MA, Russell DW, Anderson RG, Brown MS, Goldstein JL. The J.D. mutation in familial hypercholesterolemia: amino acid substitution in cytoplasmic domain impedes internalization of LDL receptors. Cell. 1986;45:15–24. doi: 10.1016/0092-8674(86)90533-7. [DOI] [PubMed] [Google Scholar]
- Davis CG, van Driel IR, Russell DW, Brown MS, Goldstein JL. The low density lipoprotein receptor. Identification of amino acids in cytoplasmic domain required for rapid endocytosis. J Biol Chem. 1987;262:4075–4082. [PubMed] [Google Scholar]
- Faundez V, Horng JT, Kelly RB. ADP ribosylation factor 1 is required for synaptic vesicle budding in PC12 cells. J Cell Biol. 1997;138:505–515. doi: 10.1083/jcb.138.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faundez V, Horng JT, Kelly RB. A function for the AP3 coat complex in synaptic vesicle formation from endosomes. Cell. 1998;93:423–432. doi: 10.1016/s0092-8674(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Feany MB, Yee AG, Delvy ML, Buckley KM. The synaptic vesicle proteins SV2, synaptotagmin and synaptophysin are sorted to separate cellular compartments in CHO fibroblasts. J Cell Biol. 1993;123:575–584. doi: 10.1083/jcb.123.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T, Broadie K. Interaction of stoned and synaptotagmin in synaptic vesicle endocytosis. J Neurosci. 2001;21:1218–1227. doi: 10.1523/JNEUROSCI.21-04-01218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T, Davis WS, Broadie K. The stoned proteins regulate synaptic vesicle recycling in the presynaptic terminal. J Neurosci. 1999;19:5847–5860. doi: 10.1523/JNEUROSCI.19-14-05847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Chacon R, Südhof TC. Genetics of synaptic vesicle function: toward the complete functional anatomy of an organelle. Annu Rev Physiol. 1999;61:753–776. doi: 10.1146/annurev.physiol.61.1.753. [DOI] [PubMed] [Google Scholar]
- Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- Galli T, McPherson PS, De Camilli P. The V0 sector of the V-ATPase, synaptobrevin, and synaptophysin are associated on synaptic vesicles in a Triton X-100-resistant, freeze- thawing sensitive, complex. J Biol Chem. 1996;271:2193–2198. doi: 10.1074/jbc.271.4.2193. [DOI] [PubMed] [Google Scholar]
- Goslin K, Banker G. Experimental observations on the development of polarity by hippocampal neurons in culture. J Cell Biol. 1989;108:1507–1516. doi: 10.1083/jcb.108.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E, Hao JC, Bennett MK, Kelly RB. A targeting signal in VAMP regulating transport to synaptic vesicles. Cell. 1995;81:581–589. doi: 10.1016/0092-8674(95)90079-9. [DOI] [PubMed] [Google Scholar]
- Grote E, Kelly RB. Endocytosis of VAMP is facilitated by a synaptic vesicle targeting signal. J Cell Biol. 1996;132:537–547. doi: 10.1083/jcb.132.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass NK, Kartenbeck MA, Leube RE. Pantophysin is a ubiquitously expressed synaptophysin homologue and defines constitutive transport vesicles. J Cell Biol. 1996;134:731–746. doi: 10.1083/jcb.134.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Hannah MJ, Schmidt AA, Huttner WB. Synaptic vesicle biogenesis. Annu Rev Cell Dev Biol. 1999;15:733–798. doi: 10.1146/annurev.cellbio.15.1.733. [DOI] [PubMed] [Google Scholar]
- Hao JC, Salem N, Peng X-R, Kelly RB, Bennett MK. Effect of mutations in Vesicle-Associated Membrane Protein (VAMP) on the assembly of multimeric protein complexes. J Neurosci. 1997;17:1596–1603. doi: 10.1523/JNEUROSCI.17-05-01596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Hartwieg E, Horvitz HR, Jorgensen EM. Mutations in synaptojanin disrupt synaptic vesicle recycling. J Cell Biol. 2000;150:589–600. doi: 10.1083/jcb.150.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V, De Camilli P. AP-2 recruitment to synaptotagmin stimulated by tyrosine-based endocytic motifs. Science. 1999;285:1268–1271. doi: 10.1126/science.285.5431.1268. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, Staunton J, Saifee O, Nonet ML, Thomas J. aex-3 encodes a novel regulator of presynaptic activity in C. elegans. Neuron. 1997;18:613–622. doi: 10.1016/s0896-6273(00)80302-5. [DOI] [PubMed] [Google Scholar]
- Jahn R, Schiebler W, Ouimet C, Greengard P. A 38,000-dalton membrane protein (p38) present in synaptic vesicles. Proc Natl Acad Sci USA. 1985;82:4137–4141. doi: 10.1073/pnas.82.12.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Südhof TC. Synaptic vesicle traffic: rush hour in the nerve terminal. J Neurochem. 1993;61:12–21. doi: 10.1111/j.1471-4159.1993.tb03533.x. [DOI] [PubMed] [Google Scholar]
- Janz R, Südhof TC. Cellugyrin, a novel ubiquitous form of synaptogyrin that is phosphorylated by pp60c-src. J Biol Chem. 1998;273:2851–2857. doi: 10.1074/jbc.273.5.2851. [DOI] [PubMed] [Google Scholar]
- Janz R, Südhof TC, Hammer RE, Unni V, Siegelbaum SA, Bolshakov VY. Essential roles in synaptic plasticity for synaptogyrin I and synaptophysin I. Neuron. 1999;24:687–700. doi: 10.1016/s0896-6273(00)81122-8. [DOI] [PubMed] [Google Scholar]
- Johnston PA, Jahn R, Südhof TC. Transmembrane topography and evolutionary conservation of synaptophysin. J Biol Chem. 1989;264:1268–1273. [PubMed] [Google Scholar]
- Kantheti P, Qiao X, Diaz ME, Peden AA, Meyer GE, Carskadon SL, Kapfhamer D, Sufalko D, Robinson MS, Noebels JL, Burmeister M. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Kedra D, Pan HQ, Seroussi E, Fransson I, Guilbaud C, Collins JE, Dunham I, Blennow E, Roe BA, Piehl F, Dumanski JP. Characterization of the human synaptogyrin gene family. Hum Genet. 1998;103:131–141. doi: 10.1007/s004390050795. [DOI] [PubMed] [Google Scholar]
- Kohn RE, Duerr JS, McManus JR, Duke A, Rakow TL, Maruyama H, Molder G, Maruyama IN, Barstead RJ, Rand JB. Expression of multiple UNC-13 proteins in the Caenorhabditis elegans nervous system. Mol Biol Cell. 2000;11:3441–3452. doi: 10.1091/mbc.11.10.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Nonet ML. Sorting and transport in C. elegans: a model system with a sequenced genome. Curr Opin Cell Biol. 2000;12:517–523. doi: 10.1016/s0955-0674(00)00125-3. [DOI] [PubMed] [Google Scholar]
- Kramer JM, French RP, Park EC, Johnson JJ. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnov PA, Enikolopov G. Targeting of synaptotagmin to neurite terminals in neuronally differentiated PC12 cells. J Cell Sci. 2000;113:1389–1404. doi: 10.1242/jcs.113.8.1389. [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Shurland DL, van der Bliek AM. Contribution of the GTPase domain to the subcellular localization of dynamin in the nematode Caenorhabditis elegans. Mol Biol Cell. 1998;9:3227–3239. doi: 10.1091/mbc.9.11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur F, Klausner RD. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- Linstedt AD, Kelly RB. Endocytosis of the synaptic vesicle protein, synaptophysin, requires the COOH-terminal tail. J Physiol. 1991;85:90–96. [PubMed] [Google Scholar]
- Littleton JT, Bai J, Vyas B, Desai R, Baltus AE, Garment MB, Carlson SD, Ganetzky B, Chapman ER. Synaptotagmin mutants reveal essential functions for the C2B domain in Ca2+-triggered fusion, and recycling of synaptic vesicles in vivo. J Neurosci. 2001;21:1421–1433. doi: 10.1523/JNEUROSCI.21-05-01421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Analysis of an Organism. San Diego: Academic Press; 1995. pp. 451–482. [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. Studying mutants that affect neurotransmitter release in C. elegans. In: Bellen H, editor. Neurotransmitter Release. New York: Oxford University Press; 1999a. pp. 265–303. [Google Scholar]
- Nonet ML. Visualization of synaptic specializations in live C. elegans using synaptic vesicle-GFP protein fusions. J Neurosci Methods. 1999b;89:33–40. doi: 10.1016/s0165-0270(99)00031-x. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Holgado AM, Brewer F, Serpe CJ, Norbeck BA, J, H, Wei L, Hartwieg E, Jorgensen EM, Alfonso A. UNC-11, a C. elegans AP180 homolog, regulates the size and protein composition of synaptic vesicles. Mol Biol Cell. 1999;10:2343–2360. doi: 10.1091/mbc.10.7.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Saifee O, Zhao H, Rand JB, Wei L. Synaptic transmission deficits in C. elegans synaptobrevin mutants. J Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Staunton J, Kilgard MP, Fergestad T, Hartweig E, Horvitz HR, Jorgensen E, Meyer BJ. C. elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 1997;17:8021–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Otsuka AJ, Jeyaprakash A, Garcia-Anoveros J, Tang LZ, Fisk G, Hartshorne T, Franco R, Born T. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron. 1991;6:113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- Perez-Clausell J, Danscher G. Intravesicular localization of zinc in rat telencephalic boutons. A histochemical study. Brain Res. 1985;337:91–98. doi: 10.1016/0006-8993(85)91612-9. [DOI] [PubMed] [Google Scholar]
- Peters C, Braun M, Weber B, Wendland M, Schmidt B, Pohlmann R, Waheed A, von Figura K. Targeting of a lysosomal membrane protein: a tyrosine-containing endocytosis signal in the cytoplasmic tail of lysosomal acid phosphatase is necessary and sufficient for targeting to lysosomes. EMBO J. 1990;9:3497–3506. doi: 10.1002/j.1460-2075.1990.tb07558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Kim E, Sheng M, Craig AM. Heterogeneity in the molecular composition of excitatory postsynaptic sites during development of hippocampal neurons in culture. J Neurosci. 1998;18:1217–1229. doi: 10.1523/JNEUROSCI.18-04-01217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier-Vigouroux A, Tooze SA, Huttner WB. Newly synthesized synaptophysin is transported to synaptic-like microvesicles via constitutive secretory vesicles and the plasma membrane. EMBO J. 1991;10:3589–3601. doi: 10.1002/j.1460-2075.1991.tb04925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifee O, Wei LP, Nonet ML. The C. elegans unc-64 gene encodes a syntaxin which interacts genetically with synaptobrevin. Mol Biol Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schivell AE, Batchelor RH, Bajjalieh SM. Isoform-specific, calcium-regulated interaction of the synaptic vesicle proteins SV2 and synaptotagmin. J Biol Chem. 1996;271:27770–27775. doi: 10.1074/jbc.271.44.27770. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Huttner WB. Biogenesis of synaptic-like microvesicles in perforated PC12 cells. Methods. 1998;16:160–169. doi: 10.1006/meth.1998.0663. [DOI] [PubMed] [Google Scholar]
- Shi G, Faundez V, Roos J, Dell'Angelica EC, Kelly RB. Neuroendocrine synaptic vesicles are formed in vitro by both clathrin-dependent and clathrin-independent pathways. J Cell Biol. 1998;143:947–955. doi: 10.1083/jcb.143.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shupliakov O, Low P, Grabs D, Gad H, Chen H, David C, Takei K, De Camilli P, Brodin L. Synaptic vesicle endocytosis impaired by disruption of dynamin-SH3 domain interactions. Science. 1997;276:259–263. doi: 10.1126/science.276.5310.259. [DOI] [PubMed] [Google Scholar]
- Smith D, Fisher PA. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: applications of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley KK. Regulation of targeting signals in membrane proteins. Mol Membr Biol. 1996;13:19–27. doi: 10.3109/09687689609160570. [DOI] [PubMed] [Google Scholar]
- Stenius K, Janz R, Südhof TC, Jahn R. Structure of synaptogyrin (p29) defines novel synaptic vesicle protein. J Cell Biol. 1995;131:1801–1809. doi: 10.1083/jcb.131.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Sugita S, Janz R, Südhof T. Synaptogyrins regulate ca2+-dependent exocytosis in PC12 cells. J Biol Chem. 1999;274:18893–18901. doi: 10.1074/jbc.274.27.18893. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Takei K, Mundigl O, Daniell L, De Camilli P. The synaptic vesicle cycle: a single vesicle budding step involving clathrin and dynamin. J Cell Biol. 1996;133:1237–1250. doi: 10.1083/jcb.133.6.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Shimuta M, Komazaki S, Ohmi K, Nishi M, Iino M, Miyata A, Kangawa K. Mitsugumin29, a novel synaptophysin family member from the triad junction in skeletal muscle. Biochem J. 1998;331:317–322. doi: 10.1042/bj3310317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan PK, Waites C, Liu Y, Krantz DE, Edwards RH. A leucine-based motif mediates the endocytosis of vesicular monoamine and acetylcholine transporters. J Biol Chem. 1998;273:17351–17360. doi: 10.1074/jbc.273.28.17351. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Thies RS, Webster NJ, McClain DA. A domain of the insulin receptor required for endocytosis in rat fibroblasts. J Biol Chem. 1990;265:10132–10137. [PubMed] [Google Scholar]
- Tsukita S, Ishikawa H. The movement of membranous organelles in axons. Electron microscopic identification of anterogradely and retrogradely transported organelles. J Cell Biol. 1980;84:513–530. doi: 10.1083/jcb.84.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqui H, Erickson JD. The cytoplasmic tail of the vesicular acetylcholine transporter contains a synaptic vesicle targeting signal. J Biol Chem. 1998;273:9094–9098. doi: 10.1074/jbc.273.15.9094. [DOI] [PubMed] [Google Scholar]
- West A, Neve R, Buckley K. Targeting of the synaptic vesicle protein synaptobrevin in the axon of cultured hippocampal neurons: evidence for two distinct sorting steps. J Cell Biol. 1997;139:917–927. doi: 10.1083/jcb.139.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann B, Franke WW. Identification and localization of synaptophysin, an integral membrane glycoprotein of Mr 38,000 characteristic of presynaptic vesicles. Cell. 1985;41:1017–1028. doi: 10.1016/s0092-8674(85)80082-9. [DOI] [PubMed] [Google Scholar]
- Winkler H. Membrane composition of adrenergic large and small dense cored vesicles and of synaptic vesicles: consequences for their biogenesis. Neurochem Res. 1997;22:921–932. doi: 10.1023/a:1022410506476. [DOI] [PubMed] [Google Scholar]
- Zhen M, Jin Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]