Abstract
Carcinomas develop in complex environments that include a diverse spectrum of cell types that influence tumor cell behavior. These microenvironments represent dynamic systems that contribute to pathological processes. Damage to DNA is a notable inducer of both transient and permanent alterations in cellular phenotypes. Induction of a DNA-damage secretory program is known to promote adverse tumor cell behaviors such as proliferation, invasion, metastasis, and treatment resistance. However, prior studies designed to identify genotoxic stress-induced factors evaluated actively proliferating in vitro cultures of cells such as fibroblasts as experimental models. Conversely, the vast majority of benign cells in a typical tumor microenvironment (TME) are not proliferating, but rather exist in quiescent (i.e., G0) or in terminally-differentiated states. In this study, the diversity and magnitude of transcriptional responses to genotoxic damage in quiescent prostate fibroblasts were assessed using gene expression profiling. The secretory damage response in quiescent cells was highly concordant with that of actively dividing cells. Quiescent human prostate stroma exposed to genotoxic agents (e.g., mitoxantrone) in vivo resulted in significant upregulation (2.7-5.7 fold; (p≤0.01) of growth factors and cytokines including: IL-1β, MMP3, IL-6, and IL-8. The paracrine effects of damaged quiescent cells consistently increased the proliferation and invasion of prostate cancer cells and promoted cell survival and resistance to apoptosis following exposure to chemotherapy.
Implications
Benign quiescent cells in the TME respond to genotoxic stress by inducing a secretory program capable of promoting therapy resistance. Developing approaches to suppress the secretory program may improve treatment responses.
Keywords: prostate, quiescence, DNA damage, senescence, microenvironment, fibroblasts
Introduction
Malignant neoplasms arise in complex biophysical environments comprised of a diverse spectrum of cell types, structural components, and biochemical constituents that have profound influences on tumor cell behavior (1-3). Of importance, organ and tissue microenvironments represent dynamic systems with shifts in the numbers and types of benign resident cells – such as fibroblasts and endothelium, and immigrating cells – including those of immune lineage, in the context of normal development and pathological processes (4, 5). The phenotypes of these cells also vary depending on responses to extrinsic factors such as paracrine signaling from other juxtaposed cell types, concentrations of hormones and systemic growth factors, pathogens, nutrients, oxygen, pH and a spectrum of other influences that can either reversibly or irreversibly alter cellular functions (6).
Damage to DNA is a notable inducer of both transient and permanent alterations in cellular phenotype. Genotoxic stress can result from a variety of events that include exposure to free radicals, telomere shortening, oncogenes, errors in DNA replication, and treatment with cancer therapeutics. Cell cycle arrest is a well-described consequence of DNA damage, with subsequent proliferation if damage is repaired, or if severe, irreversible growth arrest manifest as senescence or programmed cell death (7-9). DNA-damage response (DDR) programs provide mechanisms to avoid propagating oncogenic mutations, and also activate a secretory program that comprises a diverse spectrum of proteases, growth factors, and cytokines, collectively and somewhat synonymously termed a senescence associated secretory phenotype (SASP), senescence messaging secretome, acute stress-associated secretome, and DNA damage secretory program (DDSP) (10-13). The composite effects of these programs have been shown to contribute to wound healing, aging phenotypes, altered immune responses, and are also capable of promoting adverse tumor cell behaviors such as proliferation, invasion, metastasis, and treatment resistance (13-17).
Large scale discovery-driven efforts designed to define the spectrum of secreted proteins induced by genotoxic damage have identified several hundred growth factors, cytokines, enzymes, and matricellular proteins that are altered in benign cells following genotoxic stress or in the context of cellular senescence (13, 18, 19). However, to date the majority of these profiling studies have used actively proliferating in vitro cultures of cells such as fibroblasts as experimental models (18-20). Conversely, the vast majority of benign cells in a typical tumor microenvironment, including fibroblasts, endothelium, smooth muscle and inflammatory cells, are not proliferating, but rather exist in quiescent, G0, or terminally-differentiated states. As the cell cycle phase has been shown to influence cellular responses to genotoxic exposures and other stresses (21, 22), it is unclear to what extent damage to proliferating cells reflects that of non-dividing cells in tissue microenvironments. In this study we sought to assess the diversity and magnitude of transcriptional responses to genotoxic damage in quiescent fibroblasts, compare the secretory damage response to that of actively dividing cells, and determine if the paracrine-acting factors derived from quiescent cells promote adverse cancer cell phenotypes such as proliferation, invasion, and resistance to cancer treatment-induced cell death.
Materials and Methods
Biospecimens, cell lines and culture conditions
Tissue samples were obtained under IRB-approved biospecimen collection and handling protocols. The primary human prostate fibroblast cell line, designated PSC27, was a gift from Dr. Beatrice Knudsen. PSC27 cells were cultured in prostate stromal cell (PSC) complete medium as described previously (23). The human prostatic epithelial cell line BPH1 was a gift from Dr. Simon Hayward and was derived from nonmalignant prostatic tissue with benign hyperplasia, immortalized by SV40-LT antigen, and cultured as previously described (24). The HeLa, PC3, VCaP, LNCaP and DU145 cell lines were obtained from ATCC and routinely sub-cultured as per ATCC recommendations. Cells were either used within 4 passages after receipt from ATCC or authenticated prior to initiating the studies by genotyping at DNA Diagnostics (Fairfield, OH).
Immunohistochemistry
Prostate tissue staining for Ki-67/MIB-1 has been described previously (25). The monoclonal antibody, MIB-1 (clone MIB-1, DAKO) was used to determine the proportion of cancer epithelial, cancer-associated stromal and benign-associated stromal cells staining positive for Ki-67. Prostate cancer tissue microarray slides were scanned on Aperio ScanScope AT (Aperio Technologies, Vista, CA, USA). High-resolution 20× digital images were created for the cancer and benign cores of twenty randomly selected cases. Positive Ki-67-stained cells and the total number of cells in 20× fields were counted using ImageJ2 Cell Counter plug-in (ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA). Any nuclear staining, regardless of intensity, was considered positive for Ki-67/MIB-1. For the stromal compartment, only spindle-like cells were included in the analysis, while round small nuclei cells were not considered for immunohistochemical evaluation, thus avoiding the inclusion of inflammatory cell in the analysis. The number of Ki-67 positive cells was expressed as a percentage of immunoreactive stromal (or epithelial) cells to the total counted stromal cells (or epithelial) in a 20× field.
Laser Microdissection
Frozen sections (7 μM) from were cut from OCT embedded snap-frozen radical prostatectomy specimens into PAP-membrane slides. Approximately 1000 cells were separately microdissected for prostate cancer epithelium (CPE), benign prostate epithelium (BPE) and stroma adjacent to cancer (CAS). The corresponding benign cells for each case were microdissected from separate blocks identified as containing no adenocarcinoma cells (first choice) or, from non-neoplastic tissues at a distance >1mm from the cancer. Digital photos were taken of tissue sections before, during, and after LCM and assessed to confirm the cell type-specificity of the captured cells.
Growth arrest conditions and cell treatments
PSC27 fibroblasts were plated at a density of 2 × 104 cells per cm2 in PSC medium and allowed to attach to the tissue culture dishes. To induce quiescence by growth factor starvation, the medium was changed to DMEM with 0.1% serum and cultured for 4 days before analysis. These cells were designed PSC27-QSS. To arrest cells by contact inhibition, cells were plated at a density of 2 × 104 cells per cm2 in stromal medium and allowed to grow to confluence, usually reaching complete confluency in 7∼10 days. These cells were designed PSC27-QCI. Proliferating or quiescent cells were treated with 1 μM mitoxantrone in PSC medium, or ionizing-radiation by a 137Cssource at 743 rad/min. Media under each condition was changed every 3 days for 10 days until cells were lysed for analysis. For quiescent cells allowed to resume proliferation, quiescent cells were trypsinized, replated to cell culture vessels of larger growth area or divided into multiple vessels, with the same media applied for each subculture. For each condition, three independent replicates were performed.
Immunofluorescence analysis and quantitation of DNA-damage foci
Cells grown on coverslips were rinsed in PBS, subjected to fixation in 4% paraformaldehyde and permeabilized with 0.1%Triton-X100 prior to immunostaining. Primary mouse monoclonal anti-phospho-Histone H2A.X (Ser139) (clone JBW301) and secondary antibody Alexa Fluor® 488 (or 594)-conjugated F(ab')2 goat anti-mouse IgG were sequentially applied. Nuclei were counterstained with 2 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI) and coverslips were mounted onto glass slides. H2A.X foci were manually counted and recorded with a 4-category counting strategy: 0 foci, 1-3 foci, 4-10 foci, and >10 foci. Data from each cell line/treatment were averaged from a pool of 3 independent fields counting 100 nuclei per pool.
Bromodeoxyuridine incorporation and flow cytometry
Cells were labeled with 100-μM bromodeoxyuridine (BrdU) for 6 h before collection with trypsin; the latter was inactivated with either serum or 1 mg/ml soybean trypsin inhibitor (Sigma, St. Louis). Cells were then fixed in PBS with 67% cold ethanol. Cell membranes were lysed at 37 °C in 0.08% pepsin for 20 min, and nuclei were treated with 2M HCl for 20 min. Samples were neutralized with 0.1M sodium borate, incubated in a buffer of 10 mM Hepes, (pH7.4), 150 mM NaCl, 4% fetal bovine serum (100 mg/ml), gelatin (0.04%), and EGTA (5M), and 2 μg of anti-BrdU-FITC antibody (BD Pharmingen, San Diego) on ice for 2 h. Mouse IgG1κ was run in parallel as a negative control. For nuclear labeling, cells were incubated with 5 μg/ml Hoescht 33342 (Calbiochem-Novabiochem, San Diego) or 100 μg/ml propidium iodide (Sigma). For cells arrested by mitogen withdrawal for 4 days, Hoescht was not applied as it induced cell death. Samples were run on a Beckton Dickinson FACS Vantage SE and the data were analyzed using FACSDiva software (BD Biosciences, Palo Alto).
Genotoxic treatments and cell proliferation, invasion, and chemoresistance assays
PSC27 cells were grown until 80% confluent, or induced to arrest growth (PSC27-Q) and were treated with 1 μM mitoxantrone in PSC medium, or ionizing-radiation by a 137Cssource at 743 rad/min as previously described (13). After treatment, the cells were rinsed 3-times with PBS and left to recover 3 days in PSC medium. Following recovery, cells were designated PSC27-MIT or PSC27-Rad. Normally proliferating PSC27 cells receiving sham treatment were designated as PSC27-Pro.
To generate conditioned medium, PSC27-Pro, PSC27-Rad, PSC27-Q, and PSC27-QRad cells were rinsed three times in PBS and incubated for 3 days in DMEM with 0.5% charcoal-stripped FBS. The supernatant was harvested as conditioned media (CM) and stored frozen at −80°C. Epithelial cell lines were seeded at 20,000 cells per well in six-well plates in PSC27 conditioned medium. Cultures were incubated for 3 days and the cell numbers were indirectly determined using the CellTiter96®AQueous One Solution Cell Proliferation Assay (MTS, Promega) with signals captured using a 96-well plate reader. For trans-well invasion assays, serum-starved cells in serum-free medium were added to the top chambers of Cultrex 24-well Cell Migration Assay plates (8 μm size, Trevigen) coated with basement membrane extract (BM) prepared as 0.5× of stock solution. CM from PSC27 cells or regular epithelial media containing 10% fetal calf serum (FCS) was added to the bottom chambers. Invading cells in the bottom chambers were stained and plate absorbance was recorded at 485/520 nm emission. All assays were done in triplicate and the data are presented as the average absorbance of invading cells.
For assessing responses to chemotherapy, epithelial cells were cultured with either DMEM, or CM generated from the various PSC27 treatments. Cells received Mitoxantrone (Sigma) treatment for 3 days at concentrations near individual cell line's IC50 levels. Cell viability was then assayed, and the percentage of viable cells was calculated by normalizing absorbance of each experiment to untreated cells.
Apoptosis assays
Prostate epithelial cells were plated at a density of 2 × 104 cells per well in 6-well culture plates and cultured with CM PSC27 cells. Twelve hours later, IC50 concentrations of mitoxantrone were added to each epithelial line, with distilled water applied in parallel as control. To examine acute survival, the cell numbers were determined 12 hours after drug exposure by counting viable cells with a hemocytometer. To quantitate apoptosis, lysates were prepared 24 hours post treatment from each group, and caspase levels were measured using the Caspase-Glo 3/7 assay (Promega). For the morphological analyses, bright field pictures were taken for epithelial cells using inverted phase-contrast microscopy.
Gene expression analysis by real-time qPCR
Single-stranded cDNA for qPCR analysis was synthesized from 1 μg total RNA using a final concentration of 5 μM random hexamer priming and M-MLV RT according to Ambion's instructions. qPCR reactions were set up in a total volume of 25 μl containing 12.5 μl of 2 × Universal Master Mix, 250 nM of each primer, and 10 ng of total RNA (as hexamer-primed single-stranded cDNA). The mixtures were prepared in 96-well optical microtiter plates and amplified on the ABI7900HT Sequence Detection System using the following cycling parameters: 2 min at 50 °C, 10 min at 95 °C, and 40 alternate cycles of 15 s at 95 °C and 60 s at 60 °C. Each sample cDNA was tested in triplicate. SDS 2.4 software was used for analysis. Human RPL13A primers were used as endogenous control for normalization of signals.
Gene expression analysis by microarray hybridization
Total RNA from experimental samples was isolated using the RNeasy maxi kit (Qiagen), incorporating on-column DNase treatment using the RNase-Free DNase Set (Qiagen). A reference standard RNA for use in two-color oligo arrays was prepared as described previously (13). Total RNA was amplified one round using the Ambion MessageAmp aRNA Kit (Ambion Inc., Austin, TX). Probe labeling and hybridization was performed following the Agilent suggested protocols and fluorescent array images were collected using the Agilent DNA microarray scanner G2505C. Agilent Feature Extraction software version 10.7.3.1 was used to grid and extract the data using the GE2_105_Jan09 protocol with default settings. Data was loess normalized within arrays and quantile normalized between arrays in R using the Limma Bioconductor package. Microarray data are deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE92853. The data was reduced to unique genes using the probe with the highest average signal intensity and filtered to exclude probes with average signal intensity less than 300. The Statistical Analysis of Microarray (SAM) program (2) was used to analyze expression differences between groups using unpaired, two-sample t tests and controlled for multiple testing by estimation of q-values using the false discovery rate (FDR) method. Genes up and down-regulated with q-value <10%, >= 3-fold were considered significant and used for enrichment analysis.
We created a quiescence signature using data generated by Lemons et al (26). Raw Agilent gene expression data was downloaded from the GEO data repository accession GSE42612, and normalized as described above, then analyzed by applying a 1-sample t-test comparing 7d+14d contact inhibited vs. proliferating human neonatal dermal fibroblasts. Figure 1f shows the top 20 genes up and down-regulated dermal fibroblast quiescence genes in comparison to quiescent gene expression in prostate fibroblasts. The GSEA analyses (Figure 1g) uses the up-regulated signature, 266 genes up-regulated with q-value <10%, >= 3-fold. Pathway analysis of both quiescent datasets was performed using the Gene Ontology pathway annotations and the DAVID Functional Annotation Bioinformatics Microarray Analysis tool (https://david.ncifcrf.gov/). A summary of non-redundant pathways with modified Fisher's exact test (EASE score) < 0.05 are shown.
Figure 1. Cell proliferation and quiescence in prostate epithelium and stroma.
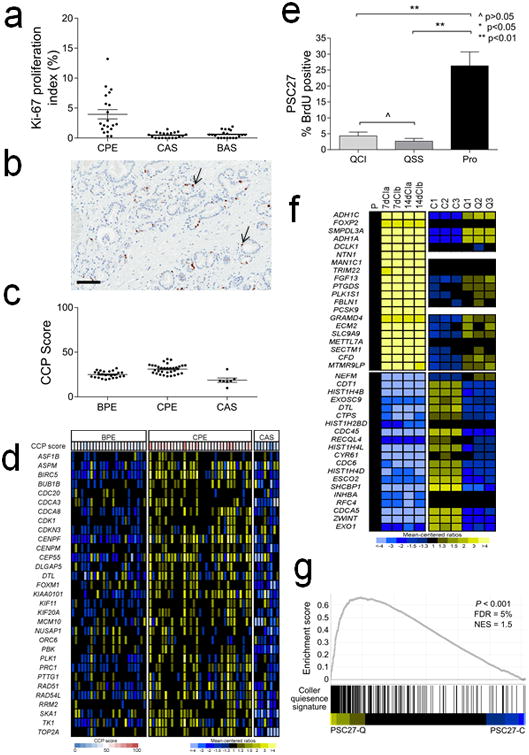
a. Ki-67 immunohistochemistry of prostate carcinoma cells (CPE), cancer adjacent stromal cells (CAS) and stromal cells adjacent to benign glands (BAS). The mean (±SEM) Ki67-index (%) for CPE, CAS and BAS was 4.0 ± 0.8% (±SEM), 0.5 ± 0.1% and 0.6 ± 0.1%, respectively. The differences were statistically different by Student's t test for CPE vs. CAS and CPE vs. BAS.
b. Image of Ki-67 immunohistochemistry. Brown chromogenic nuclear staining denotes a Ki-67 positive cell (arrow). Black line = 100 μM.
c. Cell cycle proliferation (CCP) score calculated from microarray-based quantitation of gene expession from microdissected benign prostate epithelium (BPE), prostate carcinoma (CPE) and cancer adjacent stroma (CAS).
d. Heatmap of genes comprising the cell cycle progression (CCP) score. Rows represent CCP genes and columns are tissue samples from different patients.
e. Quantitation of bromodeoxyuridine (BrdU) values from PSC27 fibroblasts proliferating (Pro) or growth arrested by contact inhibition (QCI) or serum starvation (QSS).
f. Heatmap of gene epression comparing a gene set previously shown to associate with cellular quiescence (left panels) with PSC27 prostate fibroblast quiescence (right panels). P, proliferating cells; 7dCI, cells 7 days after contact growth inhibition; 14dCI, cells 14 days after contact growth inhibition (from Lemons et al (26)). C1,C2,C3, biological replicates of proliferating PSC27 prostate fibroblasts; Q1,Q2, Q3, biological replicates of quiescent PSC27 prostate fibroblasts.
g. Gene set enrichment analysis comparing quiescent PSC27 prostate fibroblast gene expression with the quiescent fibroblast gene expression profile as determined by Lemons et al. (26).
The DNA damage signature used in the GSEA analysis of radiation-treated quiescent cells (Figure 2d) is defined as 204 genes up-regulated with q-value <0.01% >= 3-fold by 2-sample t-test comparing control vs. DNA damaging agents from Sun et al (27).
Figure 2. Effects of DNA damage on quiescent prostate fibroblast gene expression, a.

Cellular DNA damage response (DDR) foci were determined by counting H2AX foci in PSC27 prostate fibroblasts that were proliferating (Pro) or quiescent by serum starvation (QSS) or quiescent by contact inhibition (QCI).
b. Immunofluorescence detection of H2AX foci (pink; arrow) in PSC27 prostate fibroblasts. P-CON, proliferating cells sham irradiated; P-RAD, proliferating cells following irradiation; Q-SS-RAD, serum-starved quiescent cells following irradiation; Q-CI-RAD, contact inhibited quiescent cells following irradiation. Nuclei were counterstained with DAPI (blue).
c. Gene expression profiles of quiescent PSC27 prostate fibroblasts before and after exposure to ionizing radiation. Shown are heatmaps of the subset of genes altered by 3-fold or greater with an expanded view of a subset of transcripts encoding secreted proteins.
d. Gene set enrichment analysis comparing transcript alterations in irradiated quiescent PSC27 prostate fibroblasts to previously reported gene expression alterations in proliferating fibroblasts following DNA damage(27).
e. Transcript quantitation by qRT-PCR of gene expression changes following ionizing radiation. C, proliferating PSC27 cells sham irradiated; Q, quiescent PSC27 cells sham irradiated; Q+R, quiescent PSC27 irradiated; R, proliferating PSC27 cells irradiated; Q+M, quiescent PSC27 cells treated with mitoxantrone; M, proliferating PSC27 cells treated with mitoxantrone.
Results
A gene expression program associated with prostate fibroblast quiescence
Previous studies have demonstrated that benign proliferating mesenchymal cells comprising the prostate stroma sustain DNA damage following exposure to systemic genotoxic cancer therapeutics and respond with a robust DNA damage secretory program (13, 23). To determine whether stromal cells in the prostate microenvironment are proliferative or quiescent in vivo, we used immunohistochemistry to quantitate the percentage of Ki-67 positive cells in the prostate gland in different cell compartments including prostate cancer epithelium (CPE), stroma adjacent to cancer (CAS) and stroma adjacent to benign epithelium (BAS). Overall, proliferation rates were extremely low in all compartments. Compared to the average Ki-67 index of 4% in carcinoma cells (4.0 ± 0.8%; ±SEM), the Ki-67 index the other compartments was significantly lower with only rare positive cells identified:0.5 ± 0.1% in BAS (p<0.01, Student's t-test) and 0.6 ± 0.1%in CAS (p<0.01) (Figure 1a,b). We also calculated a cell cycle progression (CCP) score for cancer epithelium and stroma using a set of 31 genes previously shown to associate with adverse prostate cancer outcomes (28). The mean CCP scores for proliferating PC3 and LNCaP prostate cancer cell lines in vitro in full growth medium were 79 and 76, respectively. The mean CCP score for proliferating PSC27 fibroblasts in full growth medium was 60 and for quiescent, G0, PSC27 cells in growth medium devoid of serum the CCP score was 31. We calculated CCP scores from transcript profiles of CPE (n = 33), BPE (n = 24) and CAS (n = 7) microdissected from frozen radical prostatectomy specimens. In BPE and CPE the scores were 25 (range 19 – 32) and 31 (range 23 – 42), respectively, whereas the score for CAS was 19 (range 10 – 31) (Figure 1c,d). These data indicate that cells comprising the prostate stroma are generally quiescent, even in the context of adjacent cancerous epithelium.
To determine the effects of genotoxic exposures on non-dividing cells, we established cell quiescence using two strategies to reversibly-arrest cellular proliferation. We placed primary PSC27 prostate fibroblasts in culture conditions with serum-free growth medium deprived of mitogens, hereafter designated quiescence by serum-starvation (PSC27-QSS), or allowed PSC27 fibroblasts to grow to a high density, hereafter designated quiescence by contact inhibition (PSC27-QCI). We confirmed that that >80% of PSC27-QSS and PSC27-QCI cells were in a G0-G1 cell cycle stage by flow cytometry. Whereas non-confluent proliferating PSC27 cells growing in medium supplemented with 10% FBS (PSC27-PRO) had a cell cycle distribution of 53% G0/G1, 24% S and 22% G2, the phase distributions of PSC27-QSS were 82% G0/G1, 6% S, 10% G2 and PSC27-QCI were 88% G0/G1, 2% S, 10% G2, respectively. We confirmed the low proliferative rate of PSC27-QSS and PSC27-QCI cells by treating cultures with bromodeoxyuridine (BrdU) and assessing the percentage of BrdU positive cells which ranged from 26% of PSC27 cells grown in full medium to 5% of PSC27-QCI and 4% of PSC27-QSS (p<0.01) (Figure 1e).
We next confirmed that the gene expression program in the growth-arrested quiescent PSC27 prostate fibroblast population was concordant with previously reported assessments of gene expression in quiescence (26). We quantitated transcript levels in growth-arrested cells using genome-wide transcript microarrays. Compared to proliferating PSC27 cells, 108 transcripts were increased and 203 transcripts were decreased by 3-fold or greater (q<10%). Gene set enrichment analyses confirmed a significant enrichment of quiescence-altered transcripts between the PSC27 fibroblasts and a report by Lemons et al defining transcriptional alterations that accompany cellular quiescence (26, 29) (Figure 1f,g). Gene Set Enrichment Analyses determined that genes comprising cell-cell signaling and cell communication were significantly enhanced in quiescent cells whereas genes involved in mitotic cell cycle and cell proliferation were reduced (Supplementary Figure 1).
DNA damage in quiescent fibroblasts activates a gene expression program that encodes secreted proteins involved in wound repair, inflammatory responses and tumor growth
To ascertain differences in DNA damage sustained by proliferating fibroblasts and quiescent fibroblasts, we treated prostate fibroblast cell cultures with 10Gy ionizing radiation given in a single fraction, and 12 hours later measured DNA damage by quantitating γH2AX foci by immunofluorescence. Compared to untreated control cells, untreated PSC27-QSS and PSC27-QCI cells had no significant differences in the percentage of γH2AX foci, indicating that induced cellular quiescence is not associated with measurable DNA damage by this assay (Figure 2a). In contrast, treatment with ionizing radiation resulted in readily detectable γH2AX foci with significantly increased foci numbers in each treated cell population compared to controls (p<0.01) (Figure 2b). There were no significant differences in the number of γH2AX foci between irradiated PSC27-PRO, PSC27-QSS or PSC27-QCI cells (Figure 2a).
Having ascertained that quiescent fibroblasts respond to DNA damage by phosphorylating H2AX, indicating that DNA damage checkpoint kinases are activated, we next sought to determine if components of the downstream gene expression program induced by DNA damage are also activated. We quantitated transcript levels in quiescent PSC27 cells 7 days after exposure to 10Gy radiation using genome wide transcript microarrays. Compared to sham-treatment, radiation treatment increased the expression of 548 genes and decreased the expression of 207 genes by 3-fold or greater (q<0.01%) of which 127 of the genes with increased expression and 60 of the genes with decreased expression encode secreted or extracellular proteins (Figure 2c). We have previously reported that DNA damage induces a spectrum of growth factors, cytokines and proteases termed the DNA damage secretory program (DDSP) (13). Collectively, GSEA determined that transcripts comprising the DDSP were significantly enriched in the quiescent PSC27 fibroblasts following radiation (Figure 2d).
Quiescent PSC27 fibroblasts treated with the genotoxic chemotherapeutic agent mitoxantrone also responded by increasing the expression of DDSP components (Figure 2e). Overall, the diversity of genes with altered expression following genotoxic damage in proliferating versus quiescent fibroblasts was quite similar, though the magnitude of transcript upregulation following DNA damaging exposure was less in quiescent compared to proliferating cells (Figure 2e).
The quiescent cell DDSP is augmented by subsequent cell division
We determined that quiescent cells can activate a robust transcriptional response following genotoxic damage (Figure 2c), but the magnitude of the response was not as substantial as that when proliferating cells were exposed to genotoxic stress. In certain circumstances, the quiescent state is reversible, for example by exposure to endocrine or paracrine mitogens, inflammatory mediators, or reprogramming cues, and G0 cells can be induced to re-enter the cell cycle. To determine if cells damaged in G0, and then allowed to proliferate would further augment a secretory damage response, we treated quiescent PSC27 cells (PSC27-QCI) with radiation or mitoxantrone, and replated them in subconfluent conditions which reduced contact-inhibited growth suppression and allowed for the resumption of proliferation. Three days after replating, cells were harvested for gene expression measurements. For most DDSP-associated transcripts, the levels were significantly greater in the quiescent cells allowed to proliferate compared to cells maintained in a quiescent state (Figure 3), though the magnitude of the DDSP gene expression was still less than that produced by genotoxic exposures to proliferating cells. For IL8, a well-characterized prostate fibroblast DDSP factor, radiation exposure increased IL8 transcripts 3-fold over quiescent sham-treated PSC27 cells (p < 0.001) and 5-fold after cells were allowed to resume proliferation (p < 0.0001). In comparison, radiation treatment of proliferating PSC27 cells increased IL8 expression by 7-fold compared to sham-treated PSC27 cells (p < 0.001) and 4-fold over radiated PSC27 cells in a quiescent cell state (p < 0.001).
Figure 3. Effects of cell proliferation on the quiescent cell DNA damage secretory program.
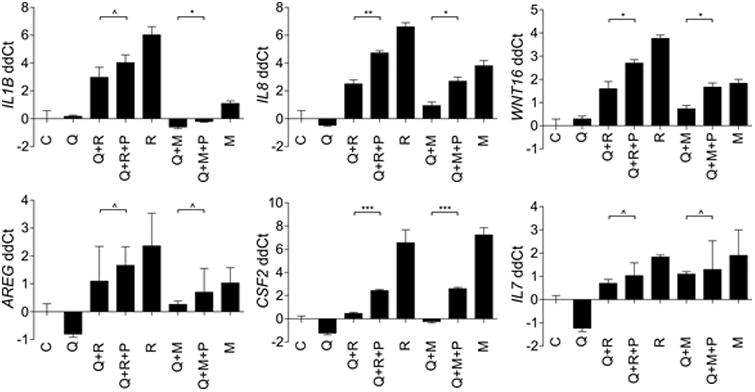
qRT-PCR measurements of gene expression alterations in quiescent PSC27 prostate fibroblasts before and after resumption of proliferation. C, proliferating PSC27 cells sham irradiated; Q, quiescent PSC27 cells sham irradiated; Q+R, quiescent PSC27 irradiated; Q+R+P, quiescent PSC27 prostate fibroblasts were irradiated, replated to allow proliferation for 3 days, and harvested for analysis; R, proliferating PSC27 cells irradiated; Q+M, quiescent PSC27 cells treated with mitoxantrone; M, proliferating PSC27 cells treated with mitoxantrone; Q+M+P, quiescent PSC27 prostate fibroblasts were treated with mitoxantrone, re-plated to allow proliferation for 3 days, and harvested for analysis
DNA damage in vivo induces the expression of DDSP components in prostate stroma
To assess the damage responses of benign cells comprising the tumor microenvironment, we examined tissues collected before and after chemotherapy exposure in men with aggressive localized prostate cancer enrolled on a clinical trial of neoadjuvant chemotherapy consisting of four cycles of the genotoxic drug mitoxantrone (MIT) and the microtubule poison docetaxel (DOC)(30, 31). We have previously shown that cells in the prostate tumor microenvironment exhibit evidence of DNA damage following chemotherapy, as determined by histone H2AX phosphorylation on Ser139 (γ-H2AX)(13).
We used laser-capture microdissection (LCM) to isolate stroma from transrectal ultrasound-guided prostate biopsies prior to chemotherapy treatment (n = 10 patients) and from radical prostatectomy tissue from the same patients after chemotherapy exposure. We quantitated transcripts by microarray hybridization and identified 65 genes encoding extracellular proteins with increased transcript levels of 1.5-fold or greater (p < 0.05) after chemotherapy exposure (Figure 4a). Genes with the most substantial induction of expression encode proteins such as WNT16, IL6, and EGF with known paracrine roles in promoting adverse tumor phenotypes (13). We further confirmed these findings using qRT-PCR to quantitate transcript levels of representative DDSP genes after and before chemotherapy and measured 2.7-fold increases in IL1β(p<0.01), 5.7-fold increases in MMP3, 4.3-fold increases in IL6 (p=0.01), and 4.5-fold increases in IL8 (p<0.01) (Figure 4b-e).
Figure 4. Gene expression alterations in quiescent prostate stroma in vivo following genotoxic chemotherapy.
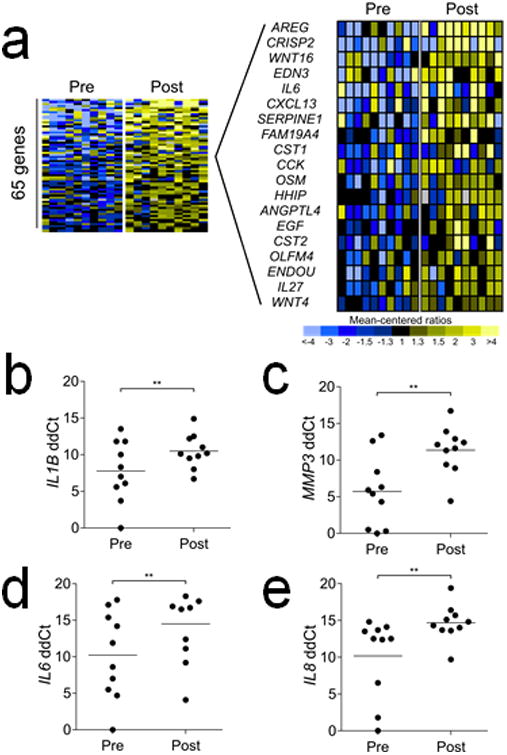
a Quantitation of gene expression in microdissected prostate stroma by microarray hybridization before (Pre) and after (Post) exposure to mitoxantrone and docetaxel (≥1.5-fold increase with p<0.05). Expanded heatmap shows a subset of the transcripts encoding extracellular proteins.
b-e. Quantitation of gene expression by qRT-PCR in microdissected prostate stroma before and after chemotherapy.
The DDSP from quiescent cells promotes tumor cell proliferation, invasion, and resistance to therapy
Previous studies have demonstrated that the SASP and DDSP resulting from damage to proliferating fibroblasts can promote adverse cancer cell phenotypes including enhanced cell proliferation, cell invasion, and resistance to cytotoxic chemotherapy (13, 32). We next sought to determine if the attenuated DDSP from quiescent fibroblasts, which more accurately reflect the proliferative state of tissue fibroblasts in vivo, could also influence tumor cell behavior. We collected conditioned growth medium (CM) from PSC27 cells that were: proliferating (PSC27-Pro), exposed to 10 Gy radiation while proliferating (PSC27-Rad), quiescent by contact inhibition (PSC27-Q) or exposed to 10 Gy radiation while quiescent (PSC27-QRad). Compared to CM from proliferating PSC27 cells, CM from quiescent PSC27 cells had no effect on tumor cell proliferation or invasion. As expected, PSC27-Rad CM significantly increased the proliferation of 5 different prostate epithelial cell lines and enhanced the invasion of cancer cells through a modified basement membrane (Figure 5a,b). For example, exposure of PC3 cells to PSC27-Rad CM increased the number of tumor cells by 2-fold compared to control medium after 5 days of culture (p<0.01) and increased the percentage of invasive tumor cells from 15% to 40% (p<0.001). CM from irradiated quiescent PSC27 cells also significantly increased the proliferation of each prostate cancer cell line tested and promoted tumor cell invasion, though the effects on these parameters was slightly less than that induced by CM from irradiated proliferating PSC27 cells (Figure 5b): compared to PSC27-Pro CM, exposure to PSC27-QRad CM increased PC3 tumor cell numbers from 40,000 to 80,000 after 5 days in culture and the percentage of invasive cells increased from 12% to 30% (Figure 5b).
Figure 5. Effects of paracrine factors from damaged quiescent fibroblasts on tumor cell proliferation, invasion and therapy resistance.
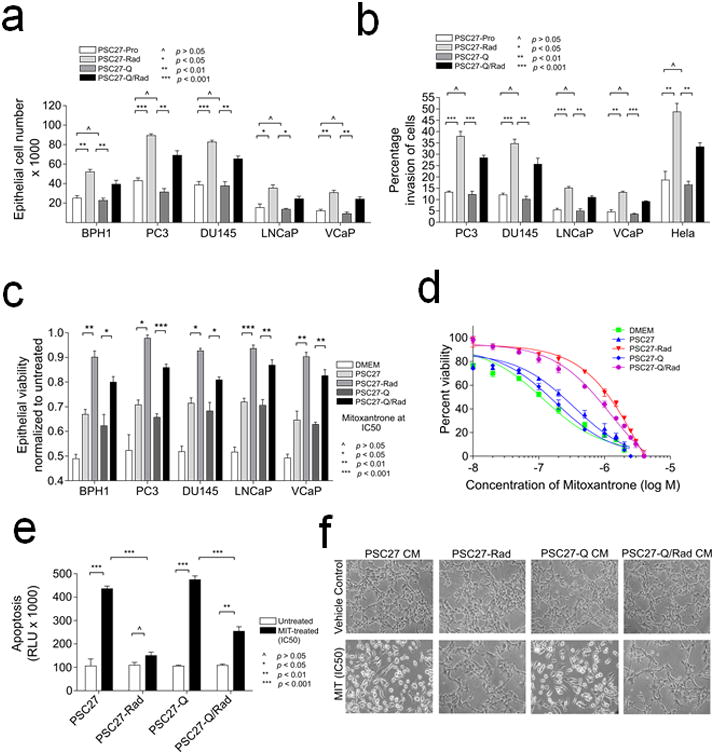
a Prostate cancer cell proliferation following exposure to conditioned medium from proliferating sham-treated (PSC27-Pro), irradiated (PSC27-Rad), quiescent (PSC27-Q), or irradiated quiescent (PSC27-QRad) fibroblast cells.
b. Assessments of cancer cell invasion following exposure to conditioned medium from proliferating sham-treated (PSC27-Pro), irradiated (PSC27-Rad), quiescent (PSC27-Q), or irradiated quiescent (PSC27-QRad) fibroblast cells.
c. Assessment of prostate cancer cell viability following exposure to IC50 concentrations of mitoxantrone in the context of conditioned medium from sham-treated proliferating fibroblasts (PSC27), irradiated proliferating fibroblasts (PSC27-Rad), quiescent fibroblasts (PSC27-Q), or irradiated quiescent fibroblasts (PSC27-QRad).
d. Assessments of PC3 prostate cancer cell viability across a range of mitoxantrone concentrations in the context of concurrent exposure to conditioned medium from sham-treated proliferating fibroblasts (PSC27), irradiated proliferating fibroblasts (PSC27-Rad), quiescent fibroblasts (PSC27-Q), or irradiated quiescent fibroblasts (PSC27-QRad). Cell viability was determined 3 days after mitoxantrone exposure.
e. Assessments of PC3 prostate cancer cell apoptosis following exposure to mitoxantrone in the context of conditioned medium from sham-treated proliferating fibroblasts (PSC27), irradiated proliferating fibroblasts (PSC27-Rad), quiescent fibroblasts (PSC27-Q), or irradiated quiescent fibroblasts (PSC27-QRad). Caspase 3 and 7 activities were measured (Glo assay of apoptosis) 24 h post exposure of PC3 cells to IC50 of mitoxantrone.
f. Bright field microscopy images of PC3 cells photographed 24 h post exposure to IC50 concentrations of mitoxantrone in the context of conditioned medium from sham-treated proliferating fibroblasts (PSC27-CM), irradiated proliferating fibroblasts (PSC27-Rad), quiescent fibroblasts (PSC27-Q CM), or irradiated quiescent fibroblasts (PSC27-QRad CM). Highly refractile cells in PSC27-CM and PSC27-Q CM conditions are indicative of apoptosis.
We have previously shown that the DDSP from proliferating fibroblasts exposed to genotoxic therapeutics can promote the resistance of prostate cancer to the effects of chemotherapy. To determine if the DDSP from quiescent cells was also sufficient to enhance chemotherapy resistance, we exposed BPH1, PC3, DU145, LNCaP and VCaP prostate cells to IC50 concentrations of mitoxantrone, which inhibits type II topoisomerase resulting in cell death by disrupting DNA synthesis and repair. After 3 days of MIT treatment, tumor cells exposed to CM from PSC27-Rad consistently demonstrated significant attenuation of chemotherapy-induced cytotoxicity across a range of MIT concentrations (p<0.05) (Figure 5c). The exposure of prostate cancer cells to CM from irradiated quiescent fibroblasts also significantly improved cell viability after MIT exposure (Figure 5c,d). For example, the percentage of surviving VCaP cells after MIT treatment increased from 63% to 83% and the percentage of surviving PC3 cells increased from 66 to 86% in a growth environment containing PSC27-QRad CM (p<0.001)(Figure 5c). To determine if the DDSP influenced tumor cell growth rates versus cell death, we measured PC3 cell apoptosis 24 hours after exposure to MIT and determined that the DDSP from quescent PSC27 cells significantly reduced MIT-induced PC3 apoptosis by 2-fold (p<0.01)(Figure 5e,f).
Discussion
The tissue microenvironments within which tumor cells exist profoundly influence a range of malignant phenotypes that include proliferation, migration, invasion, and responses to cytostatic and cytotoxic drugs. Of importance, tissue microenvironments are not static, but rather comprise a dynamic interactive system that responds to the gradual and progressive processes linked to aging, as well as those punctuated events produced by acute tissue damage including genotoxic cancer therapeutics. Molecular events associated with cellular aging processess have now been characterized in mechanistic detail. A consequence of cellular aging is the irreversible arrest of cell growth brought about DNA damage signals that culminate in the activation of cell cycle regulators such as p16 that contribute to a senescence phenotype. The physiological state of cellular senescence is accompanied by the induction of a gene expression program that comprises a spectrum of secreted growth factors and cytokines that regulate inflammatory responses and tissue repair processes. Noteably, components of this senescence-associated secretory phenotype promote tumor cell proliferation and invasion (13, 33). Though the overall burden of senescent cells in the aging host is low, they have been shown to contribute to aging–related pathologies and are hypothesized to influence the development and progression of carcinomas(14, 17).
Though DNA damage occurs contininuously at low levels due to internal metabolic processes and external exposures such as environmental radiation, cancer therapeutics have the potential to acutely and profoundly increase the levels of DNA damage far beyond the exposures a typical individual would experience during a lifetime. Such acute exposures produced by genotoxic drugs and radiotherapy overwhelm cellular repair processes and consequently result in the death of neoplastic cells. Since most therapeutics lack precise selectivity toward malignant cells, benign cells are also exposed to these insults and respond to these stresses by engaging repair processes that also include a secretory program(6, 34). Previous studies have characterized secretory damage responses in proliferating cells and the results of experiments comprising this study demonstrate that secretory responses accompany genotoxic insults in nonproliferating quiescent cells in vitro and in vivo.
The DNA damage secretory program is comprised of a complex amalgam of proteases, growth factors and cytokines that have the potential to influence different cell types within a tissue or tumor microenvironment:IL6, IL8, and IL27regulate inflammatory cell activity, AREG and EGF promote epithelial cell proliferation, MMPs modify structural extracellular matrix proteins and WNT family members influence the functions of several different mesenchymal and epithelial cell types. In addition to their roles in maintaining tissue homeostasis via remodeling and repair, these and other individual DDSP components can promote adverse tumor cell phenotypes that include proliferation, invasion, and resistance to chemotherapy-induced cell death(6, 36).
The complexity and redundancy of the DDSP suggests that while targeting the paracrine interactions of individual DDSP components may have some beneficial effects in terms of augmenting cancer directed therapeutics, a more effective strategy may involve methods to eliminate senescent cells(17), or by inhibiting key upstream nodes that propogate the initiating DNA damage signal to downstream transcription factors that regulate the expression of DDSP mRNAs. A subset of the SASP and DDSP transcriptional programs are known to be directly regulated via NFkB and indirectly via GATA4, mTOR and MAPK (33-35). As the mTOR and MAP kinases have potent pharmacological inhibitors available, clinical studies combining mTOR or MAPK inhibition in conjuction with genotoxic chemotherapy or radiotherapy could be advanced to test the concept that inhibiting a treatment-induced microenvironment-derived secretory program would augment the effectiveness of conventional cancer therapeutics.
Supplementary Material
Acknowledgments
We thank the patients and their families for their altruistic participation in this study. We thank Dr. Thomasz Beer and Celestia Higano for the development and conduct of the neoadjuvant chemotherapy clinical trial. This study was supported by NIH grants to the Fred Hutchinson Cancer Research Center P30CA015704, U01CA164188, R01CA165573, the Pacific Northwest Prostate Cancer SPORE CA097186, awards from the Department of Defense PC131820 and awards from the Canary Foundation and Prostate Cancer Foundation.
Footnotes
The authors declare they have no conflicts of interest and financial disclosures that are relevant to this publication.
References
- 1.Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet. 2009;25:30–8. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Moreno M. When neighbourhood matters: tumour microenvironment. Clin Transl Oncol. 2009;11:70–4. doi: 10.1007/s12094-009-0316-z. [DOI] [PubMed] [Google Scholar]
- 3.Hu M, Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008;18:27–34. doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi-Frias D, Vakar-Lopez F, Coleman IM, Plymate SR, Reed MJ, Nelson PS. The effects of aging on the molecular and cellular composition of the prostate microenvironment. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, et al. Leukocyte Complexity Predicts Breast Cancer Survival and Functionally Regulates Response to Chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Y, Nelson PS. Molecular pathways: involving microenvironment damage responses in cancer therapy resistance. Clin Cancer Res. 2012;18:4019–25. doi: 10.1158/1078-0432.CCR-11-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 8.Rodier F, Munoz DP, Teachenor R, Chu V, Le O, Bhaumik D, et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci. 2011;124:68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Childs BG, Baker DJ, Kirkland JL, Campisi J, van Deursen JM. Senescence and apoptosis: dueling or complementary cell fates? EMBO Rep. 2014;15:1139–53. doi: 10.15252/embr.201439245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–66. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodier F, Coppe JP, Patil CK, Hoeijmakers WAM, Munoz D, Raza SR, et al. Persistent DNA damage signaling triggers senescence-associated inflammatory cytokine secretion. Nature Cell Biol. 2009;11:973–9. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Campisi J, Higano C, Beer TM, Porter P, Coleman I, et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18:1359–68. doi: 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–6. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen DH, Oketch-Rabah HA, Illa-Bochaca I, Geyer FC, Reis-Filho JS, Mao JH, et al. Radiation acts on the microenvironment to affect breast carcinogenesis by distinct mechanisms that decrease cancer latency and affect tumor type. Cancer Cell. 2011;19:640–51. doi: 10.1016/j.ccr.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2010;29:273–83. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang J, Wang Y, Shao L, Laberge RM, Demaria M, Campisi J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–68. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–31. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 20.Herranz N, Gallage S, Mellone M, Wuestefeld T, Klotz S, Hanley CJ, et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol. 2015;17:1205–17. doi: 10.1038/ncb3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambrosio S, Di Palo G, Napolitano G, Amente S, Dellino GI, Faretta M, et al. Cell cycle-dependent resolution of DNA double-strand breaks. Oncotarget. 2015 doi: 10.18632/oncotarget.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bavik C, Coleman I, Dean JP, Knudsen B, Plymate S, Nelson PS. The Gene Expression Program of Prostate Fibroblast Senescence Modulates Neoplastic Epithelial Cell Proliferation through Paracrine Mechanisms. Cancer Res. 2006;66:794–802. doi: 10.1158/0008-5472.CAN-05-1716. [DOI] [PubMed] [Google Scholar]
- 24.Hayward SW, Wang Y, Cao M, Hom YK, Zhang B, Grossfeld GD, et al. Malignant transformation in a nontumorigenic human prostatic epithelial cell line. Cancer Res. 2001;61:8135–42. [PubMed] [Google Scholar]
- 25.Tretiakova MS, Wei W, Boyer HD, Newcomb LF, Hawley S, Auman H, et al. Prognostic value of Ki67 in localized prostate carcinoma: a multi-institutional study of >1000 prostatectomies. Prostate Cancer Prostatic Dis. 2016;19:264–70. doi: 10.1038/pcan.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemons JM, Feng XJ, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Campisi J, Higano C, Beer TM, Porter P, Coleman I, et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012 doi: 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–55. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garzotto M, Myrthue A, Higano CS, Beer TM. Neoadjuvant mitoxantrone and docetaxel for high-risk localized prostate cancer. Urol Oncol. 2006;24:254–9. doi: 10.1016/j.urolonc.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 31.Beer TM, Garzotto M, Lowe BA, Ellis WJ, Montalto MA, Lange PH, et al. Phase I study of weekly mitoxantrone and docetaxel before prostatectomy in patients with high-risk localized prostate cancer. Clin Cancer Res. 2004;10:1306–11. doi: 10.1158/1078-0432.ccr-1021-03. [DOI] [PubMed] [Google Scholar]
- 32.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–7. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laberge RM, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou L, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17:1049–61. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30(8):1536–46. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T, Notta F, Navab R, Joseph J, Ibrahimov E, Xu J, et al. Senescent Carcinoma-associated Fibroblasts Upregulate IL8 to Enhance Pro-metastatic Phenotypes. Mol Cancer Res. 2016;15(1):1–12. doi: 10.1158/1541-7786.MCR-16-0192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


