Abstract
The spatial nuclear organization of regulatory proteins often reflects their functional state. PSF, a factor essential for pre-mRNA splicing, is visualized by the B92 mAb as discrete nuclear foci, which disappeared during apoptosis. Because this mode of cell death entails protein degradation, it was considered that PSF, which like other splicing factors is sensitive to proteolysis, might be degraded. Nonetheless, during the apoptotic process, PSF remained intact and was N-terminally hyperphosphorylated on serine and threonine residues. Retarded gel migration profiles suggested differential phosphorylation of the molecule in mitosis vs. apoptosis and under-phosphorylation during blockage of cells at G1/S. Experiments with the use of recombinant GFP-tagged PSF provided evidence that in the course of apoptosis the antigenic epitopes of PSF are masked and that PSF reorganizes into globular nuclear structures. In apoptotic cells, PSF dissociated from PTB and bound new partners, including the U1–70K and SR proteins and therefore may acquire new functions.
INTRODUCTION
Splicing factors are found within the nucleus both in a diffuse form and in distinct domains termed interchromatin granules (IG) or “speckles” (Spector, 1993). These functional compartments are spatially dynamic with regard to movement and composition (Eils et al., 2000). Splicing factors within IGs are highly mobile and are constantly associating and dissociating from their compartments (Phair and Misteli, 2000). The mAb, B92, which recognizes the polypyrimidine tract binding protein (PTB)-associated splicing factor (PSF), produces typical specked nuclear pattern in immunostaining. These PSF speckles disappear during granulocytic differentiation (Lee et al., 1996; Shav-Tal et al., 2000). We recently showed that this disappearance is associated with partial degradation (Shav-Tal et al., 2000) and by the formation of alternative nuclear structures (Shav-Tal et al., 2001). The present study indicates that PSF speckles disappear also through a different mechanism, involving conformational changes that cause breakdown of PSF speckles and relocalization of the molecule in the nucleus. The functional nature of speckles is a matter of some controversy (for review, see Park and De Boni, 1999). These structures have been suggested to play a role in storage and recycling of splicing factors (Jimenez-Garcia and Spector, 1993), whereas a portion may act as reservoirs for the recruitment to active sites of transcription (Huang and Spector, 1996; Misteli et al., 1997; Zeng et al., 1997). This view does not predict a direct involvement of speckles in pre-mRNA splicing. On the other hand, it has been shown that nascent mRNA transcripts and transcription sites are closely associated with speckles, that transcription occurs at the surface of speckles (Clemson and Lawrence, 1996), and that microinjected pre-mRNAs have high affinity to speckles (Wang et al., 1991). It has thus been proposed that speckles can act coordinately in transcription and splicing and that pre-mRNA splicing can take place both at the periphery and inside speckles (Smith et al., 1999; Wei et al., 1999). A model combining both views suggests that splicing factors are recruited to transcription sites, whereas certain mRNAs are released from such sites and associate with splicing factor reservoirs (Melcak et al., 2000).
The traffic of splicing factors and their relocalization within the nucleus are thus of functional importance. This process appears to be controlled by protein phosphorylation (Misteli and Spector, 1997; Misteli, 1999); serine-arginine (SR) proteins, a major constituent of IGs (Mintz et al., 1999), are phosphoproteins containing a C-terminal serine-arginine rich RS domain and N-terminal RNA recognition motifs (RRMs; for review, see Cáceres and Krainer, 1997). Experiments monitoring GFP-tagged SF2/ASF SR protein in living cells have shown that phosphorylation controls the movement of SR proteins from speckles to sites of active transcription (Misteli et al., 1997, 1998). Similarly, in vitro and in vivo studies using several kinases indicate that phosphorylation of SR proteins releases them from IGs (for review, see Stojdl and Bell, 1999), whereas phosphatases have an opposite effect (Misteli and Spector, 1996). Therefore, it has been proposed (Cáceres et al., 1997; Misteli et al., 1998; Stojdl and Bell, 1999) that the recruitment of splicing factors from speckles occurs in two steps. First, SR proteins are physically released from speckles via RS domain phosphorylation. Subsequently, targeting to sites of transcription occurs via interactions with other proteins and association with pre-mRNA through their RRMs. Disassembly of speckles occurs also during mitosis (Spector et al., 1991), whereas Cajal bodies, nuclear lamins, nuclear pore complexes, and DNA replication sites remain intact (Gui et al., 1994). SR proteins are hyperphosphorylated during metaphase, and this opposes their retention in speckles (Gui et al., 1994).
Although PSF does not posses an RS domain, it shares some features with SR proteins. In the present study we show that hyperphosphorylation of PSF during apoptosis is associated with binding to new partner proteins, thus assigning a possible new function to this splicing factor.
MATERIALS AND METHODS
Bone Marrow Cells and Cell Lines
BM cells were obtained from 6-week-old male or female BALB/c mice (Harlan-Olac, Indianapolis, IN) and dissociated to single-cell suspensions in cold phosphate-buffered saline (PBS). HeLa and HL-60 cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and MPC-11 cells in RPMI medium supplemented with 10% FCS.
Protein Extraction, Immunoprecipitation, and Immunoblotting
Cell lines were lysed for 2 h on ice in 50 mM Tris-HCl, pH 8, containing 1% Nonidet P-40 (NP-40), 150 mM NaCl, 5 mM EDTA, 1 mM phenyl methylsulfonyl fluoride (PMSF), 3 μg/ml aprotinin, 20 μg/ml leupeptin, 10 mM iodoactetate, 1 mM sodium orthovanadate, and 10 mM sodium fluoride. BM cells were lysed by boiling immediately after addition of SDS-sample buffer (5% glycerol, 2% SDS, 62.5 mM Tris-HCl, pH 6.8, 2% 2-mercaptoethanol, 0.01% bromophenol blue). For immunoprecipitation, cell lysates were clarified by centrifugation at 15,000 × g for 10 min at 4°C. The resulting supernatants were precleared with protein A-Sepharose (Sigma, St. Louis, MO) and were immunoprecipitated at 4°C with the indicated antibody and protein A-Sepharose. Immunoprecipitates were washed (4 times) with cold SNNTE buffer (5% sucrose, 1% NP-40, 500 mM NaCl, 50 mM Tris, pH 7.4, 5 mM EDTA), boiled in SDS sample buffer, and analyzed by SDS-PAGE. Immunoblot analysis of tissue and cell lysates (50 μg/lane) was performed as described (Peled et al., 1991). Specific binding was detected with horseradish peroxidase (HRP)-coupled antibodies and enhanced chemiluminescence (ECL) reagents.
Cell-cycle Blockage and Apoptosis
MPC-11 cells were thymidine (G1/S)-blocked (5 mM, Sigma) for 24 h. The cells were then washed and incubated in medium without thymidine for 2 h before blocking at metaphase by 50 ng/ml nocodazole for 14 h. HeLa cells were thymidine-blocked (2 mM, 8 h) followed by nocodazole block (50 ng/ml, 14 h). Mitotic cells were obtained by mechanical shake-off, and purity (>90%) was checked by DNA staining. HL-60 cells were induced to undergo apoptosis with the use of 5 μg/ml actinomycin D for 4 h. Apoptotic HL-60 cells were separated from nonapoptotic cells by a Percoll density gradient as previously described (Martin et al., 1990). Presence of apoptotic cells (>50%) after 4 h of treatment was confirmed by DNA gel electrophoresis as in Park and Patek (1998), by staining with Hoechst nuclear staining for detection of nuclear condensation and fragmentation and by staining with Annexin-V-Alexa 568 (Boehringer Mannheim, Mannheim, Germany), which detects phosphatidylserine on the membranes of apoptotic cells
2-D Phosphoamino Acid Analysis and Phosphopeptide Mapping
Metabolic labeling with 32P-orthophosphate (250 μCi/ml; Amersham, Arlington Heights, IL) was performed in phosphate-free DMEM with untreated cells, mitotically arrested HeLa cells, and apoptotic HL-60 cells. PSF was immunoprecipitated from these cells, run on SDS-PAGE, and transferred to a PVDF membrane. Excision of the bands, 2-D phosphoamino acid analysis, and phosphopeptide mapping were performed as described (Van Der Geer and Hunter, 1994). For phosphopeptide mapping, phosphorylated PSF was digested by chymotrypsin and peptides were oxidized. Peptides were run in buffer, pH 4.72, and then in phosphochromatography buffer (Van Der Geer and Hunter, 1994). For phosphatase treatment, 40 μg of protein extract were incubated with 400 U of λ protein phosphatase (New England Biolabs, Beverly, MA) at 30°C for 1 h.
Antibodies
Secondary antibody used for Western blotting was goat anti-mouse HRP (Sigma). Fluorophore-labeled secondary antibody used was donkey anti-mouse FITC (Jackson, West Grove, PA). Anti-GFP mAb (JL-8) was purchased from Clontech (Palo Alto, CA). Anti-U1–70K antibody and anti-SR proteins antibody (Mab 104) were provided by Dr. Gil Ast (Tel Aviv University). Anti-PTB mAb was provided by Dr. David Helfman (Cold Spring Harbor). B92 mAb was prepared as previously described (Lee et al., 1996). Polyclonal anti-PSF 1121 antibody was prepared as previously described (Shav-Tal et al., 2000).
Immunofluorescence
Cells were fixed for 2 min in 4% paraformaldehyde with 0.5% Triton X-100 and for an additional 20 min in 4% paraformaldehyde. After washing and blocking in 5% BSA, cells were stained with the indicated antibody for 45 min and then with the appropriate fluorophore antibody for 45 min and counterstained with Hoechst. Immunofluorescence was determined in a Zeiss Axioplan microscope (Zeiss, Oberkochen, Germany).
Mouse PSF Cloning
A mouse fibroblast ZAP-Express library was divided in 20 pools and screened by PCR with the use of PSF-specific primers and vector-specific primers. Plaque lifts were prepared from positive pools containing large PSF clones and probed on filters (Schleicher & Schuell, Dassel, Germany). One large 1.7-kb clone containing the middle to C-terminal part of mouse PSF was isolated and sequenced. The N-terminal GC-rich portion of PSF was cloned with the use of RT-PCR with 3′ primers from published mouse PSF sequences and 5′ primers from mouse PSF-related sequences found in the EST database (mouse PSF sequence-GenBank accession number AY034062).
Transfections
HeLa cells were plated at 70% confluency in six-well plates and transfected with 6 μg of human GFP-PSF constructs with the use of the calcium phosphate transfection method (Graham and Eb, 1973). Cells were fixed and processed for immunofluorescence 24 h after transfection with the use of a Zeiss microscope and photographed by a CCD camera.
RESULTS
Nuclear Localization of Mouse and Human PSF: Reduced Detection by an mAb During Mitosis and Apoptosis
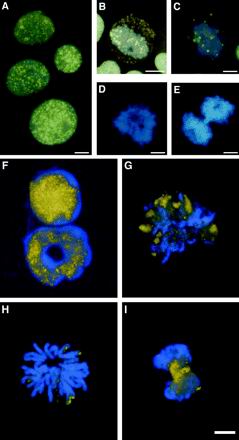
It is well documented that the speckled pattern, as visualized by use of antibodies to several splicing factors, is lost in dividing cells. Splicing factors remain intact and are diffusely dispersed throughout the cytoplasm but also concentrate in a few cytoplasmic foci (Spector et al., 1991 and references within). Speckles reassemble toward the end of mitosis. We examined the nuclear distribution of the essential splicing factor PSF during mitosis of human and mouse cells. During interphase PSF localized in nuclear speckles in immature human HL-60 myeloid leukemia cells (Figure 1A). PSF foci scattered throughout the cytoplasm in late prophase (Figure 1B), which is an intermediate stage during which transcription ceases (Gottesfeld and Forbes, 1997), the chromatin condenses, and the nuclear envelope breaks down (Newport and Spann, 1987). As mitosis proceeded, the PSF immunostaining diminished (Figure 1C) and completely disappeared during metaphase and anaphase (Figure 1, D and E). Western blot analysis clearly demonstrated that this disappearance was not due to protein degradation (Figure 2,A and B). Thus, although PSF remains intact in mitosis, it becomes inaccessible to the B92 antibody. In mitotic cells from mouse bone marrow, PSF-speckled staining diminished, yet in contrast to the findings with the human HL-60 cells, some staining was concentrated at the rim of the metaphase chromosomes (Figure 1, G and H). During anaphase PSF staining reappeared and speckles began to reassemble (Figure 1I). Differences between human and mouse in immunostaining with antibodies to PSF have been documented in our previous studies on maturing granulocytes from mouse bone marrow. The common speckled distribution observed in immature granulocytes disappears in mature and segmented granulocytes (Lee et al., 1996), although the protein remains intact (Shav-Tal et al., 2001). On the other hand, in human peripheral blood granulocytes immunostaining with anti-PSF antibodies produces a clear specked pattern (Shav-Tal and Zipori, unpublished results). These data suggested possible structural differences between the human and mouse proteins. The middle region of mouse PSF has previously been cloned (Chanas-Sacréet al., 1999; Shav-Tal et al., 2000), and its amino acid sequence is 98% identical to human PSF (Patton et al., 1993). However, the GC-rich N-terminus and the C-terminus remained unidentified. We obtained the entire sequence of mouse PSF mRNA by cloning from a mouse fibroblast library (Figure 3A). Mouse and human PSF cDNAs were found to be highly homologous (Figure 3B), and in total, mouse PSF protein is 8 amino acids shorter than human PSF. The C-terminal region containing the functional domains is almost identical in mouse and human, whereas in the N-terminus several differences are observed, although the structural elements (P and Q stretches) are conserved (Figure 3B). It has been suggested (Patton et al., 1993) that the N-terminal region of PSF, which has high proline and glutamine content, has the characteristics of domains involved in protein–protein interactions (Courey and Tjian, 1988; Tanese et al., 1991). Thus, the differences in sequences may lead to different interactions and to differences in localization.
Figure 1.
PSF localization during cell cycle in human leukemic cells and mouse hemopoietic cell. HL-60 cells were stained for immunofluorescence with the B92 antibody. (A) Interphase nuclei with PSF staining (B92 in yellow) on the background of Hoechst DNA stain (blue). (B) A nucleus at late prophase showing chromatin condensation, in which nuclear membrane breakdown has occurred and PSF speckles are found in the cytoplasm. (C) Nucleus in prometaphase; (D) metaphase; (E) anaphase. Bone marrow cells were stained for immunofluorescence with the B92 antibody. (F) Interphase nuclei of an immature cell and a mature granulocyte exhibit a speckled pattern. (G) Prometaphase; (H) metaphase; (I) anaphase. Bar, 5 μm. (Artificial coloration was achieved by computer coloring of overlapping black and white pictures of FITC staining and Hoechst staining taken from the same field).
Figure 2.
Phosphorylation of PSF during mitosis and apoptosis. HeLa cells were blocked at G1/S by a thymidine block followed by a nocodazole block causing cells to stop at metaphase. (A) Protein extracts were made from untreated and treated cells, and blots were probed with the B92 antibody. (B) Extracts were made from HeLa cells treated with nocodazole for the indicated time periods. (C) The cells were then released from the mitotic block, and extracts were made after a few hours. (D) Mouse MPC-11 cells underwent the same treatments. (E) Mitotically blocked HeLa protein extracts were treated with λ protein phosphatase at 30°C for 1 h. (F) HL-60 apoptotic cells were separated from nonapoptotic cells with the use of a Percoll gradient. The shift in the apoptotic cells was compared with mitotic cells. All the shifts observed were reproducible.
Figure 3.
Cloning of mouse PSF cDNA. (A) The cDNA sequence of mouse PSF was obtained with the use of library screening and RT-PCR. (B) Comparison of mouse and human amino acid sequences.
Lack of detection of PSF by the B92 antibody, such as that observed in mouse metaphase cells, was also evident and was a more striking occurrence in HL-60 cells undergoing spontaneous apoptosis (Figure 4, A and B). We considered that this could be due to complete degradation of the protein in apoptosis because of the high sensitivity of PSF to proteases. However, Western blotting indicated that PSF remains intact in apoptotic cells, whereas PTB is cleaved (Shav-Tal et al., 2000). Thus, both in apoptotic cells and in mitotic cells PSF speckles disappear; however, no detectable diffuse staining is observed, as previously described for other splicing factors (Spector et al., 1991). These data imply a possible change occurring in PSF that modifies the antigenic epitope recognized by the B92 antibody.
Figure 4.
PSF nuclear localization during apoptosis and mitosis. (A) Untreated HL-60 cells stained with the B92 antibody. (B) An apoptotic nucleus is seen by Hoechst DNA counterstaining. HeLa cells were transfected with GFP-PSF (C); DNA counterstain (D). Transfected cells were synchronized to metaphase by a nocodazole block (E); DNA counterstain (F). Apoptotic cells were identified by membranal Annexin V staining (G) and GFP-PSF was seen to localize in large aggregates (H); DNA counterstain (I). GFP-PSF was transfected into cells that were then blocked at G1/S (J) and double stained with the B92 antibody (K); DNA counterstain (L). Apoptotic cells from a culture transfected with GFP-PSF (M) were double-stained with B92 (N), DNA (O). Bar, 5 μm.
Overexpressed GFP-tagged PSF Localizes in Apoptotic Bodies Inaccessible to Anti-PSF Antibodies
Overexpression of tagged recombinant PSF could provide a tool to identify the protein, regardless of any conformational changes it may undergo during the cell cycle or in apoptosis. We therefore used human GFP-PSF (Dye and Patton, 2001) that localized in speckles in interphase nuclei and also produced a diffuse nucleoplasmic staining (Figure 4, C and D). In mitotic cells part of the GFP-PSF protein was concentrated in several rounded structures, whereas some was diffusely distributed throughout the cytoplasm (Figure 4, E and F). In apoptotic cells, identified by Annexin-V staining, GFP-PSF concentrated in several large round aggregates in each cell, and no diffuse staining was observed outside of these structures (Figure 4, G–I). In addition, we found in G1/S thymidine-blocked cells that cannot enter into mitosis, that GFP-PSF localized in stellate-like structures rather than in speckles (Figure 4J). To assess the availability of GFP-PSF structures to the B92 antibody, G1/S-blocked and apoptotic cells expressing GFP-PSF, were stained with this anti-PSF antibody (B92). In G1/S-blocked cells the B92 antibody stained the GFP-PSF structures and showed an image identical to that produced by GFP (Figure 4, J–L). Thus, in G1/S-blocked cells PSF retained the antigenic epitope recognized by the B92 antibody. On the other hand, in apoptotic cells, B92 did not stain the large GFP-PSF structures (Figure 4, M–O). It is thus evident that during apoptosis PSF is modified to the extent that it is totally unrecognized by the B92 antibody. This is not occurring in mitotic cells in which antigenicity is preserved.
It has been shown that phosphorylated SR proteins are associated with the U1-snRNP complex during apoptosis (Utz et al., 1998). We thus decided to check whether endogenous PSF might bind to different partners during apoptosis. Part of nuclear PSF is normally found in complex with the PTB protein (Patton et al., 1993). PTB is proteolytically cleaved during apoptosis (Shav-Tal et al., 2000) and indeed is not found in complex with PSF in these cells (Figure 5A). On the other hand, immunoprecipitation experiments showed that PSF is associated with the U1–70K protein and SR proteins in apoptotic cells (Figure 5, B–D). Thus, the masking of antigenic sites during apoptosis may be due to interactions with new proteins.
Figure 5.
Analysis of binding partners of PSF. HL-60 cells were incubated with actinomycin D for 5 h when more than 50% of the cells become apoptotic. Lysates from apoptotic (Apo) and nonapoptotic (−) cells were immunoprecipitated with an anti-PTB antibody and Western blotted with the B92 antibody (A), immunoprecipitaed with anti–U1–70K and blotted with B92 (B), immunoprecipitated with anti–U1–70K and blotted with anti-SR (C), or immunoprecipitated with the B92 antibody and Western blotted with anti-SR antibodies (D).
PSF Is Hyperphosphorylated during Apoptosis
One possible modification of PSF that may account for acquisition of new protein binding capabilities could be its degree of phosphorylation. Treatment of HL-60 cells with actinomycin D for >4 h leads to apoptosis (Martin et al., 1990). Apoptotic cells were identified and separated from nonapoptotic cells as described in MATERIALS AND METHODS (data not shown). We found that PSF from apoptotic protein cell extracts, presented retarded migration of PSF compared with cells possessing a normal nucleus (Figure 2F). Human HeLa and mouse plasmacytoma (MPC-11) cells were synchronized to G1/S and then blocked at M phase. Retarded migration of the PSF band was seen in these states. The 100-kDa PSF band from nocodazole-blocked HeLa cells (metaphase) migrated more slowly than that from untreated cells, and both of the above ran slower than PSF from thymidine-blocked cells (interphase; Figure 2A). The shift in mitotic cells was best observed after a 14-h block (Figure 2B). When the mitotic block was released, the PSF band was shifted back (Figure 2C). The same features were seen in the mouse MPC-11 cells (Figure 2D). Treatment of the mitotic protein extracts with Lambda phosphatase showed a faster migrating PSF band (Figure 2E), thus indicating that the change in PSF migration is probably due to phosphorylation and that PSF can be found in different phosphorylation states. Although we observed hyperphosphorylation in both apoptosis and mitosis, a comparison of the shifted bands implies different levels or sites of PSF phosphorylation in these two different cellular states.
Phosphoamino acid analysis of untreated cells vs. apoptotic or mitotic cells showed that in all cases the majority of phosphorylation on PSF was found on serine residues and to a lesser extent on threonine residues (Figure 6A). Phosphoamino acid analysis of the 68-kDa degradation product of PSF found in human cells showed high reduction in threonine phosphorylation, thus mapping the sites of threonine phosphorylation to the degradable part of PSF, i.e., the C terminus (Shav-Tal et al., 2000).
Figure 6.
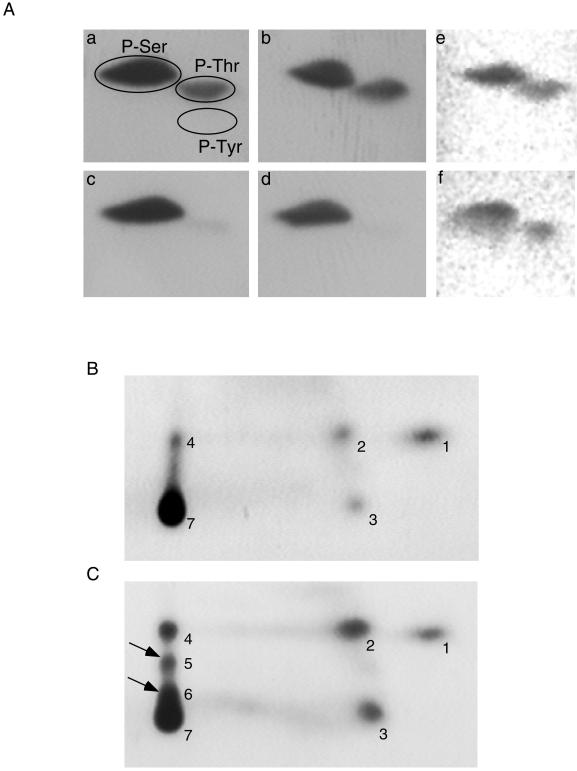
Phosphoamino acid analysis and phosphopeptide mapping of PSF. Phosphoamino acid analysis of PSF immunoprecipitated by B92 antibody from 32P metabolically labeled HeLa and HL-60 cells shows phosphorylation on serine and threonine residues (A): untreated HL-60 cells (a), apoptotic HL-60 cells (b). This analysis of the 68-kDa product of PSF shows only phosphoserine in untreated HL-60 cells (c) and apoptotic HL-60 cells (d). Phosphoamino acid analysis of untreated HeLa cells (e) and mitotic HeLa cells (f). Phosphorylated PSF was immunoprecipitated by B92 antibody, and PSF bands were digested with chymotrypsin and subjected to phosphopeptide mapping. Peptide mixture was loaded on the bottom left corner and the run by electrophoresis in pH 4.72 and then by ascending TLC. Untreated HL-60 cells (B). Apoptotic HL-60 cells (C). Arrowheads, additional phosphorylated peptides.
Because no major differences were observed between the different forms of PSF in phosphoamino acid analysis, phosphorylated PSF was subjected to phosphopeptide mapping. This was performed in HL-60 apoptotic cells only, because PSF in HeLa cells did not incorporate 32P efficiently. As can be seen by comparing Figure 6B to 6C, two additional peptides (arrows) appeared in apoptotic cells. These are hydrophobic and large peptides because they migrate only in the organic phase and are probably derived from the same peptide that is additionally phosphorylated in apoptotic cells, thus multiply phosphorylated in a cluster of sites. Because the chymotrypsin used to produce the peptides cannot digest the N-terminal, proline-rich core of PSF, we predict that the large, nonmigrating peptide (No. 7 in Figure 6B) is the N-terminus of PSF. The additional phosphorylated peptides (Nos. 5 and 6 in Figure 6C) resulting from this peptide may therefore be the cause of the changes leading to the lack of detection by our antibody.
To verify that the hyperphosphorylation of PSF takes place in the N-terminus, different GFP-PSF vectors were used in transfections. Figure 7C shows that both full-length GFP-PSF (amino acids 1–707) and endogenous PSF are detected by the B92 antibody in Western blots, and retarded migration of both proteins is observed in nocodazole-arrested cells (metaphase). Because phosphorylation is thought to control the localization of splicing factors in speckles, we checked whether the removal of RRM2 of PSF, which is responsible for PSF targeting to speckles (Dye and Patton, 2001; Figure 7, D and F), is the region hyperphosphorylated during mitosis. However, although GFP-PSF ΔRRM2 does not localize in speckles, it was also hyperphosphorylated during metaphase (Figure 7F). We then proceeded to check the behavior of the C-terminal half of PSF during mitosis. GFP-PSF 338–707, which can localize in the nucleus (Figure 7, G and H), did not show any shift in mitotic cells (Figure 7I), indicating that hyperphosphorylation occurs on the N-terminal half of PSF. In this case anti-GFP antibody was used for Western blotting because our antibody recognizes only the N-terminus of PSF.
Figure 7.
Retarded migration of GFP-PSF constructs. GFP-PSF was transfected into HeLa cells (A), DNA counterstain (B), and endogenic PSF and GFP-PSF were identified in untreated and in metaphase-blocked cells in Western blots with the use of the B92 antibody (C). GFP-PSF ΔRRM2 that does not localize in speckles was transfected into cells (D), DNA (E), and both PSFs were identified as above (F). GFP-PSF aa338–707 was transfected into cells (G), DNA (H), and extracts as above were immunoblotted with an anti-GFP antibody. Bar, 5 μm.
DISCUSSION
Apoptosis entails proteolysis of several regulatory molecules and subsequent protein degradation as part of the process that lends to cessation of cellular functions and cell death (Stroh and Schulze-Osthoff, 1998). Splicing factors are particularly sensitive to proteolytic degradation (La Branche et al., 1991; Casciola-Rosen et al., 1994; Waterhouse et al., 1996; Shav-Tal et al., 2000). In this context it is intriguing that PSF remains intact at a time point in the apoptotic process in which PTB, which is a binding partner to PSF, is cleaved (Shav-Tal et al., 2000). We show in the present study that the B92 mAb failed to immunostain apoptotic cells despite the fact that the relative amount of the PSF protein in the cells remained unchanged. One possible explanation of this phenomenon was that the protein is diffusely distributed within the cell and the local concentration is therefore below the detection power of the immunofluorescence method used. This seems to be the case with mitotic cells. Most of the endogenous PSF protein becomes undetectable with the exception of distinct structures, whereas overexpression of GFP-PSF allowed to visualize the protein both in a diffuse form and in a few round bodies at the margin of chromosomes. Similarly, GFP-PSF–expressing cells blocked at G1/S exhibit unique PSF stellate structures, as well as diffuse green fluorescent staining, both of which are exactly duplicated by staining with the B92 antibody. By sharp contrast, in apoptotic cells, GFP-PSF aggregates in large rounded structures with no diffuse staining in the rest of the cell. These apoptotic PSF-containing structures were completely inaccessible to the B92 antibody, implying that the antigenic epitope had been blocked. The inaccessibility of PSF in immunofluorescence is not restricted to the B92 mAb only and occurs also with the use of a polyclonal antiserum directed against an antigenic region in the N-terminus, different from the epitopes recognized by B92. These changes are associated with hyperphosphorylation of PSF. However, the B92 antibody efficiently binds to denatured PSF from apoptotic cells, as evidenced by Western blotting of cellular proteins. This indicates that it is not the hyperphosphorylation per se that masks the antigenic sites. Thus, PSF seems to undergo a major conformational change or massive interactions with new partner molecules during apoptosis. The latter possibility is probably the main cause of this antigenic masking, because during apoptosis PSF dissociated from PTB while associating with U1–70K and SR proteins. It should be considered that during apoptosis PSF assumes a new function and serves as a docking site for several regulatory proteins, which upon recruitment into the PSF apoptotic globules, are removed from the active regulatory machinery.
The PSF protein, both in human and in mouse, is composed of two major regions. The functional C-terminal region contains the two RRMs needed for interactions with RNA (Patton et al., 1993) and for the localization of the protein in speckles (Dye and Patton, 2001). This region is highly conserved between mouse and human. The N-terminal region (first 110 residues) is unusually rich in proline, glycine, and glutamine residues. In parallel with properties of other proline-rich segments in proteins, this segment is believed to play a role in interactions recruiting other molecules. This region is less conserved between mouse and human in comparison to the C terminus, yet, the major structural elements are evolutionarily conserved. The B92 monoclonal anti-PSF antibody interacts with the N-terminal portion of PSF (Shav-Tal et al., 2000). The antigenic epitope that this antibody recognizes is the tetrapeptide PPXH (Zipori, unpublished observations) that appears three times in the N-terminus of PSF and is conserved between mouse and human, although the amino acids in-between these epitopes vary between the species. Interestingly, in 50% of the cases the unconserved amino acids in the N-terminal region are either serine or threonine residues, i.e., putative phosphorylation sites. This may explain some of the differences that we observed between mouse and human PSF. It is likely that protein interactions occurring via this region might mask the antigenic epitope during apoptosis by the phosphorylation of residues in this portion of the protein.
Phosphoamino acid analysis of phosphorylated PSF from mitotic, apoptotic, and untreated cells shows constitutive phosphorylation on serine and threonine residues. The same analysis with the 68-kDa degradation product of PSF maps the phosphorylated threonine residues to the C terminus of PSF. Phosphopeptide mapping of phosphorylated PSF shows that there are several additional phosphorylated residues on PSF from apoptotic cells compared with controls possessing an intact nucleus and that the hyperphosphorylation probably occurs on the N-terminus of the protein. This was further verified by the use of different GFP-PSF constructs: GFP-PSF shows retarded migration in mitotically blocked cells and so does GFP-PSF ΔRRM2, which lacks RRM2, the speckle localization signal. This indicates that the dissociation of PSF from speckles during mitosis probably does not occur because of phosphorylation in this region. GFP-PSF amino acids 338–707 containing only the C terminus did not show any retarded migration, thus implying that hyperphosphorylation of PSF occurs on the N-terminus.
The levels of phosphorylation of PSF as seen by its retarded migration in Western blots are different in mitotic, G1/S-blocked and apoptotic cells. In both mitosis and apoptosis, transcription and pre-mRNA splicing are inactive. This would explain why splicing factors are found in the hyperphosphorylated form, which inactivates their splicing activity. It seems that hyperphosphorylation of PSF occurs at two different levels during mitosis and apoptosis. Untreated cells have intermediate levels of phosphorylation whereas G1/S-blocked cells are hypophosphorylated. Because the higher and lower levels of phosphorylation occur during cell states in which splicing is inhibited, it indicates that both hyper- and hypophosphorylation can be a means for the negative regulation of the PSF protein in pre-mRNA splicing. A similar phenomenon was observed with SR proteins. During early development in nematodes, SR proteins change from a hyperphosphorylated and inactive form to an intermediately phosphorylated and active form (Sanford and Bruzik, 1999). Further dephosphorylation reduces their activity. Similarly, in mammalian cells, both hyper- and hypophosphorylation inhibit the splicing activity of SR proteins (Prasad et al., 1999) and disassemble nuclear speckles (Misteli and Spector, 1996), thus suggesting that intermediate levels of phosphorylation are needed for the functional and structural integrity of these proteins. An alternative interpretation of the differential phosphorylation of PSF, during mitosis and particularly in apoptosis, would be that this protein has functions that are unrelated to splicing. Interestingly, topoisomerase I was shown to specifically phosphorylate SR proteins (Rossi et al., 1996). It therefore may be speculated that the removal of PSF from speckles via hyperphosphorylation is a means for its activation in other cellular functions. Indeed, PSF appears to function in negative regulation of transcription (Urban et al.,2000) in the stimulation of topoisomerae I activity (Straub et al., 1998; 2000), in DNA pairing processes that occur during DNA recombination and repair (Akhmedov and Lopez, 2000), and, as shown here, in binding to U1–70K and SR proteins during apoptosis.
The differential phosphorylation may also point to the involvement of different kinases and/or phosphatases during the various cell states. A number of mammalian kinases have been shown to cause disassembly of nuclear speckles and relocalization of splicing factors, and to subsequently affect splicing factor activity. SRPK-1/2, Clk-1/2/3/4, cdc2-kinase, cyclin E-cdk2, topoisomerase I, U1 70-kDa associated kinase, cGMP-dependent kinase, and lamin B-receptor kinase were all shown to phosphorylate SR proteins and other splicing factors in vitro (Woppmann et al., 1993; Gui et al., 1994; Colwill et al., 1996; Nikolakaki et al., 1996; Rossi et al., 1996; Nayler et al., 1997; Duncan et al., 1998; Kuroyanagi et al., 1998; Okamoto et al., 1998; Seghezzi et al., 1998; Wang et al., 1998; Koizumi et al., 1999; Wang et al., 1999). In addition, some of these kinases were shown to affect the splicing activity of such factors (Mermoud et al., 1994; Cao et al., 1997; Xiao and Manley, 1998; Prasad et al., 1999). How all of these enzymes work in concert in vivo, if at all, is unknown. Cell- and tissue-specific differences in expression of these kinases (Nayler et al., 1997; Kuroyanagi et al., 1998; Wang et al., 1998; Papoutsopoulou et al., 1999) can only provide a partial explanation. A more likely hypothesis is that different kinases are recruited to nuclear speckles during different cell states and phosphorylate splicing factors in a manner specific to their biochemical properties. For example, in the case of the SF2/ASF SR protein, SRPK1 and cdc2 kinase were shown to phosphorylate the protein on different residues (Okamoto et al., 1998). In addition, phosphorylation can differentially modulate protein–protein interactions of several splicing factors (Xiao and Manley, 1998; Yeakley et al., 1999). Thus, although PSF does not contain an RS domain, its cellular localization is controlled in a manner similar to that of SR proteins.
The functional significance of PSF phosphorylation during apoptosis remains unresolved because of the lack of data concerning the fate of splicing factors in programmed cell death. Even though protein phosphorylation is commonly observed in apoptosis (Gjertsen and Doskeland, 1995), the specific roles for this are unknown. It has been suggested that protein phosphorylation targets proteins for cleavage during apoptosis. Such a connection seems to hold in the case of lamin B (Shimizu et al., 1998). Several RNA processing proteins are cleaved by caspases during apoptosis (Wolf and Green, 1999); however, no connection to phosphorylation has been indicated. It was shown that phosphorylated SR proteins associate with the U1-snRNP complex and snoRNPs during apoptosis (Utz et al., 1998; Overzet et al., 2000), and our results show that PSF is another component of this complex during apoptosis. Another study suggests that splicing factors regulate apoptosis, perhaps by regulating caspase activity (Jiang et al., 1998). It is speculated (Prasad et al., 1999) that the phosphorylation of splicing factors at unique residues facilitates the inactivation of these proteins either by rendering them inactive or by irreversibly complexing them with other proteins of the splicing machinery and thus titrating them out of the splicing reaction.
Our data substantiate the concept of the dynamic nucleus that reorganizes its components through a mechanism of phosphorylation, depending on the functional stage of the cell. Our data together with the data accumulated on SR proteins, point to the involvement of several different kinases and phosphatases in the phosphorylation status of nuclear factors. Regulation by phosphorylation is also an end to modulate the activity of the components of the transcription and polyadenylation complexes (Hunter and Karin, 1992; Colgan et al., 1996) and provides a linkage between processes of transcription and splicing (Hirose et al., 1999).
ACKNOWLEDGMENTS
The authors are grateful to Dr. Tony Hunter for his advice on phosphopeptide mapping. The authors thank Dr. Gil Ast and Dr. David Helfman for providing us with antibodies. Dov Zipori is an incumbent of the Joe and Celia Weinstein professorial chair at the Weizmann Institute of Science. The work in Ghent was supported by grant Interuniversity Attraction poles, Services of the Prime Ministre IUAP nr. P4/23.
Abbreviations used:
- BM
bone marrow
- IG
interchromatin granules
- PSF
PTB-associated splicing factor
- PTB
polypyrimidine tract binding protein
REFERENCES
- Akhmedov AT, Lopez BS. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF, and promotes DNA strand invasion. Nucleic Acids Res. 2000;28:3022–3030. doi: 10.1093/nar/28.16.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Krainer AR. Mammalian pre-mRNA splicing factors. In: Krainer AR, editor. Eukaryotic mRNA processing. New York: Oxford University Press; 1997. pp. 174–212. [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Chanas-Sacré G, Mazy-Servais C, Wattiez R, Pirard S, Rogister B, Patton JG, Belachew S, Malgrange B, Moonen G, Leprince P. Identification of PSF, the polypyrimidine tract-binding protein-associated splicing factor, as a developmentally regulated neuronal protein. J Neurosci Res. 1999;57:62–73. doi: 10.1002/(SICI)1097-4547(19990701)57:1<62::AID-JNR7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Casciola-Rosen LA, Miller DK, Anhalt GJ, Rosen A. Specific cleavage of the 70-kDa protein component of the U1 small nuclear ribonucleoprotein is a characteristic biochemical feature of apoptotic cell death. J Biol Chem. 1994;269:30757–30760. [PubMed] [Google Scholar]
- Clemson CM, Lawrence JB. Multifunctional compartments in the nucleus: insights from DNA and RNA localization. J Cell Biochem. 1996;62:181–190. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C181::AID-JCB6%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Duncan PI, Stojdl DF, Marius RM, Scheit KH, Bell JC. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;241:300–308. doi: 10.1006/excr.1998.4083. [DOI] [PubMed] [Google Scholar]
- Dye BT, Patton JG. An RNA Recognition Motif (RRM) is required for the localization of PTB–associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- Eils R, Gerlich D, Tvaruskó W, Spector DL, Misteli T. Quantitative imaging of pre-mRNA splicing factors in living cells. Mol Biol Cell. 2000;11:413–418. doi: 10.1091/mbc.11.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjertsen BT, Doskeland SO. Protein phosphorylation in apoptosis. Biochim Biophys Acta. 1995;1269:187–199. doi: 10.1016/0167-4889(95)00117-b. [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gui J-F, Lane WS, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. Dynamic organization of pre-mRNA splicing factors. J Cell Biochem. 1996;62:191–197. doi: 10.1002/(sici)1097-4644(199608)62:2<191::aid-jcb7>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Jiang Z-H, Zhang W-J, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci USA. 1998;95:9155–9160. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia LF, Spector DL. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer AR, Hagiwara M. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs) J Biol Chem. 1999;274:11125–11131. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi N, Onogi H, Wakabayashi T, Hagiwara M. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem Biophys Res Commun. 1998;242:357–364. doi: 10.1006/bbrc.1997.7913. [DOI] [PubMed] [Google Scholar]
- La Branche H, Frappier D, Chabot B. Proteolysis of splicing factors during rat and monkey cell fractionation. Nucleic Acids Res. 1991;19:4509–4514. doi: 10.1093/nar/19.16.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BC, Shav-Tal Y, Peled A, Gothelf Y, Jiang W, Toledo J, Ploemacher RE, Haran-Ghera N, Zipori D. A hematopoietic 49-kD nuclear antigen: predominance in immature normal and tumor granulocytes and detection in hematopoietic precursor cells. Blood. 1996;87:2283–2291. [PubMed] [Google Scholar]
- Martin SJ, Lennon SV, Bonham AM, Cotter TG. Induction of apoptosis (programmed cell death) in human leukemic HL-60 cells by inhibition of RNA or protein synthesis. J Immunol. 1990;145:1859–1867. [PubMed] [Google Scholar]
- Melcak I, Cermanova S, Jirsova K, Koberna K, Malinsky J, Raska I. Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs. Mol Biol Cell. 2000;11:497–510. doi: 10.1091/mbc.11.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Cohen PTW, Lamond AI. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. RNA splicing: what has phosphorylation got to do with it? Curr Biol. 1999;9:R198–R200. doi: 10.1016/s0960-9822(99)80128-6. [DOI] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 1997;7:135–138. doi: 10.1016/S0962-8924(96)20043-1. [DOI] [PubMed] [Google Scholar]
- Nayler O, Stamm S, Ullrich A. Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem J. 1997;326:693–700. doi: 10.1042/bj3260693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987;48:219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- Nikolakaki E, Simos G, Georgatos SD, Giannakouros T. A nuclear envelope-associated kinase phosphorylates arginine-serine motifs and modulates interactions between the lamin B receptor and other nuclear proteins. J Biol Chem. 1996;271:8365–8372. doi: 10.1074/jbc.271.14.8365. [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Onogi H, Honda R, Yasuda H, Wakabayashi T, Nimura Y, Hagiwara M. cdc2 kinase-mediated phosphorylation of splicing factor SF2/ASF. Biochem Biophys Res Comm. 1998;249:872–878. doi: 10.1006/bbrc.1998.9247. [DOI] [PubMed] [Google Scholar]
- Overzet K, Gensler TJ, Kim SJ, Geiger ME, Van Verooij WJ, Pollard KM, Anderson P, Utz PJ. Small nucleolar RNP scleroderma autoantigens associate with phosphorylated serine/arginine splicing factors during apoptosis. Arthritis Rheum. 2000;43:1327–1336. doi: 10.1002/1529-0131(200006)43:6<1327::AID-ANR15>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Papoutsopoulou S, Nikolakaki E, Chalepakis G, Kruft V, Chevaillier P, Giannakouros T. SR protein-specific kinase 1 is highly expressed in testis and phosphorylates protamine 1. Nucleic Acids Res. 1999;27:2972–2980. doi: 10.1093/nar/27.14.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DJ, Patek PQ. Detergent and enzyme treatment of apoptotic cells for the observation of DNA fragmentation. Biotechniques. 1998;24:558–560. doi: 10.2144/98244bm07. [DOI] [PubMed] [Google Scholar]
- Park PC, De Boni U. Dynamics of structure-function relationships in interphase nuclei. Life Sci. 1999;64:1703–1718. doi: 10.1016/s0024-3205(99)00003-x. [DOI] [PubMed] [Google Scholar]
- Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Peled A, Zipori D, Abramsky O, Ovadia H, Shezen E. Expression of α-smooth muscle actin in murine bone marrow stromal cells. Blood. 1991;78:304–309. [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F, Laborier E, Forne T, Divita G, Derancourt J, Riou JF, Antoine E, Cathala G, Brunel C, Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–82. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Bruzik JP. Developmental regulation of SR protein phosphorylation and activity. Genes Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Lee BC, Bar-Haim S, Vandekerckhove J, Zipori D. Enhanced proteolysis of pre-mRNA splicing factors in myeloid cells. Exp Hematol. 2000;29:1029–1038. doi: 10.1016/s0301-472x(00)00510-5. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Lee BC, Bar-Haim S, Schori H, Zipori D. Reorganization of nuclear factors during myeloid differentiation. J Cell Biochem. 2001;81:379–392. doi: 10.1002/1097-4644(20010601)81:3<379::aid-jcb1052>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Cao C-X, Shao R-G, Pommier Y. Lamin B phosphorylation by protein kinase Cα and proteolysis during apoptosis in human leukemia HL60 cells. J Biol Chem. 1998;273:8669–8674. doi: 10.1074/jbc.273.15.8669. [DOI] [PubMed] [Google Scholar]
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl DF, Bell JC. SR protein kinases: the splice of life. Biochem Cell Biol. 1999;77:293–298. [PubMed] [Google Scholar]
- Straub T, Grue P, Uhse A, Lisby M, Knudsen BR, Tange TO, Westergaard O, Boege F. The RNA-splicing factor PSF/p54nrb controls DNA-topoisomerase I activity by a direct interaction. J Biol Chem. 1998;273:26261–26264. doi: 10.1074/jbc.273.41.26261. [DOI] [PubMed] [Google Scholar]
- Straub T, Knudsen BR, Boege F. PSF/p54nrb stimulates “jumping” of DNA topoisomerase I between separate DNA helices. Biochemistry. 2000;39:7552–7558. doi: 10.1021/bi992898e. [DOI] [PubMed] [Google Scholar]
- Stroh C, Schulze-Osthoff K , Death by a thousand cuts: an ever increasing list of caspase substrates. Cell Death Differ. 1998;5:997–1000. doi: 10.1038/sj.cdd.4400451. [DOI] [PubMed] [Google Scholar]
- Tanese N, Pugh BF, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol Endocrinol. 2000;14:774–782. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- Utz PJ, Hottelet M, van Venrooij WJ, Anderson P. Association of phosphorylated serine/arginine (SR) splicing factors with the U1-small ribonucleoprotein (snRNP) autoantigen complex accompanies apoptotic cell death. J Exp Med. 1998;187:547–560. doi: 10.1084/jem.187.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis. 1994;15:544–554. doi: 10.1002/elps.1150150173. [DOI] [PubMed] [Google Scholar]
- Wang H-Y, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu X-D. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao L-G, Wang Y-L, Pederson T. Localization of pre-messenger RNA at discrete nuclear sites. Proc Natl Acad Sci USA. 1991;88:7391–7395. doi: 10.1073/pnas.88.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Kramer A, Robinson PJ. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J. 1999;18:4549–4559. doi: 10.1093/emboj/18.16.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse N, Kumar S, Song Q, Strike P, Sparrow L, Dreyfuss G, Alnemri ES, Litwack G, Lavin M, Watters D. Heteronuclear ribonucleoproteins C1 and C2, components of the spliceosome, are specific targets of interleukin 1beta-converting enzyme-like proteases in apoptosis. J Biol Chem. 1996;271:29335–29341. doi: 10.1074/jbc.271.46.29335. [DOI] [PubMed] [Google Scholar]
- Wei X, Somanathan S, Samarabandu J, Berezney R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J Cell Biol. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf BB, Green DR. Suicidal tendencies: apoptotic cell death by caspase family proteinases. J Biol Chem. 1999;274:20049–20052. doi: 10.1074/jbc.274.29.20049. [DOI] [PubMed] [Google Scholar]
- Woppmann A, Will CL, Kornstädt U, Zuo P, Manley JL, Lührmann R. Identification of an snRNP-associated kinase activity that phosphorylates arginine/serine rich domains typical of splicing factors. Nucleic Acids Res. 1993;21:2815–2822. doi: 10.1093/nar/21.12.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S-H, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factors ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Tronchere H, Olesen J, Dyck JA, Wang H-Y, Fu X-D. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Kim E, Warren SL, Berget SM. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]