Abstract
The development of human metastatic cancer is a multistep process, involving the acquisition of several genetic mutations, tumour heterogeneity, and interactions with the surrounding microenvironment. Due to the complexity of cancer development in mammals, simpler model organisms, such as the vinegar fly, Drosophila melanogaster, are being utilized to provide novel insights into the molecular mechanisms involved. In this review, we highlight recent advances in modelling tumorigenesis using the Drosophila model, focusing on the cooperation of oncogenes or tumour suppressors, and the interaction of mutant cells with the surrounding tissue in epithelial tumour initiation and progression.
1. Introduction: Drosophila as a Model for Understanding Human Cancer
For over 100 years, research utilizing the powerful genetics of the vinegar fly, Drosophila melanogaster, has contributed to the understanding of fundamental cellular and developmental processes relevant to the medical field (reviewed in [1, 2]). Indeed, research using the Drosophila model had now been granted five Nobel Prizes for Medicine or Physiology. Moreover, the Drosophila model has proven to be a highly suitable system for understanding cancer and in developing cancer therapies (reviewed in [3–15]). Use of Drosophila as a model organism for cancer research was pioneered by genetic screens, conducted in the late 1900s, which identified many Drosophila tumour-causing mutations (reviewed in [16, 17]). Many of these were novel tumour-suppressor genes or oncogenes, which were subsequently shown to also have tumourigenic properties in mammalian systems and to be involved in human cancer (reviewed in [8, 9, 11, 18, 19]).
The strengths of the Drosophila model for cancer research lie in the evolutionary conservation of genes and signalling pathways between flies and humans, its lower genetic redundancy, simpler biology, rapid life cycle, and powerful genetics (reviewed in [1, 2, 15]). Due to the sophisticated genetic tools available, cancer-causing mutations can be studied in a tissue-specific or mosaic context. In the study of tumorigenesis in Drosophila, the developing epithelial tissues of the Drosophila larval imaginal discs that generate the adult eye-antenna or wing-thorax or the epitheliums of the adult intestine are commonly used (reviewed in [7, 20–22]). Indeed, it is mosaic (clonal) analyses using these epithelial tissues that have enabled new insights into the initiation and progression of cancer. In this review, we highlight recent studies focusing primarily on Drosophila epithelial tissues, showing how cooperating interactions between cells, and between mutations in oncogenes or tumour-suppressor genes, drive cancer initiation and progression.
2. Cell Competition and Cooperating Interactions between Cells in Tumorigenesis
Epithelial tumours can be initiated by multiple molecular lesions, including deregulation of signalling pathways and the perturbation of cell polarity/morphology, such as those generated by loss of function of the cell polarity regulator, Scribbled (Scrib) [15, 23–25]. The clonal-analysis approach has enabled the molecular interactions between the developing epithelial tumour and the surrounding normal tissue, the innate immune system, or distant organs to be revealed (reviewed in [6, 26–30]). The interaction between a tumour cell and the surrounding normal cells in an epithelium is important in determining whether the tumour cell survives and proliferates or is eliminated. The phenomenon of “cell competition,” a surveillance mechanism that compares the fitness of cells in an epithelium, is critical for the active elimination of cells of lower fitness (losers) by cells of greater fitness (winners) within an epithelial tissue (reviewed in [29, 31–33]) (Figure 1). Cell competition involves the interaction of cells and cell-surface molecules or a modified innate immune signalling pathway, leading to caspase-mediated apoptosis of the loser cells by the winner cells. The mechanism of cell competition depends upon the molecular lesion. Cells with low levels of the cell growth regulator, dMyc, or of ribosomal proteins, which reduce cellular growth, are recognized and eliminated differently from those where cell polarity is impaired [34–39] (Figure 1(a)). Differentially expressed cell-surface receptor isoforms of the Flower protein [37, 38] or modified innate immune signalling involving Toll-Like Receptor-NfκB (TLR-NfκB) signalling are involved in the elimination of low dMyc or ribosomal protein expressing cells [35].
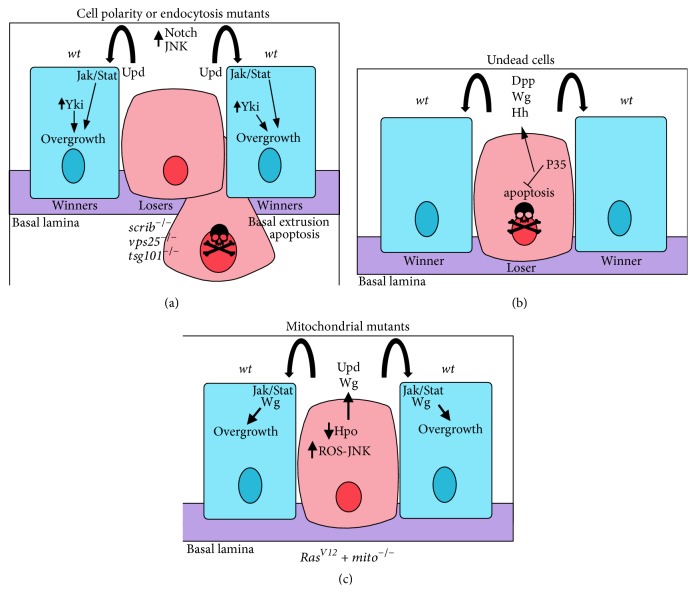
Figure 1.
Cell competition mechanisms. The three main types of cell competition are shown. Mutant cells are in pink, wild-type cells are in blue, hemocytes are in grey, and the basement membrane (basal lamina) is in purple. (a) Classical cell competition: within an epithelium, cells with reduced levels of dMyc, ribosomal subunits mutants (minutes), Jak-Stat or Wg signalling, or high levels of Hippo signalling (losers) are eliminated by apoptosis, induced by the surrounding wild-type cells (winners). The loser cells express on their cell surface the Flower-Lose (FweLose) isoform (red dots), which marks them for elimination when in contact with the surrounding wild-type cells that express the Flower-Ubi (FweUbi) isoform (green dots). Additionally, signalling via the Spätzle ligand and Toll-Like Receptors (TLRs) in the loser cells triggers cell death via upregulation of cell death inducers, Rpr or Hid. Cells with upregulated Hippo signalling (or yki mutants) exhibit decreased dMyc levels, but cells with decreased ribosomal function, Jak-Stat, or Wg signalling undergo dMyc-independent cell competition. (b) Supercompetition: cells with high levels of dMyc, Jak-Stat, increased Wg signalling, or decreased Hippo signalling show “supercompetitor” behaviour and induce apoptosis in neighbouring wild-type cells. This occurs via the Flower-code or via Spätzle-TLR signalling in the loser cells. (c) Cell polarity mutant cell competition: cell polarity-impaired mutant cells are recognized by their epithelial neighbours or hemocytes (grey) and the TNFR-JNK signalling ligand, Egr (TNF), which is secreted by the wild-type epithelial cells or hemocytes. Mutant cells are removed by JNK-dependent and caspase-dependent apoptosis. JNK activation in neighbouring wild-type cells together with PVR, ELMO, and Mbc signalling is required in the wild-type cells for the removal of the dying cells. Hemocytes play the predominant role in engulfment and removal of the dead cells. The interaction of PTP10D in the mutant cell with SAS in the wild-type cell is important for “loser” cell fate of the polarity-impaired mutant cell. The Slit-Robo-Ena signalling pathway plays an important role in basal extrusion of the mutant cell, where the hemocytes are localized.
Clonal alterations in signalling pathways such as Wingless (Wg/Wnt), Jak-Stat, and the Hippo negative tissue-growth control pathways can also induce cell competition (reviewed in [33, 36, 40]). Impairment of Hippo signalling, in addition to upregulating cell cycle and cell survival genes, leads to the upregulation of dMyc and results in a supercompetitor phenotype, where the surrounding wild-type cells are actively eliminated [41, 42] (Figure 1(b)). However, the cell competition mechanism that occurs upon differences in the Wg or Jak-Stat signalling occur by dMyc-independent mechanisms, which are currently not well defined [43, 44].
By contrast, scrib mutant cell competition requires the interaction of a membrane tyrosine phosphatase, PTP10D, on the loser cell and a membrane protein, Sas, on the winner cell, which results in repression of Epidermal Growth Factor Receptor- (EGFR-) Ras small-GTPase signalling and the activation of the Jun N-terminal Kinase (JNK) signalling in the loser cell [34] (Figure 1(c)). Additionally, JNK signalling activates the Slit-Robo-Ena signalling pathway leading to downregulation of E-Cadherin (E-Cad) and the basal extrusion of scrib mutant cells, where they die [45, 46]. Indeed, downregulation of E-Cad appears to be important in scrib mutant cell extrusion and elimination, since overexpression of E-Cad in scrib mutant clones reduced cell extrusion and promoted clonal overgrowth [45]. JNK signalling also overrides the impaired Hippo signalling in scrib mutant cells in a clonal context, preventing their overgrowth [47, 48]. Furthermore, differing levels of dMyc or Jak-Stat signalling between the polarity-impaired mutant cells and the surrounding wild-type cells has also been implicated in the elimination of the mutant cells in particular contexts [49–51].
In addition to cell competition, the interactions between the tumour and its microenvironment are critical for whether the tumour cells will undergo apoptosis or overgrow and eliminate the normal tissue (Figures 1(c) and 2). Interactions between the surrounding wild-type epithelial cells, mesenchymal cells (myoblasts), or macrophage-like innate immune system cells (hemocytes) contribute to the fate of the tumour cells [52–62]. Mechanistically, the emerging picture from the study of neoplastic tumours generated in imaginal epithelial tissues (such as with mutants in the neoplastic tumour-suppressor gene (nTSG), scrib), is that tumour development occurs through the cooperative interaction of factors produced from surrounding epithelial cells or hemocytes and feed-forward mechanisms within the tumour cell amplifying this loop (Figure 2). Hemocytes are attracted to sites of cell competition by the secretion of fragments of the Tyrosyl-tRNA synthetase protein (dminiTyr and dEMAP), which is triggered by JNK activation and Metalloproteinase (MMP) dependent cleavage in dying loser cells [63] (Figure 2(a)). Mechanistically, dEMAP upregulates PI3K signalling in the hemocytes, which is required for hemocyte chemotaxis [64] and may be important in engulfment of the dying cells [63].
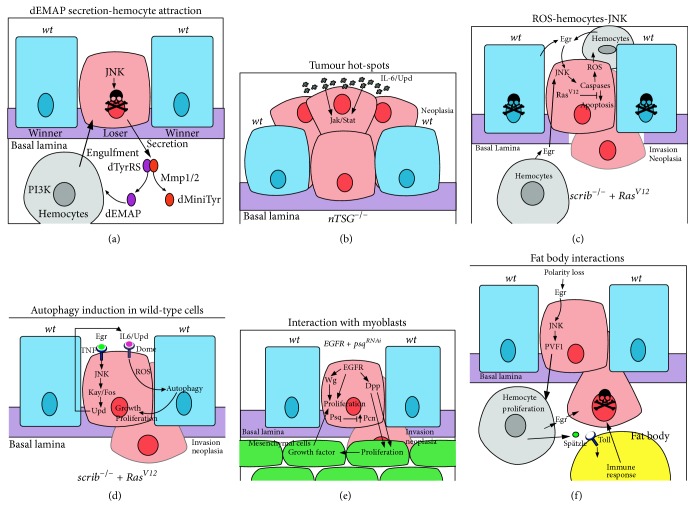
Figure 2.
Cooperative interactions between the tumour and surrounding cells in tumorigenesis. Interactions between cells are shown that result in either the death of the mutant cell or cell survival, proliferation, and neoplastic transformation. Mutant cells are in pink, wild-type cells are in blue, hemocytes are in grey, myoblasts (mesenchymal cells) are in green, a fat body adipocyte is in yellow, and the basement membrane (basal lamina) is in purple. (a) dEMAP secretion-hemocyte attraction: JNK signalling in a cell polarity-impaired loser cell transcriptionally upregulates MMP1, which acts to cleave secreted dTyrRS to form dEMAP and dminiTyr. dEMAP attracts hemocytes to the loser cell by upregulating PI3K signalling in the hemocytes, which is required for chemotaxis and possibly engulfment of the loser cell. (b) Tumour hot-spots: neoplastic tumour-suppressor mutants (nTSGs) induce tumours more preferably, in regions where there is a stiff basal lamina and there are developmentally high levels of the Upd (IL-6) ligand to elevate Jak-Stat signalling, which promotes cell survival and proliferation of the tumour cells. (c) ROS-hemocytes-JNK: in scrib mutant RasV12-expressing tumour cells, a feedback loop between the hemocytes and the mutant cells promotes tumorigenesis. In the mutant cells, Ras signalling and caspase activation leads to ROS production that is released from the cells and promotes hemocytes to produce Egr (TNF). Egr signals via the TNFR-JNK pathway in the mutant cell leading to the upregulation of caspase activity, and some apoptosis, which is required for tumour overgrowth and invasion. Due to the disruption of the peripodial epithelium in large scrib mutant RasV12-expressing tumours, hemocytes most likely interact with the tumour on both apical and basal sides. (d) Induction of autophagy in surrounding wild-type cells: scrib mutant RasV12-expressing tumour cells are metabolically stressed, which leads to ROS production. Egr-JNK signalling leads to the transcriptional upregulation of Upd, ligands for the Dome-Jak-Stat signalling pathway, which is elevated in the mutant cells. Jak-Stat signalling and ROS production are required for the induction of autophagy in the surrounding wild-type cells, and also at distant sites, such as the fat body, muscle, and gut (not shown), which facilitates tumour growth and neoplastic transformation, possibly through supplying amino acids, glucose, and other nutrients to the tumour cells. (e) Interactions with myoblasts: in EGFR-overexpressing psq-knockdown tumours cooperative interactions are observed between the tumour cells and the surrounding myoblasts (mesenchymal cells). EGFR induces Wg and Dpp expression, and psq knockdown leads to increased levels of the extracellular matrix protein, Perlecan (Pcn). Wg acts to promote proliferation of the tumour cells, whilst Dpp, facilitated by Pcn in the basal lamina, stimulates proliferation of the myoblast cells. In turn, the myoblast cells provide unidentified growth factors that drive proliferation and neoplastic transformation of the tumour cells. Myoblasts also supply Egr (not shown), which would be expected to activate the TNFR-JNK signalling pathway in the tumour cells. (f) Interactions with the fat body: polarity-impaired tumours through Egr-JNK signalling upregulate PVF1, a ligand for the PVR receptor on hemocytes, which promotes hemocyte proliferation. Hemocytes, in turn, supply Egr to the tumour cells, and the Toll Receptor ligand, Spätzle, to the fat body, which induces innate immune system signalling in the fat body. These interactions are required to induce apoptosis of tumour cells.
A highly important pathway in cell-cell interactions that triggers tumour cell death is the Tumour Necrosis Factor (TNF), Eiger (Egr), pathway (Figure 1(c)). Egr signals via the TNF receptor (TNFR), Grindenwald (Grnd), and leads to the activation of the JNK signalling pathway in the tumour cell, which, through the activation of caspases, results in caspase-mediated apoptosis of initiating tumour cells [143]. Egr can be produced from the adjacent wild-type epithelial cells, myoblasts, or the hemocytes [52, 55, 61, 89, 144]. The wild-type cells on the border of the mutant clone also require JNK signalling, though in a nonapoptotic role, and the induction of PVR (PDGF/VEGF receptor homolog)-ELMO (Ced-12 homolog)-Mbc (Dock180 homolog) signalling to induce engulfment of the mutant cells [54] (Figure 1(c)). Whilst there is evidence that wild-type epithelial cells engulf the scrib mutant dying cells [54], hemocytes play the predominant role in this process, as well as in cell competition due to variations in dMyc or ribosomal protein levels [145, 146]. Furthermore, in tumour development, microenvironmental “hot-spots” have been revealed where the tumour has a greater chance of progressing, which has parallels with mammalian systems [27, 147]. Molecularly, the “hot-spots” are due to endogenously higher levels of Jak-Stat signalling and the presence of a stiff basement membrane extracellular matrix, resulting in extrusion of the tumour cells apically, where they survive (Figure 2(b)). Conversely, in “cold-spots,” tumour cells extrude basally from the epithelium and die, perhaps due to exposure to hemocytes (see below). Molecularly, the level of Slit-Robo-Ena signalling is important for the direction of cell extrusion and therefore dictates whether the aberrant cells will be eliminated by basal extrusion, remain in the epithelium and overgrow, or are apically extruded into the lumen and progress to invasive tumours [45, 46].
By contrast, if cell death is prevented in the mutant cells by blocking caspase activity or upregulation of a cell survival pathway, such as the EGFR-Ras signalling pathway, then the cells survive and form invasive tumours [23, 65, 66, 89–91, 144]. This occurs since TNFR-JNK signalling is repurposed to promote cell morphology changes and migratory cell behaviour (reviewed in [143]). Ras signalling prevents caspase-mediated cell death, and instead caspase activity induces the formation of reactive oxygen species (ROS) within the cell and promotes their secretion [57] (Figure 2(c)). Extracellular ROS, in turn, attracts hemocytes, which secrete TNF and amplify the JNK signalling pathway in the tumour cell [57]. Interestingly, a recent report revealed that ROS, released from the scrib mutant RasV12-expressing tumour cells, promotes autophagy (a catabolic process that degrades cellular macromolecules and organelles to provide energy) in the surrounding wild-type cells, as well as systemically in gut, muscle, and adipose tissues [60] (Figure 2(d)). The induction of autophagy may serve to provide glucose, amino acids, and other nutrients that facilitate tumour growth. In the scrib mutant RasV12-expressing cells, Egr-JNK-Fos (Kay) signalling together with Ras-MAPK signalling generates metabolic stress, leading to ROS production [60, 101]. JNK and impaired Hippo signalling in these tumour cells also result in the transcription of unpaired 1–3 (upd1–3), which encode IL-6-related ligands for the Domeless (Dome) receptor of the Jak-Stat pathway, thereby activating this signalling pathway and promoting tumour growth [148]. Interestingly, Upd1–3 acts in an autocrine manner in the tumour cells to promote autophagy in the neighbouring wild-type cells, most likely by stimulating ROS production or secretion [60].
Furthermore, myoblast cells are thought to provide growth factors, which are currently unidentified, to the epithelial tumour cells to stimulate proliferation and survival [52] (Figure 2(e)). In an EGFR-driven pipsqueak knockdown neoplastic tumour model, EGFR signalling induces upregulation of Wg, which promotes epithelial tumour cell proliferation, but tumour growth is dependent on the neighbouring myoblast cells. Interestingly, a codependency occurs between the epithelial neoplastic tumour cells and the mesenchymal cells, whereby the TGFβ/Bone Morphogenetic Protein- (BMP-) family morphogen, Decapentaplegic (Dpp), produced in the epithelial cells promotes the expansion of mesenchymal cell compartment, and, in turn, the myoblast cells are required for epithelial cell tumorigenesis [52]. Recent studies have shown that the myoblast cells also produce Egr [61], which, via TNFR signalling, promotes tumorigenesis when cell death is blocked in the epithelial tumour cells. Despite studies showing the importance of Egr in inducing JNK signalling in neoplastic tumours [89], an intrinsic mechanism also exists to elevate JNK signalling in the tumour cells, involving Rho1-GTPase signalling and activation of the JNKKK, Wallenda [61, 149]. Thus, initially, impairment of cell polarity may trigger JNK activation through the Rho1-Wallenda pathway, and, subsequently, myoblasts and hemocytes in the tumour microenvironment are stimulated to produce Egr, thereby amplifying JNK activation in the tumour.
In addition to interactions between the epithelial tumour cells and their local microenvironment, there is also evidence for communication between the hemocyte and the fat body adipocytes [56] (Figure 2(f)). In polarity-impaired neoplastic tumour-bearing larvae, hemocytes supply the Toll ligand, Spätzle, to the fat body adipocytes, which leads to induction of the Toll-NFκB innate immune response signalling pathway in the adipocytes and the production of immune peptides. Egr-JNK signalling in the tumour cells also contributes to the cellular crosstalk, since it results in the transcriptional upregulation of the ligand, PVF1, which, through the PVR signalling pathway, stimulates hemocyte proliferation, thereby elevating Spätzle production from the hemocytes and innate immune signalling in the fat body. This mechanism is required to restrain tumour growth, since knockdown of Spätzle expression in the hemocytes results in reduced Toll pathway signalling in the fat body and reduced tumour cell death. However, whether the fat body-induced immune response only functions to activate the hemocytes, or also by secretion of diffusible signals, to promote tumour cell death, is presently unclear. Moreover, since a Spätzle-modified Toll signalling pathway leading to caspase activation has been observed in dMyc and ribosomal protein cell competition mechanisms [35], the hemocytes might also supply Spätzle to the tumour cells to contribute to their death. Consistent with this, crosstalk between the Toll and JNK signalling pathways in triggering cell death occurs in eye-antennal and wing epithelial tissues [150]. In these tissues, JNK signalling in the epithelial cells induces Spätzle upregulation in the surrounding peripodial membrane cells by an unknown mechanism, which, in turn, activates Toll-NfκB signalling in the epithelial cells. Thus, Spätzle production by hemocytes or peripodial membrane cells, together with Egr-JNK signalling and signals from the fat body, may all be involved in triggering tumour cell death.
To summarize, in cell competition within epithelial tissues, signals from the myoblasts, the extracellular matrix, the cellular innate immune system, and systemic responses all influence whether the tumour cells will be eliminated or survive and progress to form overgrown invasive tumours. Moreover, if cell death of the tumour cells is blocked, tumour intrinsic and cell-cell signalling pathways that are normally antitumorigenic can instead become tumour-promoting (see below). Cell competition mechanisms are conserved in mammalian systems (reviewed in [36, 39, 151, 152]), and the tumour microenvironment plays a key role in mammalian tumorigenesis (reviewed in [153–156]). Thus, the findings from these Drosophila studies of cellular interactions in tumorigenesis are likely to provide new insights into the understanding of human cancer initiation and progression.
3. Cooperation Interactions between Oncogenic or Tumour-Suppressor Mutations in Tumour Initiation and Progression
The development of malignant cancer requires the deregulation of many processes, including increased cell proliferation, reduced differentiation and apoptosis, increased invasion, and altered metabolism (reviewed in [157]). There are only a few tumour-causing genes that when individually knocked down or overexpressed in whole epithelial tissues or large domains, are capable of inducing all the hallmarks of cancer that can be modelled in Drosophila (reviewed in [3, 11, 15, 158]). Many genes, when deregulated, can cause hyperplastic tumours, characterized by increased tissue growth that are still capable of differentiating, but only a few result in neoplastic tumours, in which the tissue overgrows and shows reduced differentiation and a loss of tissue architecture (reviewed in [159]). Genes capable of conferring many hallmarks of cancer when knocked down or mutated in large domains in epithelial tissues are the junctional (cell polarity regulators, Scrib, Dlg, and Lgl) and endocytic (such as Rab5) neoplastic tumour suppressors. Moreover, recent studies have shown that lgl mutant tumours, in addition to possessing other cancer hallmarks, are able to induce an angiogenesis-like process in Drosophila, tracheogenesis, in order to obtain an increased oxygen supply [160, 161]. A gene capable of conferring neoplastic overgrowth when expressed in large epithelial tissue domains is the activated version of the receptor tyrosine kinase gene, PVR [159, 162, 163]. Additionally, a recent study has shown that expression of the oncogenic fusion between the KIF5B kinesin motor protein and the Ret tyrosine kinase, KIF5B-Ret, promotes many hallmarks of cancer in tracheal epithelial cells [164]. However, as cancer arises from mutations that occur in single cells surrounded by normal tissue, it is uncommon for perturbations in any one gene to confer all the properties that are required for a normal cell to transform into a proliferative-invasive cancer within the context of a wild-type epithelium, since cell competition leads to the elimination of aberrant cells. Even with potent tumour-causing mutations, when generated clonally or by induction in a tissue domain, growth of the tumour beyond a certain size is required to overcome apoptosis induced by cell competition [49, 165]. Thus, the phenomenon of cell competition is one reason why at least two mutations are required for tumour progression when initiated in single cells or small patches of cells, particularly concerning mutants in cell polarity or endocytosis regulators. We will now highlight various cooperative tumorigenesis mechanisms that have been modelled in Drosophila, focusing primarily on epithelial tissues (summarized in Table 1), and discuss the important insights these studies have revealed. We will first cover the genes/pathways involved in cell death, caspases (cysteine proteases), and the JNK signalling pathway, since they can have context-dependent roles in tumorigenesis.
Table 1.
Cooperating genes in Drosophila tumorigenesis.
| Cell-autonomous cooperative tumorigenesis | ||
|---|---|---|
| 1st mutation/mechanism | 2nd mutation/mechanism | Phenotype/references |
| Cell polarity gene perturbations | ||
|
| ||
|
Loss of function in apicobasal polarity regulators Results in cell polarity loss, JNK activation, mild Hippo pathway impairment |
Neoplastic overgrowth in whole tissue context and cell polarity loss and apoptosis in clonal context (reviewed in [19]) | |
|
| ||
|
Scribble (scrib, dlg, lgl) and Par and Crb polarity module gene loss of function Scribble module loss of function phenotypes dependent on aPKC activation |
Ras
V12 overexpression Dependent on ROS production, TNF (Egr)-JNK signalling, caspase (Dronc) activity Dependent on impairment of Hippo signalling Dependent on PI3K signalling and glutamate utilization |
Invasive neoplastic tumours of the larval eye-antennal epithelium [23, 48, 57, 60, 65–68] |
|
| ||
| scrib loss of function, aPKC-CA overexpression, crb overexpression | Inhibition of JNK signalling | Neoplastic tumour overgrowth in eye-antennal epithelium [66] |
|
| ||
|
lgl or scrib loss of function Results in JNK activation |
Ras
V12 overexpression Requires Hippo pathway impairment |
Neoplastic tumours in the larval wing epithelium [49] |
|
| ||
|
scrib loss of function Results in JNK activation |
Notch intra (Act) overexpression | Invasive neoplastic tumours in the larval eye-antennal epithelium [23, 66, 69] |
|
| ||
|
scrib loss of function Results in JNK activation |
Abrupt (BTB-POZ Zn finger transcription factor) overexpression Results in JNK activation Results in Hippo pathway impairment Results in downregulation of differentiation and Ecdysone response genes |
Invasive neoplastic tumours in the eye-antennal and wing epithelial tissues [70] |
|
| ||
|
scrib loss of function |
Taiman (Ecdysone coactivator) Results in reduced differentiation |
Invasive neoplastic tumours in the eye-antennal and wing epithelial tissues [70] |
|
| ||
|
scrib loss of function Results in JNK activation |
Slit-Robo2-Ena loss of function | Overgrown tumours in the eye-antennal epithelial tissues [45] |
|
| ||
|
scrib loss of function Results in JNK activation |
Slit-Robo overexpression Requires Ena Results in JNK activation and activation of a positive feedback loop |
Excessive extrusion and luminal tumour overgrowth in larval eye-antennal epithelial tissues [45] |
|
| ||
|
lgl loss of function Results in Hippo pathway impairment |
Inhibition of JNK signalling | Invasive neoplastic tumours of the larval/pupal eye neural-epithelium [71] |
|
| ||
| lgl loss of function | Myc overexpression | Invasive neoplastic tumours of the larval wing epithelium [50] |
|
| ||
| lgl loss of function | Hippo pathway impairment | Neoplastic tumours of the larval wing epithelium [49] |
|
| ||
| lgl loss of function | Notch intra (Act) overexpression | Neoplastic tumours of the larval wing epithelium [72] |
|
| ||
|
scrib, dlg or lgl depletion |
cno mutants Results in activation of Ras-MAPK signalling |
Enhanced neoplastic tumours of the antennal epithelium [73] |
|
| ||
|
Par-1 overexpression Results in cell polarity loss and Hippo pathway impairment |
Eye-antennal and wing tissue overgrowth [74] | |
|
| ||
| Par-1 overexpression | Notch intra (Act) overexpression | Hyperplastic eye-antennal epithelium [75] |
|
| ||
| Actin cytoskeletal regulators | ||
|
| ||
| Activation of Actin cytoskeletal regulators Rac1, RhoGEF2, Pbl, RhoV14, Rho1, RokCAT, sqhEE Results in activation of JNK signalling and cell morphology changes |
RasV12 (Raf gain-of-function) overexpression | Invasive neoplastic tumours of the larval eye-antennal epithelium [76, 77] |
|
| ||
| RhoGEF2 overexpression Results in JNK activation and cell morphology changes |
Abrupt (BTB-POZ Zn finger transcription factor) overexpression Results in reduced expression of differentiation gene, Dac |
Neoplastic tumours of the larval eye-antennal epithelial tissue [78] |
|
| ||
| Src64B overexpression Results in JNK activation and cell morphology changes |
Blocking JNK Rac1-Dia, Ras-MAPK, Hippo pathway impairment |
Eye-antennal epithelial tissue overgrowth [79] |
|
| ||
|
csk loss of function (Src activation) Depends on Actin cytoskeleton regulators, JNK activation, STAT activation, Hippo pathway impairment, Wingless (Wnt) expression/signalling and insulin-PI3K signalling |
RasV12 overexpression Promotes cell proliferation and survival |
Invasive neoplastic tumours of the larval eye-antennal epithelium [80–85] |
|
| ||
| Src42A or Src64B overexpression Results in Egr independent activation of JNK and Jak-Stat signalling |
Notchintra (Act) overexpression |
Neoplastic tumours of the larval eye-antennal and wing epithelium [86] |
|
| ||
| Src64B overexpression Results in cell morphology changes |
Abrupt overexpression Reduces expression of differentiation gene and Dac and Dll |
Neoplastic tumours of the larval eye-antennal epithelial tissue [78] |
|
| ||
| Troponin I overexpression |
Ras
V12 overexpression Notchintra (Act) overexpression lgl mutant RasV12 overexpression |
Tumour overgrowth or neoplastic tumour overgrowth in wing epithelial tissue [87] |
|
| ||
| Signalling pathway deregulation | ||
|
| ||
|
Ras
V12
overexpression
Results in tissue overgrowth, which depends upon EGF-EGFR activation and Arf6 mediated Hedgehog signalling |
Eye-antennal and wing epithelial tissue overgrowth [88] | |
|
| ||
| TNF-JNK signalling | Invasive neoplastic tumours in the larval eye-antennal epithelium [76, 89–91] | |
|
| ||
| Immune signalling and activation of JNK | Invasive neoplastic tumours of the adult hindgut epithelium [92] | |
|
| ||
| Ben/dUev1a E2 ubiquitin ligase overexpression Results in JNK activation (via binding Traf2) |
Invasive neoplastic tumours in the larval eye-antennal epithelium [93–95] | |
|
| ||
|
sds22 (PP1) loss of function Results in cell morphology/polarity loss Results in Myosin II, JNK activation |
Invasive neoplastic tumours of the larval eye-antennal epithelium [96] | |
|
| ||
| PP6 phosphatase (FMT, PpV) knock down Results in Tak1-JNK activation |
Invasive neoplastic tumours of the eye-antennal epithelium [97] | |
|
| ||
| Infection/inflammation Results in Imd-dTab2-dTak1-JNK signalling and MMP1 expression |
Hindgut epithelial tumour invasion [92, 98] | |
|
| ||
| Impaired Hippo pathway signalling Results in upregulation of Ras pathway genes, Upd-Jak-Stat signalling |
Eye-antennal and wing tissue overgrowth [65, 99] | |
|
| ||
| Lysosomal protein loss of function—deep orange, carnation, vps16A | Invasive neoplastic tumours of the larval eye-antennal epithelium [100] | |
|
| ||
| Autophagy loss of function—e.g., Atg8a, Atg7, Atg9, Atg1, Atg13, Syx17 Requires ROS and JNK upregulation |
Invasive neoplastic tumours of the larval eye-antennal epithelium [101] | |
|
| ||
| Chromosome remodelling complex mutation polyhomeotic Depends on ectopic Notch activation |
Invasive neoplastic tumours of the larval eye-antennal epithelium [102] | |
|
| ||
| Chinmo (BTB-POZ Zn finger transcription factor) overexpression | Overgrown tumours in the eye-antennal epithelial tissues [69] | |
|
| ||
| Fruitless (BTB-POZ Zn finger transcription factor) overexpression | Overgrown tumours in the eye-antennal epithelial tissues [69] | |
|
| ||
|
PTEN knockdown (Elevated PI3K signalling) |
Larval-Pupal tracheal epithelial tissue invasive tumours [103] | |
|
| ||
| apc (Wingless/Wnt) signalling | Adult midgut epithelial tissue overgrowth [104, 105] | |
|
| ||
|
p53, apc, pten knockdown dSmad4, apc, pten knockdown |
Adult hindgut epithelial tissue invasive tumours [106] | |
|
| ||
|
pico (MRL) overexpression chickadee (Profilin) overexpression mal (SRF cofactor gene) overexpression Requires JNK-MMP1 activity |
Glial cell overgrowth and invasion [107] | |
|
| ||
|
EGFR activation/overexpression
Depends on Ras and Hh signalling |
Eye-antennal and wing epithelial tissue overgrowth [88] | |
|
| ||
|
fat loss of function (Hippo pathway impairment) |
Eye-antennal and wing epithelial tissue overgrowth [108] | |
|
| ||
|
bantam micro-RNA expression Results in downregulation of Socs36E Leads to increased Jak-Stat signalling |
Invasive overgrowth of the larval wing epithelium [109] | |
|
| ||
|
miR-10 or miR-375 Micro-RNA expression Results in downregulation of Psq transcription factor |
Invasive overgrowth of the larval eye-antennal and wing epithelium [52] | |
|
| ||
|
miR-8 Micro-RNA expression Results in downregulation of Peanut protein expression, cytokinesis blockage, formation of polyploid cells |
Invasive overgrowth of the larval wing epithelium [110] | |
|
| ||
| PI3K pathway activation Requires Tor, Sin1, Rictor, Myc, Cyclin D-Cdk4, Rb-E2F and Cdc25 Requires RIOK1, RIOK2 |
Glia cell invasive brain tumours and eye neural-epithelium tumours [111–113] | |
|
| ||
|
Notch
intra (Act)
/Delta overexpression
Results in tissue overgrowth |
Eye-antennal and wing tissue overgrowth (reviewed by [114]) | |
|
| ||
| Notchintra (Act) overexpression | Mef2 overexpression | Invasive neoplastic tumours of the larval eye neural-epithelium [115] |
|
| ||
| Notchintra (Act) overexpression | Chinmo (BTB-POZ Zn finger transcription factor) overexpression | Overgrown tumours in the eye-antennal epithelial tissues [69] |
|
| ||
| Notchintra (Act) overexpression | Fruitless (BTB-POZ Zn finger transcription factor) overexpression | Overgrown tumours in the eye-antennal epithelial tissues [69] |
|
| ||
| Delta overexpression | Overexpression of transcription factors Psq/Lola (eyeful model) | Invasive tumours larval/pupal eye neural-epithelium, which are capable of differentiation to express ELAV [116] |
|
| ||
| Delta overexpression | Overexpression of Akt or PI3K (Dp110) | Invasive tumours larval/pupal eye neural-epithelium, which are capable of differentiation (ELAV expression) [117] |
|
| ||
| Delta overexpression Results in repression of boi gene expression and reduced Hedgehog signalling |
Overexpression of mir-7 micro-RNA Results in downregulation of ihog translation and reduced Hedgehog signalling |
Eye-antennal disc overgrowth and invasive cells cable of differentiation [118] |
|
| ||
| Delta overexpression | Overexpression of Zfh1 (Zeb1 family transcription factor gene) | Invasive tumours larval/pupal eye neural-epithelium, which are capable differentiation to express ELAV [119] |
|
| ||
| Delta overexpression Delta with Pipsqueak/Lola overexpression (eyeful model) |
Knockdown of atonal (transcription factor gene) Results in reduced JNK signalling |
Invasive tumours larval/pupal eye neural-epithelium, which are capable differentiation to express ELAV [120] |
|
| ||
| Delta overexpression Delta with Pipsqueak/Lola overexpression (eyeful model) |
Knockdown of cut (transcription factor gene) Results in increased reaper expression and elevated PI3K-Akt signalling |
Invasive tumours in larval/pupal eye-antennal epithelium, which are capable differentiation to express ELAV [121] |
|
| ||
|
Hippo pathway impairment (Yki overexpression)
Results in increased tissue growth through upregulation of cell growth (Myc), proliferation (CycE) and antiapoptotic genes (Diap1), elevation of Upd-Jak-Stat signalling |
Increased tissue growth (reviewed by [122]) | |
|
| ||
| BAP (Brahma) complex knockdown (brm, Snr1, mor, Bap111, osa) Results in upregulation of Wingless (Wnt) and Dpp signalling |
Neoplastic tumour overgrowth in larval wing epithelial tissue [123] | |
|
| ||
| Taiman (Ecdysone Receptor coactivator) overexpression Results in expression of germline stem cell factors |
Hyperplastic tumour overgrowth in larval wing epithelial tissue [124] | |
|
| ||
| Guidance receptors | ||
|
| ||
|
Frazzled (Dcc) loss of function and expression of the Caspase inhibitor P35
Results in an invasive phenotype in eye epithelial cells, but cells can differentiate |
Inhibition of JNK signalling Requires Rho1 |
Invasive, but differentiated, tumours in larval/pupal eye-antennal epithelial tissues [125] |
|
| ||
| Mitotic checkpoint, chromosome instability, DNA damage repair genes | ||
|
| ||
|
Nek2 (centrosomal kinase) overexpression |
Ret
MEN2B overexpression—elevated Ras, PI3K, Src, JNK signalling Csk− RasV12 Results in Rac1, Rho1, Wg signalling and elevated expression of Diap1, MMP1 Results in PI3K signalling |
Invasive tumours in larval eye-antennal epithelial tissue [126] |
|
| ||
|
bub3 knockdown
Results in aneuploidy |
p35 overexpression to block effector caspase activity |
Neoplastic overgrowth of wing epithelial cells [127] |
|
| ||
|
DNA repair or DNA damage checkpoint mutants
Depletion of okra (DmRAD54) or spnA (DmRAD51) (Homologous recombination of DNA double strand-breaks in G2) grp (chk1) and mei-41 (ATR) knockdown (DNA damage checkpoint) |
Ionizing irradiation and p35 overexpression Results in JNK activation, which leads to MMP1 and Wg upregulation |
Overgrowth and cell delamination/migration in the wing epithelial tissue [128] |
3.1. Caspases in Cooperative Tumorigenesis: Context Dependency
Blocking cell death in the mutant tissue (via blockage of effector caspase activity by overexpressing p35) can, in some cases, enable the survival of the mutant cells, thereby revealing their tumourigenic properties. Examples of caspases acting in a tumour-suppressor role occur in epithelial tissues containing scrib, rok, mud, Sin3a, Snr1, Csk, or frazzled mutant cells [125, 141, 166–168] or overexpressing a subunit of the Vacuolar ATPase (V-ATPase) complex, Vha44 [169]. However, caspases can also be oncogenic in some contexts. Indeed, activating certain caspases at low levels, insufficient to induce cell death (at least not rapidly), can promote an invasive phenotype [168, 170]. Similarly, caspase activity within the tumour is also required for growth of tumours generated by mutations of the endocytosis regulator, Rab5 [165]. Caspase activity is also observed in wing epithelial tumours generated by mutation of the cell polarity regulator gene, lgl, which correlates with JNK pathway activation and is important for tumour invasion [161]. Additionally, in polarity-impaired RasV12 epithelial tumours, described above, reducing cell death by knocking down caspase activity reduces tumorigenesis [57]. Thus, caspase activity can be tumour promoting or tumour suppressing, depending on context. These findings have implications for cancer therapy, which is designed to induce caspase-mediated cell death, since mild-to-moderate activation of caspases may instead promote tumour growth and invasive behaviour.
3.2. The JNK Signalling Pathway in Cooperative Tumorigenesis: Context Dependency
The JNK signalling pathway can also have context-dependent roles in tumorigenesis in Drosophila and in mammalian systems (reviewed in [143, 171–174]). In some types of cell competition, such as that induced by polarity impairment, the JNK pathway is required to promote apoptosis and therefore is inhibitory for tumour progression (acting as a tumour suppressor) [23, 91, 136]. In these cases, when JNK signalling is blocked using a kinase-dead dominant-negative JNK transgene (bskDN), tumour cells delaminate from the epithelium, overgrow, and invade into the surrounding epithelium. This occurs in clones for cell polarity regulators, such as scrib or lgl mutants, but also occurs upon overexpression of an activated version of aPKC or wild-type crb in clones in the developing eye epithelia when bskDN is coexpressed [66, 71] (Table 1). The mechanism by which JNK-independent cell invasion occurs in these cases is unknown. Interestingly, in lgl mutant clones expressing bskDN, large GFP-marked tumours are observed in the eye, and clumps of GFP-marked cells occur elsewhere in the head and also in body of the pupae/pharate adult [71]. Cooperative interactions also occur upon blocking JNK and activating other signalling pathways to promote tumorigenesis. Overexpression of the Src tyrosine protein kinase gene, Src64B, activates JNK signalling and leads to cell death in the eye-antennal epithelium, but when bskDN is coexpressed, tumour overgrowth occurs, in a mechanism involving upregulation of the actin-cytoskeletal regulators, Rac1 and Dia, as well as Ras signalling, which inhibit the Hippo pathway, thereby promoting tumour growth [79]. Similarly, in mutants affecting endocytosis, such as Vps4, blocking JNK signalling promotes the formation of neoplastic tumours in epithelial tissues, by an unknown mechanism [175].
In another model of tumorigenesis in the developing eye, mutants in frazzled (an ortholog of mammalian Deleted in Colorectal Cancer, DCC, a regulator of axon guidance), combined with the blockage of apoptosis by expression of the effector caspase inhibitor, p35, results in elevated JNK and Rho1 activity and promotes cell invasion [125]. However, photoreceptor differentiation still occurs, leading to the migration of differentiated photoreceptor cells to distant sites. Blockage of JNK signalling in frazzled mutant p35 expressing cells enhances the invasive phenotype in a Rho1-dependent manner (Table 1).
Another tumour type, where blocking JNK promotes an invasive phenotype, is the eyeful model [116] (Table 1). In this model, overexpression of the Notch ligand, Delta, combined with overexpression of the transcription factor genes, lola and pipsqueak, in the developing eye, promotes an invasive phenotype but does not affect differentiation, resulting in differentiated photoreceptor cells located at distant sites. This phenotype is dependent on the Polycomb group chromatin-remodelling factor, histone deacetylases, and reduced expression of Rbf1 (the Drosophila ortholog of the retinoblastoma tumour suppressor) [116]. Using this model, another group found that overexpression of atonal (a transcription factor gene, involved in eye differentiation) reduces the eyeful invasive phenotype, whereas knockdown of atonal enhances it [120] (Table 1). Atonal functions by inducing JNK activity and possibly enhances cell death and therefore blocking JNK results in restoration of the invasive phenotype [120]. Mammalian atonal, ATOH1, also acts as a tumour suppressor, which may also involve JNK activation [176]. Consistent with the involvement of JNK as a tumour suppressor in this context, in Delta-expressing Drosophila eye epithelial cells, blocking JNK activity also enhances the invasive phenotype [120]. How invasion occurs upon blocking JNK activity in Delta-expressing cells is unknown. Altogether, these examples indicate that blocking JNK can promote cell survival of tumourous cells and that alternate mechanisms promote cell invasion. In studies where mechanistic insights were obtained, these have indicated the involvement of Rho1 or Rac1, which are known regulators of the actin cytoskeleton in cell migration (reviewed in [177]), and the activation of these small-GTPase may very well be involved in other cases of JNK-independent cell invasion.
In contrast to the above examples that highlight a tumour suppressive role for JNK signalling, in other contexts, the JNK pathway can function as a tumour promoter, by altering cell morphology, driving cell invasion, and blocking differentiation. For example, in lgl mutant wing epithelial tissue, JNK activation promotes cell morphology changes that potentiates the loss of apicobasal cell polarity and enables tumour formation [178, 179]. Furthermore, in scrib, dlg, or lgl mutant RasV12-expressing clones in the developing eye (see below), inhibition of JNK prevents invasive behaviour of cells into the brain lobes-ventral ganglion and promotes differentiation and pupariation [66, 89–91, 93]. Similarly, in wing epithelial tissues overexpressing the Vha44 component of the V-ATPase, which activates JNK signalling and results in invasive tumours, blocking JNK suppresses the invasive phenotype [169]. Additionally, in eye epithelial tissue activation/overexpression of the Rho1 or Rac1 small GTPases (which regulate actin polymerisation and F-actin/Myosin II contractility) also cooperate with RasV12 to promote invasive overgrowth, dependent upon increased JNK activity [76, 77] (Table 1). In another model, impairment of the Sds22/PP1 phosphatase in RasV12-expressing cells in the anterior-posterior boundary of the developing wing epithelium, in a JNK-dependent manner, leads to invasive tumours [96] (Table 1). Here, Myosin II activation is also required for invasion, which mechanistically may involve regulation of the JNK pathway by Rho1-Rok-Myosin II signalling, as has been observed in other contexts [61, 76, 77, 149, 180]. Indeed, JNK's oncogenic role in cooperative tumorigenesis is evident in experiments showing that overexpressing JNK pathway genes in combination with RasV12 in the developing eye epithelium induces invasive tumour growth [76, 90, 91, 149]. Moreover, overexpression of the E2 ubiquitin ligase, Ben/dUev1a, which activates JNK signalling, also cooperates with RasV12 to promote invasive tumour growth [94] (Table 1). More recently, loss of function mutations in the PP6 phosphatase have been shown to act upstream of the Tak1 protein kinase, a JNKKK, to induce invasive tumorigenesis in RasV12-expressing eye-antennal epithelial cells [97]. Furthermore, in the adult Drosophila hindgut epithelium, JNK activation through the Egr (TNF) pathway, in response to bacterial infection, also cooperates with RasV12 to promote invasive overgrowth [92, 98] (Table 1).
In summary, the JNK pathway is an important player in cooperative tumorigenesis but dependent on context it can have a tumour-suppressing or tumour-promoting role. Due to this context dependency, which is also observed in mammalian systems [173, 174], the activation of JNK alone in a tumour is not a clear diagnostic or prognostic marker of outcome, and knowledge of other molecular defects is required to predict tumour behaviour.
3.3. Cooperation between Cell Polarity Impairment and Oncogenes
Impairment of cell polarity is a powerful force in tumorigenesis (reviewed in [15, 19, 181]). When cell polarity genes (scrib, dlg, and lgl) are mutated or knocked down in a clonal context, aberrant mitotic spindle orientation, cell polarity impairment, ectopic cell proliferation, and aberrant differentiation occur, but, despite this, malignant tumours do not form, and the mutant tissue is mostly eliminated by JNK-mediated cell death [23, 90, 91, 136, 141, 182]. However, in arguably the first demonstration of Drosophila cooperative tumorigenesis, expression of oncogenic Ras (RasV12) or Notch (Notchintra (Act)) in scrib mutant clones prevents their elimination by cell death and instead promotes cell proliferation to produce overgrown undifferentiated and invasive tumours [23, 65, 90, 91] (Table 1, Figure 2(c)). Similar cooperative tumourigenic interactions were also observed for dlg and lgl mutants and RasV12 [65] and also for lgl and NotchAct [72]. In these cooperative interactions, RasV12 and NotchAct promote cell survival and proliferation, whilst scrib mutation leads to aPKC activation, which results in impairment of the Hippo negative tissue-growth pathway, leading to the activation of the downstream cotranscriptional activator, Yki, and tissue overgrowth [47, 48, 183]. Additionally, scrib mutation promotes JNK activation, which blocks differentiation and progression to the pupal stage and leads to an invasive cell phenotype through upregulation of MMP1 (a metalloprotease, involved in degradation of the extracellular matrix), Paxillin (a regulator of integrin signalling), Robo (a guidance receptor), and various actin-cytoskeletal regulators [45, 66, 90, 184]. More recently, global expression analyses of scrib mutant tissue [148], and scrib mutant RasV12-expressing or scrib mutant NotchAct-expressing epithelial tissues [69, 99, 185–187], has revealed the spectrum of deregulated genes that contribute to cooperative tumorigenesis. In addition to members of the JNK and Hippo pathways, these include Polycomb chromatin-remodelling complex components, the BTB-POZ zinc-finger transcription factor genes, chinmo and fruitless, the Ets-family transcription factor, Ets21c, and the nuclear receptor transcription factor gene, ftz-F1. These transcription factors contribute to the switching of the differentiation state of the tissue towards a progenitor cell-like fate, deregulation of signalling pathways, and the promotion of cell proliferation, survival, and invasion. Additionally, genetic screens of scrib mutant RasV12-expressing tumours have revealed the importance of the PI3K signalling pathway [67], and chemical screens have revealed the importance of glutamate utilization enzymes, the TCA cycle, and pyrimidine synthesis [68] for tumour growth. scrib mutant RasV12-expressing tumours, in a JNK-dependent manner, upregulate the diffusible Insulin-like peptide, dILP8 [69]. This, in turn, in the prothoracic gland, leads to the downregulation of the secreted steroid hormone, Ecdysone, which is required for metamorphosis, and therefore pupariation is delayed/prevented, thereby leading to the formation of oversized (giant) larvae [188–190]. In addition, scrib mutant RasV12-expressing tumours secrete the insulin growth factor binding protein, ImpL2, which is an antagonist of Insulin signalling that results in wasting of adipose, muscle, and gonadal tissues in the larvae [191]. Thus, polarity impairment together with the Ras oncogene leads to a plethora of gene expression changes and perturbed signalling pathways, which together promote the tumourigenic phenotype, as well as affecting other tissues in the larvae. Expression profiling and functional analyses of lgl mutant epithelial tissue have revealed that, similar to scrib mutants, signalling pathways (Hippo and JNK) and cell fate genes are deregulated [72, 160, 178, 179, 192–194]. However, other signalling pathways, such as Notch, PI3K, and Wingless, are also elevated in lgl mutant tissue [72, 160, 194–196], but they have not been reported to be so in scrib mutant tissue. Therefore, the cooperative tumorigenesis mechanisms of scrib and lgl mutants with RasV12 might be slightly different.
Many features of the cooperative tumourigenic interaction between scrib mutants and oncogenic Ras are conserved in mammalian epithelial systems, both in vitro [197] and in vivo in epithelial cells of the mouse prostate, lung, breast, and skin tissue [198–201]. Whilst a complete mechanistic picture is lacking, studies in mammalian cell lines have revealed that Scrib depletion in EGF-stimulated epithelial cells elevates ERK as well as JNK signalling [197], and cell polarity perturbation leads to Hippo pathway impairment [202, 203]. Thus, at least some aspects of the mechanism of cooperation between oncogenic Ras and cell polarity genes mutations have proven to be conserved between Drosophila and mammals, and further studies are needed in mammalian systems to reveal whether other downstream events are also conserved. Furthermore, lgl mutants cooperate with overexpression of the dMyc transcription factor in the wing epithelium [50], which has also been observed for scrib downregulation and Myc in mouse mammary epithelial tissue [204], but whether similar mechanisms are involved is currently not known. Additionally, in the wing epithelial tissue, lgl mutant cells that are undergoing cell competition-mediated elimination cooperate with impaired Hippo signalling to generate overgrown neoplastic tumours [49]. However, whether this also occurs in mammalian systems is currently unknown.
Subsequent studies using polarity-impaired epithelial tumour models have revealed novel cooperating genes (see Table 1), which provide insight into mechanisms of tumorigenesis relevant to human cancer. Notable recent examples of these include overexpression of the BTB-POZ transcription factor gene, abrupt, which was discovered in a genetic screen to cooperate with scrib loss to induce neoplastic tumours in the eye-antennal epithelium [70]. Through target gene identification, abrupt overexpression was shown to cooperate with scrib mutants in tumorigenesis by downregulation of multiple differentiation genes and deregulation of the Hippo and JNK signalling pathways. Moreover, genes responsive to the steroid hormone, Ecdysone, were downregulated, which contributes to the developmental block at the larval stage, enabling the continuation of invasive tumour growth [70]. Interestingly, the Ecdysone Receptor- (ER-) associated factor, Taiman, which binds to Abrupt in ovarian tissues [205], is required for the growth of scrib mutant abrupt-overexpressing tumours, and overexpression of taiman in scrib mutant cells also leads to invasive neoplastic tumours [70]. More recently, Taiman was shown to bind to the Hippo pathway cotranscription factor, Yki, and to control the transcription of a novel set of genes that regulate germ-line stem cell identity [124], although whether these genes are also deregulated in taiman or abrupt-overexpressing scrib mutant tumours has not been investigated.
Another signalling pathway involved in scrib mutant tumorigenesis is the Slit-Robo-Ena pathway. This pathway is involved in the basal extrusion of scrib mutant tissue from the epithelium, where they die, and downregulation of this pathway results in overgrown (but noninvasive) tumours within the eye-antennal epithelium [45]. Conversely, hyperactivation of the Slit-Robo-Ena pathway in scrib mutant or wild-type cells results in a hyperextrusive phenotype, with the apically (lumenally) extruded cells forming overgrown tumours, which might occur by the peripodial membrane epithelium preventing access of the innate cellular immune system cells to the tumour [45, 46]. However, it is also possible that the lumenal microenvironment is conducive to tumour cell growth and survival, which may be dependent on morphogens, such as Dpp, produced from the peripodial epithelium [206].
Scribble module genes, but not other apical-basal cell polarity genes, were identified as Drosophila neoplastic tumour-suppressor genes; however the downregulation of Crb and Par modules cell polarity genes together with RasV12 expression in the eye-antennal epithelial also results in neoplastic tumour formation [65]. Furthermore, overexpression of Par1 cell polarity regulator, which inactivates Hippo signalling [74], cooperates with activated Notch signalling in promoting tumourous overgrowth in the eye-antennal epithelium [75], similar to that which occurs with activated Notch and scrib mutants [23] or lgl mutants [72]. It is likely that similar mechanisms are involved in the cooperation of Crb and Par module gene mutants with RasV12, as well as with Par1 and NotchAct, as occurs with scrib mutants with RasV12 or NotchAct; however formal demonstration is currently lacking. Interestingly, canoe (cno, afadin/AF-6 in mammals), a gene involved in another type of cell polarity, asymmetric cell division [207], has been recently shown to cooperate with scrib, dlg, or lgl depletion in epithelial tumorigenesis [73]. Mechanistically, this synergistic interaction involves the activation of Ras-MAPK signalling, which implicates the wild-type function of Cno as well as Scrib, Dlg, and Lgl in the repression of Ras signalling [73], as occurs with the mammalian Cno (Afadin/AF-6) and Scrib [197, 208].
3.4. Cooperative Tumorigenesis Involving Actin-Cytoskeletal Regulators
Deregulation of the actin cytoskeleton leads to cell morphology changes, increased tissue growth through impairment of the Hippo pathway, and reduced cell-cell adhesion and can promote invasive phenotypes [76, 77, 177, 209–212]. However, in a clonal context, tissue growth due to deregulated actin-cytoskeletal gene expression is restrained by JNK-mediated cell death, and therefore cell death blockage or oncogenic activation is required for tumorigenesis. Indeed, the activated small GTPases Rho1 and Rac1, which regulate the actin cytoskeleton, cooperate with RasV12 in tumorigenesis in a JNK-dependent manner [76, 77] (Table 1). Furthermore, downstream of the Rho1-GTPase, the Rok protein kinase, and activated Myosin II, which regulate F-actin filament contractility, cooperates with RasV12 to promote tumorigenesis [77]. Mechanistically, the contribution of the Rho1-Rok-Myosin II pathway to RasV12-driven tumorigenesis most likely involves JNK activation [77], and also Hippo pathway impairment, as increased F-actin contractility leads to Yki activation-induced tissue growth (reviewed in [211, 213]). Activation of Rho1, by RhoGEF2 overexpression, also cooperates with overexpression of the Abrupt BTB-POZ transcription factor in inducing neoplastic tumours of the eye-antennal epithelium by blocking expression of differentiation genes [78] (Table 1). It is likely that JNK activation and Hippo impairment are also involved in this cooperative interaction; however this remains to be confirmed.
The Src nonreceptor tyrosine protein kinase, a key regulator of the actin cytoskeleton as well as adherens junctions [214], cooperates with several oncogenes to promote tumorigenesis in Drosophila (Table 1). Activation of Src through knockdown of its negative regulator, Csk, together with RasV12 also results in invasive overgrown tumours of the eye-antennal epithelium [80–84]. Mechanistically, Src activates JNK and Stat signalling, modulates the actin cytoskeleton, and impairs Hippo signalling to promote invasive overgrowth in cooperation with RasV12 [79–83, 85]. Moreover, on a high sugar diet, Src-activated RasV12-driven tumours have an altered metabolism and elevate Wg signalling, which leads to upregulation of the Insulin-Receptor gene expression, enabling the tumour cells to become insulin-responsive and aggressively overgrow, whilst other larval tissues are insulin-resistant and hypoplastic [81, 84]. Overexpression of Src64B or Src42A also cooperates with activated Notch signalling to promote tumorigenesis in eye-antennal and wing epithelial tissue, in a mechanism requiring JNK activation in a TNF-independent manner [86]. Given the link between Src and actin-cytoskeletal regulators [85], and the discovery of a mechanism linking Rho1 to JNK activation via the JNKKK, Wallenda [149], a similar mechanism might be involved in the activation of JNK in Src NotchAct cooperative tumorigenesis. Additionally, Src64B overexpression cooperates with overexpression of the Abrupt BTB-POZ transcription factor in the eye-antennal epithelium by blocking differentiation genes and promoting a progenitor-like cell fate [78]. Although Src expression changes the repertoire of Notch target gene transcription in the NotchAct tumours [86], whether differentiation blockage is also involved in this tumour type and other Src-driven tumours remains to be determined.
Similarly, when induced in a clonal setting, overexpression of the actin-cytoskeletal regulator, Troponin I, cooperates with NotchAct expression, RasV12 expression, and lgl mutant RasV12 expression to promote tumour overgrowth by altering gene transcription [87]. Genes upregulated included those encoding the Insulin Receptor (InR), Rap1 (a Ras-related protein), and Dilp8 (insulin-related peptide), which are likely to affect tumour growth by promoting cell proliferation and in the case of Dilp8 by delaying pupariation through downregulation of Ecdysone production in the prothoracic gland.
3.5. Deregulation of Signalling Pathways in Cooperative Tumorigenesis
EGFR-Ras-MAPK. The mitogenic EGFR-Ras-MAPK signalling pathway is a powerful inducer of tissue growth but also induces differentiation in Drosophila (reviewed in [215, 216]). Moreover, this pathway is important in cancer, as mutations in Ras signalling pathway genes that elevate pathway activity are present in ~30% of human cancers (reviewed in [217–219]). Although oncogenic Ras is a potent inducer of tissue growth, high level of pathway flux leads to senescence or differentiation, thereby limiting tumorigenesis (reviewed in [220, 221]). Thus, additional mutations are required for Ras-driven malignant cancer development.
Oncogenic Ras requires EGFR signalling to potently induce tissue overgrowth in both Drosophila and human cells [88]. Mechanistically, this occurs through the endocytosis regulator, Arf6, which is important for the trafficking of the Hedgehog morphogen and activation of the Hedgehog signalling pathway. In Drosophila, activated Ras signalling cooperates with many pathways to promote tumorigenesis (Table 1). In addition to mutations/overexpression of cell polarity, actin cytoskeletal, and JNK pathway genes that cooperate with oncogenic Ras in tumorigenesis in Drosophila (discussed above), many other cooperative interactions have been revealed in various Drosophila epithelial tissues that confer either hyperplastic or neoplastic overgrowth (Table 1). Hyperplastic tumourigenic interactions include the cooperation of RasV12 with the overexpression of chinmo or fruitless BTB-POZ domain transcription factor genes [69], and with impaired Hippo pathway signalling [65, 99], which results in enhanced hyperplastic overgrowth of eye-antennal epithelial tissue. In the cooperation of Hippo pathway impairment with Ras activation, a global transcriptome analysis has provided insight into how the differentiation function of Ras signalling is reprogrammed to promote tumorigenesis, by showing that Yki elevates the expression of the Ras target gene, pointed, which is crucial for the synergistic tissue growth [99, 222].
Conversely, in the eye-antennal epithelial tissue, RasV12 cooperates with lysosomal gene loss of function to cause neoplastic overgrowth [100]. Additionally, mutations in the Polycomb complex chromatin-remodelling gene, polyhomeotic (ph), cooperate with RasV12 in a clonal context to induce eye-antennal tissue neoplastic tumours, which depends on Notch pathway activation [102]. However, loss of ph and other Polycomb complex genes, when generated in a whole eye-antennal epithelial tissue, results in neoplastic tumours, which in this context is dependent on ectopic Upd-Jak-Stat signalling [223]. These differences might depend on the level of expression and the region of the tissue affected, but, additionally, in the clonal context, the induction of cell competition might affect the cooperative mechanism involved in neoplastic tumour formation.
Interestingly, autophagy gene knockdown cooperates with RasV12 to produce different outcomes depending on context [101]. Knockdown of autophagy genes using UAS-RNAi lines via the eyeless-GAL4 driver, or, clonally, within the developing eye epithelium, enhances RasV12 hyperplastic overgrowth, whereas using the eyeless-FLP-out Tubulin-GAL4 system, which results in the strong expression of the transgenes throughout the whole eye-antennal epithelium, autophagy gene knockdown together with RasV12 expression results in neoplastic overgrowth and death at the larval-pupal stage. Mechanistically, the cooperation of RasV12 with autophagy gene knockdown, in both the hyperplastic and neoplastic tissue overgrowth effects, occurs because oncogenic Ras signalling induces autophagy in imaginal disc epithelial tissues, and consequently the blockage of autophagy at any step of the pathway results in ROS accumulation and activation of JNK signalling [101]. This finding may also be relevant to human cancer, since in human pancreatic cancers, where K-RasG12V mutations are common, downregulation of several autophagy genes correlates with poor prognosis [101]. Since autophagy inhibitors are being considered for cancer therapy (reviewed in [224]), this study highlights the need for caution with Ras-driven cancers, where inhibiting autophagy might inadvertently exacerbate cancer development.
Overexpression/activation of EGFR also cooperates with several genes in tumorigenesis in Drosophila epithelial tissues (Table 1). EGFR cooperates with impaired Hippo pathway signalling, leading to tissue overgrowth [108] and also with the overexpression of the bantam micro-RNA (which is a downstream target of Yki [225, 226] and also of EGFR signalling [227]), leading to overgrown invasive tumours [109]. EGFR cooperates with the bantam micro-RNA by elevating Jak-Stat signalling due to bantam repressing the translation of the Jak-Stat signalling inhibitor, Socs36E. Activated Ras together with knockdown of Socs36E causes similar cooperative effects, showing that, downstream of EGFR, Ras signalling is crucial for this cooperation. Elevated expression of the Snail transcription factor, a driver of the epithelial-to-mesenchymal transition (EMT), also occurred in these tumours, as well as expression of the JNK target, MMP1, suggesting that JNK activation is also involved. This group also discovered that the overexpression of micro-RNAs, mir-10, or mir-375, cooperates with overexpression of EGFR in promoting invasive overgrown tumours [52]. This cooperation occurs by downregulation of the transcription factor, Pipsqueak (Psq), which leads to increased expression of the extracellular matrix protein, Perlecan (Pcn), resulting in tumour overgrowth by a non-cell-autonomous mechanism involving the surrounding myoblast cells [52, 228] (Figure 2(e), see above). Perlecan promotes Dpp signalling in the myoblasts, supporting their proliferation, and, in turn, the myoblasts provide growth factors that promote epithelial tumorigenesis. More recently, the same group found that overexpression of another micro-RNA and the miR-200 family member, miR-8, cooperates with EGFR overexpression to result in clonal overgrowth, cell polarity loss and invasive phenotypes in the wing epithelial tissue [110]. Curiously, these tumours became polyploid, which was attributed to miR-8 repressing the translation of the Septin, Peanut, which is required for cytokinesis. However, although Peanut downregulation was required, it was not sufficient for tumorigenesis with EGFR overexpression, suggesting other miR-8 targets are also involved. These tumours also acquire a supercompetitor phenotype and are able to induce cell death of, and engulf, their neighbours. In mammalian systems, miR-200 family downregulation induces an EMT in some settings [229]; however its overexpression occurs in ovarian cancers where cells commonly exhibit polyploidy [230–233]. Thus, this unusual cooperative behaviour, identified in Drosophila, might have relevance to certain types of human cancer.
Several studies have also focused on directed modelling in Drosophila of EGFR-Ras-driven human cancers, such as lung, colorectal, and glioblastoma cancers (Table 1). In a model of Ras-driven lung cancer, RasV12 coexpression with PTEN knockdown (which elevates PI3K signalling) in the larval-pupal tracheal epithelial cells results in tracheal cell overgrowth and invasive tumours [103]. Colorectal cancer was modelled by knocking down the adenomatous polyposis coli (apc) gene and overexpressing RasV12 in the adult midgut [104, 105], which resulted in hyperplasia. In another study, the adult hindgut was used and RasV12 was expressed together with p53, apc, pten knockdown or dSmad4, apc, and pten knockdown (commonly observed mutations in human colorectal cancers), which resulted in invasive tumours [106]. Glioblastoma was modelled in Drosophila by expressing constitutively active forms of EGFR and PI3K, which is commonly observed in human glioblastomas [111, 112, 234]. Genetic analysis revealed that dMyc, Cdc25, Cdk4, and the TORC2 regulatory subunits, Sin1 and Rictor, were important in glial cell tumorigenesis in the brain and eye tissue [111]. Moreover, a genetic screen of the kinome led to the identification of RIOK1 and RIOK2 kinases, which promote mTORC-Akt signalling to drive glial tumour growth [113]. A recent study has also revealed cooperative tumorigenesis in glial tumour growth and invasion between RasV12 and overexpression of pico (a MRL family gene), chickadee (profilin, encoding an actin-cytoskeletal regulator) or Mal (encoding a cofactor of Serum Response Factor (SRF)) [107], suggesting that SRF signalling might be a novel pathway to investigate in human glioblastomas.
Delta-Notch. The Delta-Notch signalling pathways play multiple roles in tissue growth and development in Drosophila, and ectopic activation leads to overgrowth phenotypes (reviewed in [114, 235, 236]). For neoplastic tumour formation, additional gene mutations are required together with Notch-Delta overexpression/activation, as detailed below. Activated Notch was shown to cooperate with overexpression of the transcription factor Mef2, leading to disruption to the actin cytoskeleton and apicobasal cell polarity [115] (Table 1). This cooperative interaction is JNK dependent, requiring upregulation of Egr [115]. In the eyeful model, in which Delta is overexpressed with the psq and lola transcription factor genes [116], cooperative tumorigenesis occurs upon downregulation of the cut transcription factor gene, which leads to a disruption to adherens junction-mediated cell-cell adhesion and cell-basement membrane β-integrin-mediated adhesion, causing increased invasion [121] (Table 1). In these cooperative interactions of cut downregulation with Delta overexpression, or with the eyeful model, upregulation of the cell death gene, reaper (rpr), and elevated PI3K-Akt signalling are involved [121]. The invasive phenotype of these tumours required MMPs, which is a JNK target, but whether JNK was also involved was not determined. Since caspase activation and JNK signalling have been previously linked to invasive cell behaviour in the wing epithelium [168], it is possible that JNK and caspase activation are also involved in the invasive phenotype of cut downregulation in the eyeful model. Additionally, PI3K or Akt overexpression has been previously shown to cooperate with Delta overexpression in the eye epithelial tissue to induce an overgrown invasive phenotype, which might be relevant to human cancer, particularly T cell acute lymphoblastic leukaemia, where Notch and Akt pathway activation often occurs [117, 237]. The Delta-driven invasive phenotype of the eyeful model was suppressible by overexpression of the miR-200 family micro-RNA, miR-8 [119]. This tumour-suppressor role for miR-8 is in contrast to its oncogenic role observed in another study [110] and highlights that, like JNK and caspases, miR-8 also has a context-dependent role in tumorigenesis. Whilst human miR-200 family micro-RNAs are considered regulators of the epithelial phenotype and tumour suppressors (reviewed in [238]), these discoveries in Drosophila highlight that more research is needed to determine whether the miR-200 family also have context-dependent effects in human cancer.
Mechanistically, in the eyeful model, miR-8 blocks the invasive phenotype by repressing the translation of the Notch ligand, Serrate, and the zinc-finger transcription factor, Zfh1 (an ortholog of mammalian ZEB1, which is an EMT inducer), and, consistent with this, coexpression of Delta, or Serrate, with Zfh1 cooperatively promotes an invasive phenotype [119]. This mechanism might be important in mammalian cancer, since JAGGED1 (mammalian ortholog of Delta/Serrate) is regulated by the miR-8 orthologs, miR-200c, and miR-141, in colorectal cancer cell lines [119], and reduced miR-200 expression is associated with coupregulation of JAGGED1 and ZEB1 proteins in pancreatic and basal-type breast cancer cell lines [239].
The same group also found that another micro-RNA, mir-7, when overexpressed, enhances Delta-driven tumour overgrowth and promotes invasion in the eye-antennal epithelium, although the cells were still capable of differentiating [118] (Table 1). In this case, the cooperation occurred via blocking Hedgehog pathway signalling, which normally acts to restrict Delta/Serrate-Notch signalling during eye development. mir-7 reduced translation of the Hedgehog receptor mRNA, ihog (interference hedgehog), whereas Notch signalling blocked transcription of the coreceptor gene, boi (brother of ihog), thereby leading to reduced Hedgehog signalling and enhancing Delta-Notch-driven tumour growth and invasion. Consistent with the mechanism, blocking Hedgehog signalling by knocking down expression of the Hh pathway transcription factor, Ci, also cooperated with Delta overexpression to phenocopy the effect of overexpression of mir-7 and Delta [118]. These studies may provide insights into some forms of human cancer, where the mir-7 ortholog is overexpressed and oncogenic, such as lung and skin cancers [240], or the Ihog orthologs (BOC and CDO) are downregulated or have a tumour-suppressor functions, such as in pancreatic cancer [241] or rhabdomyosarcoma [242].
Hippo. The Hippo tissue-growth control pathway consists of a protein kinase cascade involving Hippo and Warts protein kinases, which when activated, leads to the Warts-mediated phosphorylation and inactivation of the Yki cotranscriptional activator, thereby limiting tissue growth (reviewed in [243]). Hippo is regulated by multiple upstream inputs, including signalling pathways, cell polarity, and mechanical cues (reviewed in [122, 183, 244, 245]). Due to its powerful effect in controlling tissue growth, downregulation of the Hippo pathway is commonly observed in Drosophila cooperative tumorigenesis, as well as in human cancers (reviewed in [18, 122, 245, 246]). In addition, to the examples described above, which reveal the cooperation of Hippo pathway impairment in cooperative tumorigenesis with cell polarity impairment and oncogenic Ras, Yki overexpression has also been shown to cooperate with the knockdown of the Brahma (Brm) chromatin-remodelling complex [123] and overexpression of the Taiman transcription regulator [124] (Table 1). Impairment of the Brm-BAP chromatin-remodelling complex (using brm, snr1, or osa mutants) in epithelial tissues promotes cell cycle entry, alters Ras, Notch, and Dpp signalling, and deregulates Ecdysone responsive genes [247–254]. Brm complex knockdown also deregulates the Hippo pathway in epithelial tissues [255, 256], and therefore it is perhaps surprising that Brm downregulation cooperates with Yki overexpression [123]. Cooperation might occur, due to Brm complex knockdown downregulating the Ras signalling pathway, which decreases cell proliferation and survival [251, 254], and since Yki overexpression provides a strong cell proliferation and survival signal, it would be expected to override decreased cell survival exhibited by Brm complex knockdown alone. However, the recent study showed that Brm-BAP complex depletion, together with Yki overexpression, results in upregulation of Dpp and Wg morphogens leading to neoplastic tumour overgrowth in the larval wing epithelial tissue [123]. Cooperation with Yki and Taiman overexpression occurs by a unique mechanism involving the ectopic expression of germ-line stem cell genes in wing epithelial tissue [124]. This occurs because Yki and Taiman can form a complex leading to upregulation of a new spectrum of Yki targets normally not expressed in imaginal disc epithelial tissue, which alters differentiation.
Mitotic Checkpoints, Chromosome Instability and DNA Damage Repair Genes. Genes important in mitotic checkpoints, DNA repair, and genomic integrity play important tumour-suppressor functions in preventing cancer (reviewed in [157]). Indeed, knockdown of the spindle-assembly checkpoint (SAC) gene, bub3, which leads to chromosome instability (CIN) and aneuploidy, results in neoplastic tumorigenesis in the wing epithelial tissue when cell death is blocked [127, 140] (Table 1). The results from one group suggested that the mechanism by which this occurs is a SAC-independent function of Bub3 [127], but the second study revealed a novel mechanism that was induced by aneuploidy and cell delamination [140] (see below). A role for the DNA damage checkpoint and DNA repair after exposure to ionizing radiation (IR) has also been revealed in cooperative tumorigenesis [128] (Table 1). Here, IR together with apoptosis inhibition results in overgrowth and cell delamination/migration in the wing epithelial tissue, which is enhanced by knockdown of the DNA repair genes, okra (DmRAD54) or spnA (DmRAD51), which are involved in homologous recombination of DNA double-strand breaks, as well as by knockdown of the DNA damage checkpoint genes, grp (chk1) and mei-41 (ATR).
An unusual example of cooperative tumorigenesis concerns the Nek2 (NimA related kinase 2) centrosome kinase, which is also involved in the SAC, and its loss of function leads to CIN (reviewed in [257]). However, overexpression of Nek2, which is not expected to cause CIN, cooperates with oncogenic pathways to drive neoplastic tumour formation without apparent effects on CIN [126] (Table 1). Here, expression of the activated form of the Ret tyrosine kinase (RetMEN2B), which mimics oncogenic mutations in human thyroid cancer, and RasV12 together with Nek2 overexpression, leads to invasive overgrowth of the eye-antennal epithelial tissue. Similar cooperativity occurs with mutant csk, together with RasV12 and Nek2 overexpression. The Ret oncogene results in increased signalling through the Ras-MAPK, PI3K, Src, and JNK pathways [258–260]. Nek2 overexpression results in increased Wg signalling and altered expression of Rho1, Rac1, and E-Cadherin, leading to altered cell morphology [126]. Coexpression of Nek2 and oncogenic Ret lead to enhanced local invasion and distant metastases. Mechanistically, Nek and Ret result in elevated expression of MMP1, which is expected to promote extracellular matrix degradation. Additionally, Nek and Ret lead to elevated expression of Diap1 (an antiapoptotic protein, and target of Hippo and Jak-Stat signalling), as well as Wg expression and PI3K signalling, which together are expected to drive tumour growth. A similar cooperative invasive phenotype was observed with elevated Src activity (csk mutant) with RasV12 [126]. PI3K signalling was critical for the cooperative invasive phenotype, since inhibiting PI3K suppressed the cooperative behaviour. Although Nek2 is thought to have a tumour-suppressor function due to its role in the SAC and chromosome stability, it is also oncogenic and drugs are being developed to inhibit its function in cancer therapy (reviewed in [257]). Thus, this study in Drosophila provides insight into how Nek2 alone, as well as when combined with oncogenic mutations, promotes invasive properties [126], which is relevant to the understanding of Nek2-overexpressing human cancers.
In summary, the above examples of cooperative tumorigenesis in Drosophila tissues, and the delineation of the mechanisms involved, provide insights towards the understanding of various human cancers where these pathways are deregulated and present possible novel avenues for therapeutic intervention.
4. The Effect of the Tumour on Normal Tissue Growth and Intertumoural Cooperation
Not only does the mutant cell depend on the surrounding microenvironment for its proliferation and neoplastic transformation, but there are also examples in Drosophila where the tumour induces overgrowth of the genetically wild-type surrounding epithelial cells (termed non-cell-autonomous overgrowth) (Figure 3). The sophisticated genetics of Drosophila have enabled modelling of complex intertumoural cooperation, through generating genetically different populations of epithelial cells. In mammalian systems, tumours also exert non-cell-autonomous effects on cells in their microenvironment that affect tumour development (reviewed in [153–156]). Additionally, cancer heterogeneity, where different populations of tumour cells interact to promote tumorigenesis, is a recognized phenomenon in mammalian cancer (reviewed in [53, 261–264]). We will now discuss Drosophila models of cooperative tumorigenesis, where non-cell-autonomous cell proliferation or tumour heterogeneity occurs.
Figure 3.
Non-cell-autonomous overgrowth. Examples of different types of non-cell-autonomous overgrowth. Mutant cells are in pink, wild-type cells are in blue, hemocytes are in grey, and the basement membrane (basal lamina) is in purple. (a) Cell polarity or endocytosis mutant cells are induced by JNK signalling to undergo cell death and induce non-cell-autonomous overgrowth of the surrounding wild-type cells. In vps25 or tsg101 (ept) endocytic mutants, which also show apicobasal cell polarity defects, ectopic activation of Notch signalling leads to the expression and secretion of the Dome-Jak-Stat pathway ligand, Upd, which promotes non-cell-autonomous proliferation and overgrowth of surrounding tissue. In scrib mutant cells, elevated JNK signalling, and impaired Hippo signalling, leads to transcriptional upregulation of Upd, which activates Dom-Jak-Stat signalling in the surrounding wild-type cells, thereby inducing their proliferation. (b) Undead cells, where apoptosis is initiated, but effector caspase activity is blocked, emit morphogens (such as Wg, Dpp, and Hh) that promote proliferation of their wild-type epithelial neighbours, thereby leading to non-cell-autonomous overgrowth. (c) Mitochondrial mutants expressing RasV12 lead to non-cell-autonomous overgrowth. The mitochondrial impairment results in the production of ROS, which induces JNK activation, which, in turn, results in Hippo pathway impairment, leading to expression of the Yki targets, Upd and Wg. Upd elevates Jak/Stat signalling and Wg induces Wg pathway signalling in the surrounding wild-type cells to promote their overgrowth.
4.1. Non-Cell-Autonomous Cell Proliferation
In clonal settings, the initiation of cell death within mutant tissue results in signalling events that lead to non-cell-autonomous cell proliferation of the surrounding wild-type epithelium (reviewed in [28, 33, 265–269]) (Figure 3). In scrib mutant clones, non-cell-autonomous proliferative effects on the surrounding wild-type epithelial tissue occurs due to JNK-mediated expression of Upd, which in turn induces the Dome-Jak-Stat signalling pathway in surrounding cells [47, 137] (Figure 3(a)). Additionally, there is also evidence that the Hippo pathway is impaired in wild-type cells surrounding scrib mutant clones [270]. Likewise, clones mutant for the endocytic trafficking genes, vps25 or tsg101 (ept), also induce proliferation of the surrounding wild-type cells, due to impairment of the Hippo pathway and/or upregulation of the Dome-Jak-Stat signalling pathway [47, 137, 270, 271] (Figure 3(a)), which occurs through the aberrant activation of Notch signalling, leading to upregulation of Upd (Dome ligand) in the mutant cells [271, 272]. vps25 mutant cells also lead to Hippo pathway impairment in the surrounding wild-type cells [270, 271], which may partially involve signalling through the Fat atypical cadherin [270]. Ectopic Notch signalling alone also leads to non-cell-autonomous tissue growth, as well as cell-autonomous proliferation [271, 273]. Similarly, activation of the Hh pathway in clones, in a Notch-dependent manner, also results in non-cell-autonomous cell proliferation and expression of the Diap1 cell death inhibitor in the surrounding wild-type cells [274, 275]. Since Diap1 is a transcriptional target of Stat and Yki [276, 277], Jak-Stat or Hippo pathway deregulation may be involved. However, in all these examples, cell proliferation is limited. By contrast, in settings where cell death of the mutant cells is blocked by decreasing or preventing caspase activity, substantial overgrowth of the wild-type tissue occurs (Table 2). For example, elevated Hh signalling in clones blocked for cell death induces the Dpp morphogen production in the mutant cells, which activates the Dpp signalling pathway in the surrounding wild-type cells to induce non-cell-autonomous tissue growth [135]. In this setting, Yki was also upregulated non-cell-autonomously, and together with the Dpp signalling-activated transcription factor, Mad (Smad), induces expression of the bantam micro-RNA to promote non-cell-autonomous tissue growth [135] (Table 2). Another example of non-cell-autonomous tissue overgrowth occurs with “undead cells” [267–269]. Undead cells are generated when cell death is initiated by upregulation of an apoptosis regulator (such as Hid or Rpr), but apoptosis is blocked by expression of the p35 effector caspase inhibitor [59, 129–134] (Figure 3(b)). The undead cells continually express and secrete the morphogens, Wg, Dpp, or Hh, which act to elevate these signalling pathways in the surrounding wild-type cells, thereby inducing their uncontrolled proliferation. The induction of non-cell-autonomous tissue overgrowth by undead cells is dependent on the activation of JNK signalling in the undead cells, which transcriptionally upregulates the expression of the morphogen genes [129, 268] (Figure 3(b), Table 2). Consistent with this, strong activation of JNK together with Raf (protein kinase that functions downstream of Ras signalling) results in non-cell-autonomous overgrowth of surrounding wild-type cells [136]. Additionally, overexpression of RasV12 together with the actin-cytoskeletal genes, RhoGEF2, Rac1, or activated alleles of Rho1 (Rho1V14), can initially induce varying degrees of non-cell-autonomous tissue growth; however tumour growth predominates over time [76, 77]. Similar effects are also observed with overexpression of the abrupt transcription factor gene with RhoGEF2 or Src64B [78]. Conversely, overexpression of abrupt with Rac1 leads to a strong non-cell-autonomous tissue overgrowth [78]. The mechanism by which this non-cell-autonomous overgrowth is induced is currently unknown but may involve the cells acquiring an undead-like state.
Table 2.
Tumour-wild-type tissue and intertumoural interactions.
| Non-cell autonomous overgrowth/tumorigenesis | ||
|---|---|---|
| 1st mutation/mechanism | 2nd mutation/mechanism | Phenotype/references |
| hid/rpr overexpression | p35 expression (blocks effector caspase activity) generates “undead cells” | Eye-antennal or wing epithelial tissue non-cell autonomous overgrowth [59, 129–134] |
| Initiator caspase (Dronc) activation | Dependent on ROS production, JNK activation, Dpp, Wg upregulation/secretion | |
|
| ||
| ptc mutant | ark (apaf1) mutant | Eye-antennal and wing epithelial tissue non-cell autonomous overgrowth [135] |
| Hh pathway upregulation |
Results in upregulation of Dpp secretion from mutant cells and elevated Dpp signalling and Yki activity in the wild-type cells | |
|
| ||
| Raf GOF overexpression | Strong activation of JNK (hepACT) | Non-cell autonomous overgrowth and morphology changes of eye-antennal epithelial tissue [136] |
|
| ||
| Rac1 overexpression | Abrupt (BTB-POZ Zn finger transcription factor) | Eye-antennal epithelial tissue non-cell autonomous overgrowth [78] |
|
| ||
| Intertumoural cooperation | ||
|
| ||
|
Ras
V12
overexpression
|
scrib mutant cells next to RasV12 overexpressing cells | Invasive neoplastic tumours of the larval eye neural-epithelium [137] |
| Activation of JNK and Upd upregulation and Jak-Stat signalling | ||
|
| ||
| Mitochondrial dysfunction in RasV12 overexpressing cells next to RasV12 overexpressing cells | Invasive neoplastic tumours of the larval eye neural-epithelium [138, 139] | |
| Results in ROS production, upregulation of JNK, deregulation of Hippo, secretion of Wg and Upd | ||
|
| ||
| Chromosome instability | Cell death blockage (p35 expression) | Invasive tumours in the wing epithelium [61, 140] |
| Induced by spindle assembly or spindle-assembly checkpoint mutants leading to cell delamination (rod, bub3, asp) |
Results in Metabolic stress, ROS induced-JNK activation in epithelial cells promoting cell delamination | |
| Results in Secretion of Wg, Upd from delaminated cells, promoting the proliferation of epithelial tumour cells | ||
|
| ||
| Spindle orientation defects | Cell death blockage (p35 expression) with mud knockdown | Invasive tumours in the eye-antennal or wing epithelium [61, 141, 142] |
| Due to mutants/knockdown of genes involved in spindle alignment (mud, scrib, dlg) leading to cell delamination |
Ras V12 or p35 expression with scrib or dlg | |
| Results in Rho1-Wnd induced JNK activation in epithelial cells promoting cell delamination | ||
| Results in secretion of Wg, Upd from delaminated cells, promoting the proliferation of epithelial tumour cells | ||
|
| ||
| Dosage compensation mechanism mutants | p35 overexpression or deletion of rpr, hid, grim to block cell death | Invasive tumours in the wing epithelium [142] |
| Knockdown of msl1 or msl2 in males or Sxl in females | Results in ROS induced-JNK activation | |
| Results in MMP1 expression in delaminating cells | ||
A different form of non-cell-autonomous cell proliferation occurs without cell death of the mutant cells but instead results in the cells acquiring a senescent secretory phenotype [138, 139] (Figure 3(c); Table 2). This new mechanism was discovered in a genetic screen for mutations that cooperate with RasV12 in the developing eye, which revealed that mutations in mitochondrial oxidative phosphorylation genes together with RasV12 lead to non-cell-autonomous overgrowth of the surrounding wild-type tissue. Mechanistically, this involves the generation of reactive oxygen species (ROS) by the mitochondrial-impaired cells, which then lead to JNK pathway activation. In turn, JNK activation results in impairment of Hippo pathway signalling, and, consequently, elevated Yki induces expression of Upd and Wg, which, respectively, induce signalling through the Dome-Jak-Stat and Wg signalling pathways in surrounding wild-type cells, leading to tissue overgrowth. Similar mechanisms of non-cell-autonomous tissue growth or tumorigenesis induced by senescent secretory cells are also observed in other settings in Drosophila and in human cancers (reviewed in [28, 53]).
4.2. Intertumoural Cooperation
Recent studies in Drosophila have revealed mechanisms by which cells of different populations can cooperate to generate neoplastic tumours [61, 137–140, 278, 279] (Figure 4, Table 2). Remarkably, RasV12 cells generated next to scrib mutant cells (interclonal), rather than in the same cells (intraclonal, Figure 4(a)), became neoplastically transformed [137] (Figure 4(b)). This occurred by upregulation of JNK signalling and Upd expression in the scrib− cells, which induces Dome-Jak-Stat signalling in the neighbouring RasV12 cells, thereby promoting neoplastic overgrowth. A similar mechanism involving Dome-Jak-Stat signalling, together with Wg signalling, induces neoplastic overgrowth of RasV12 cells when generated next to mitochondrial respiratory chain gene mutant cells that were also overexpressing RasV12 [138] (Figure 4(c)). The RasV12-expressing mitochondrial gene mutant cells exhibit properties of cellular senescence and acquire a secretory phenotype, through ROS, p53, and JNK upregulation, leading to JNK signalling amplification, similar to that which occurs in response to cellular stress [280], which leads to the transcriptional upregulation of Upd and Wg expression [139].
Figure 4.
Different modes of cooperative tumorigenesis. Examples of different modes of cooperative tumorigenesis. Mutant cells are in pink, RasV12-expressing cells are in green, wild-type cells are in blue, delaminated mutant cells are in dark pink, and the basement membrane (basal lamina) is in purple. (a) Intraclonal cooperation with cell polarity mutants and RasV12: JNK activation in the tumour cells cooperates with oncogenic Ras signalling to promote tumour overgrowth and invasion. (b) Interclonal cooperation with cell polarity mutants and RasV12: JNK signalling and Hippo pathway impairment in the scrib mutant cells lead to the production of Upd, which induces Dome-Jak-Stat signalling in the surrounding RasV12-expressing cells, thereby inducing their overgrowth and invasion. (c) Interclonal cooperation with a mitochondrial mutant overexpressing RasV12 and RasV12-expressing surrounding cells: Upd and Wg are produced by the mitochondrial mutant RasV12-expressing surrounding cells (see Figure 3(c)), which induce upregulation of Dome-Jak-Stat and Wg signalling, respectively, in the RasV12 cells to induce their neoplastic overgrowth and invasion. (d) Delaminating cells cooperation: in tumours generated by chromosome instability (CIN) mutants (rod, bub3, and asp) or mutants that effect spindle orientation (scrib, dlg, and mud), some cells delaminate, resulting in two populations of cells, which in the case of spindle orientation mutants are not genetically different. The delaminated cell population produces the Wg and Upd ligands to upregulate Wg and Dome-Jak-Stat pathways, respectively, in the nondelaminated cells, thereby inducing their proliferation.
Aneuploidy, generated by mutations in spindle-assembly checkpoint (SAC) genes (asp, rod, and bub3) together with blocking apoptosis, also results in cooperative tumorigenesis involving tumour heterogeneity [61, 140, 142, 278] (Figure 4(d)). In this case, CIN induces metabolic stress leading to ROS production, which, via the JNKKK, Ask, activates JNK, leading to cell delamination and basal extrusion of the aneuploid cells. JNK activity also induces Upd and Wg upregulation and secretion from the aneuploid cells, which act on the mutant epithelial cell population to drive their proliferation through the Dome-Jak-Stat and Wg signalling pathways, respectively [61, 140, 142]. The delaminated cells (mesenchymal-like cells) are unable to proliferate but contribute to tumorigenesis by secreting Upd and Wg. Thus, two cellular populations, with the same original genotype, one epithelial and the other mesenchymal-like (which contains aneuploid cells), cooperate to generate the neoplastic tumour. Strikingly, spindle orientation defective mutants, such as mud (an ortholog of mammalian Numa, which is important for the localization of Dynein/Dynactin motor proteins in spindle orientation), or the cell polarity mutants, scrib and dlg, also lead to the generation of two cellular populations, which, upon blocking cell death, cooperate to promote neoplastic tumour formation [61, 141]. In this case, spindle misorientation causes the extrusion of cells from the epithelium, where they lose cell-cell adhesion and their epithelial morphology. Thus, these two populations are not genetically different, although, due to the loss of cell polarity and altered signalling pathways in the delaminated cells, they are likely to have different transcriptomes. Mechanistically, cooperation involves induction of JNK signalling, through a Rho1-Wallenda pathway leading to transcriptional upregulation of Upd and Wg [61]. Additionally, knockdown of dosage compensation genes (msl1 or msl2 in males, or Sxl in females), which result in genome-wide expression changes on the X chromosome similar to aneuploidy, also result in tumour heterogeneity-induced cooperative tumorigenesis when cell death is blocked [142] (Table 2). Here ROS and JNK signalling are induced and the delaminating cells upregulate MMP1. Thus, dosage compensatory gene mutants, when cell death is blocked, show a similar mechanism to CIN, due to SAC gene knockdown, in inducing tumour heterogeneity.
In summary, the analysis of cooperative tumorigenesis in Drosophila has revealed several different mechanisms by which tumour heterogeneity is generated and elucidated the mechanism by which two populations of cells can cooperate in promoting neoplastic tumours. Since heterogeneity is a common phenomenon and an important factor in human cancer (reviewed in [53, 261–264]), the findings in Drosophila may provide insight into understanding how heterogeneity arises and contributes to human cancer progression.
5. Conclusions and Future Perspectives
We have highlighted in this review how damaged cells are recognized and eliminated in epithelial tissue (cell competition) and the dreadful consequences of the failure of these surveillance mechanisms or of cell death. The persistence of damaged cells, by blocking cell death or by the activation of various oncogenes, drives hyperplastic or neoplastic tumorigenesis by various mechanisms. In cooperative tumorigenesis, which can occur in a myriad of ways, signalling pathways are deregulated to promote tumorigenesis, of which the JNK, Upd (IL6)-Dome-Jak-Stat, and Wg pathways are highly prominent. Interestingly, tissue regeneration also requires these pathways [281], suggesting that normal tissue repair mechanisms are usurped during neoplastic tumorigenesis. As with human cancer, the interaction of the epithelial tumours with their microenvironment plays an important role in neoplastic tumour development in Drosophila models. Moreover, non-cell-autonomous cell proliferation induced by the mutant cells affects the surrounding wild-type tissues, causing aberrant tissue overgrowth when mutant cell apoptosis is blocked. Additionally, Drosophila studies have revealed how tumour heterogeneity arises and has delineated novel mechanisms by which different cell populations are involved in cooperative tumorigenesis. These include the interplay between scrib mutant cells juxtaposed to oncogenic Ras-expressing cells and the senescence-induced secretory phenotypes generated by mitochondrial respiratory chain mutations together with oncogenic Ras, where each induces neoplastic tumours non-cell-autonomously. Furthermore, cell polarity, spindle orientation, and spindle-assembly checkpoint mutants, which cause delamination of cells, together with blockage of cell death or the activation of oncogenic pathways, lead to tumour heterogeneity and the crosstalk between two cellular populations to promote neoplastic tumorigenesis. Taken together, Drosophila studies have revealed novel cooperative gene interactions in tumorigenesis and the mechanisms by which this occurs, which is of relevance to human cancer. In the Omics age of human cancer research, the plethora of information that is being generated is often difficult to fathom, and functional studies are required to reveal the important cancer drivers and how they cooperate. Due to its sophisticated genetics, Drosophila will continue to play an important role in revealing the function of cancer-causing genes in vivo and elucidating their mechanisms of action in cooperative tumorigenesis. Moreover, Drosophila is now emerging as a highly suitable model organism for the discovery of anticancer compounds against various cancer types, which can be then developed for clinical use, with reduced need for animal models (reviewed in [3, 10, 13, 14, 282, 283]). Thus, in the new age of pharmacogenetics, Drosophila will continue to play a fruitful role in elucidating new cooperative gene interactions in cancer and identifying anticancer compounds that then can be harnessed for anticancer therapy.
Acknowledgments
The authors thank John E. La Marca for comments on the manuscript. Helena E. Richardson is supported by funds from the La Trobe Institute of Molecular Science and La Trobe University, and Marta Portela was supported by Juan de la Cierva-Incorporacion postdoctoral fellowship (IJCI-2014-19272) from the Spanish Ministerio de Ciencia e Innovacion (MICINN). The authors apologize to the Drosophila community, if they have inadvertently missed any relevant literature in the writing of this review.
Conflicts of Interest
The authors declare that they have no conflicts of interest regarding the publication of this article.
References
- 1.Ugur B., Chen K., Bellen H. J. Drosophila tools and assays for the study of human diseases. Disease Models & Mechanisms. 2016;9(3):235–244. doi: 10.1242/dmm.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nature Reviews Genetics. 2005;6(1):9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- 3.Sonoshita M., Cagan R. L. Modeling human cancers in Drosophila. Current Topics in Developmental Biology. 2017;121:287–309. doi: 10.1016/bs.ctdb.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Bennett D., Lyulcheva E., Cobbe N. Drosophila as a Potential Model for Ocular Tumors. Ocular Oncology and Pathology. 2015;1(3):190–199. doi: 10.1159/000370155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L., Kounatidis I., Ligoxygakis P. Drosophila as a model to study the role of blood cells in inflammation, innate immunity and cancer. Frontiers in Cellular and Infection Microbiology. 2014;3, article 113 doi: 10.3389/fcimb.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tipping M., Perrimon N. Drosophila as a Model for Context-Dependent Tumorigenesis. Journal of Cellular Physiology. 2014;229(1):27–33. doi: 10.1002/jcp.24427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell G. P., Thompson B. J. Colorectal cancer progression: Lessons from Drosophila? Seminars in Cell & Developmental Biology. 2014;28:70–77. doi: 10.1016/j.semcdb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez C. Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nature Reviews Cancer. 2013;13(3):172–183. doi: 10.1038/nrc3461. [DOI] [PubMed] [Google Scholar]
- 9.Rudrapatna V. A., Cagan R. L., Das T. K. Drosophila cancer models. Developmental Dynamics. 2012;241(1):107–118. doi: 10.1002/dvdy.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson H. E., Willoughby L., Humbert P. O. In Encyclopedia of Life Sciences. Wiley; 2015. Screening for anti-cancer drugs in Drosophila. [Google Scholar]
- 11.Cheng L., Parsons L. M., Richardson H. E. Modelling cancer in Drosophila–The next generation (version 2.0) In Encyclopedia Life Sciences: Wiley; 2013. [Google Scholar]
- 12.Rosales-Nieves A. E., González-Reyes A. Genetics and mechanisms of ovarian cancer: Parallels between Drosophila and humans. Seminars in Cell & Developmental Biology. 2014;28:104–109. doi: 10.1016/j.semcdb.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 13.Yadav A. K., Srikrishna S., Gupta S. C. Cancer Drug Development Using Drosophila as an in vivo Tool: From Bedside to Bench and Back. Trends in Pharmacological Sciences. 2016;37(9):789–806. doi: 10.1016/j.tips.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Das T. K., Cagan R. L. A Drosophila approach to thyroid cancer therapeutics. Drug Discovery Today: Technologies. 2013;10(1):e65–e71. doi: 10.1016/j.ddtec.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brumby A. M., Helena E. R. Using Drosophila melanogaster to map human cancer pathways. Nature Reviews Cancer. 2005;5(8):626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- 16.Watson K. L., Justice R. W., Bryant P. J. Drosophila in cancer research: The first fifty tumor suppressor genes. Journal of Cell Science. 1994;107(18):19–33. doi: 10.1242/jcs.1994.Supplement_18.4. [DOI] [PubMed] [Google Scholar]
- 17.Gateff E. Tumor suppressor and overgrowth suppressor genes of drosophila melanogaster: Developmental aspects. The International Journal of Developmental Biology. 1994;38(4):565–590. [PubMed] [Google Scholar]
- 18.Harvey K. F., Zhang X., Thomas D. M. The Hippo pathway and human cancer. Nature Reviews Cancer. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 19.Elsum I., Yates L., Humbert P. O., Richardson H. E. The Scribble-Dlg-Lgl polarity module in development and cancer: From flies to man. Essays in Biochemistry. 2012;53(1):141–168. doi: 10.1042/BSE0530141. [DOI] [PubMed] [Google Scholar]
- 20.Herranz H., Eichenlaub T., Cohen S. M. Cancer in Drosophila. Imaginal Discs as a Model for Epithelial Tumor Formation. Current Topics in Developmental Biology. 2016;116:181–199. doi: 10.1016/bs.ctdb.2015.11.037. [DOI] [PubMed] [Google Scholar]
- 21.Beira J. V., Paro R. The legacy of Drosophila imaginal discs. Chromosoma. 2016;125(4):573–592. doi: 10.1007/s00412-016-0595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou S. X., Singh S. R. Stem-cell-based tumorigenesis in adult Drosophila. Current Topics in Developmental Biology. 2017;121:311–337. doi: 10.1016/bs.ctdb.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Brumby A. M., Richardson H. E. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO Journal. 2003;22(21):5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilder D., Perrimon N. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 2000;403(6770):676–680. doi: 10.1038/35001108. [DOI] [PubMed] [Google Scholar]
- 25.Bilder D., Li M., Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289(5476):113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 26.Droujinine I. A., Perrimon N. Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annual Review of Genetics. 2016;50:539–570. doi: 10.1146/annurev-genet-121415-122024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamori Y., Deng W.-M. Tissue-Intrinsic Tumor Hotspots: Terroir for Tumorigenesis. Trends in Cancer. 2017;3(4):259–268. doi: 10.1016/j.trecan.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enomoto M., Vaughen J., Igaki T. Non-autonomous overgrowth by oncogenic niche cells: Cellular cooperation and competition in tumorigenesis. Cancer Science. 2015;106(12):1651–1658. doi: 10.1111/cas.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston L. A. Socializing with MYC: Cell competition in development and as a model for premalignant cancer. Cold Spring Harbor Perspectives in Medicine. 2014;4, article a014274(4) doi: 10.1101/cshperspect.a014274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastor-Pareja J. C., Xu T. Dissecting social cell biology and tumors using Drosophila genetics. Annual Review of Genetics. 2013;47:51–74. doi: 10.1146/annurev-genet-110711-155414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merino M. M., Levayer R., Moreno E. Survival of the Fittest: Essential Roles of Cell Competition in Development, Aging, and Cancer. Trends in Cell Biology. 2016;26(10):776–788. doi: 10.1016/j.tcb.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Di Gregorio A., Bowling S., Rodriguez T. A. Cell Competition and Its Role in the Regulation of Cell Fitness from Development to Cancer. Developmental Cell. 2016;38(6):621–634. doi: 10.1016/j.devcel.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Morata G., Ballesteros-Arias L. Cell competition, apoptosis and tumour development. The International Journal of Developmental Biology. 2015;59(1-3):79–86. doi: 10.1387/ijdb.150081gm. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto M., Ohsawa S., Kunimasa K., Igaki T. The ligand Sas and its receptor PTP10D drive tumour-suppressive cell competition. Nature. 2017;542(7640):246–250. doi: 10.1038/nature21033. [DOI] [PubMed] [Google Scholar]
- 35.Meyer S. N., Amoyel M., Bergantinos C., et al. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science. 2014;346, article 1258236(6214) doi: 10.1126/science.1258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levayer R., Moreno E. Mechanisms of cell competition: Themes and variations. The Journal of Cell Biology. 2013;200(6):689–698. doi: 10.1083/jcb.201301051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casas-Tinto S., Torres M., Moreno E. The flower code and cancer development. Clinical and Translational Oncology. 2011;13(1):5–9. doi: 10.1007/s12094-011-0610-4. [DOI] [PubMed] [Google Scholar]
- 38.Rhiner C., López-Gay J. M., Soldini D., et al. Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila. Developmental Cell. 2010;18(6):985–998. doi: 10.1016/j.devcel.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Di Giacomo S., Sollazzo M., Paglia S., Grifoni D. MYC, cell competition, and cell death in cancer: The inseparable triad. Gene. 2017;8(4, article 120) doi: 10.3390/genes8040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston L. A. Competitive Interactions between Cells: Death, Growth, and Geography. Science. 2009;324(5935):1679–1682. doi: 10.1126/science.1163862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziosi M., Baena-López L. A., Grifoni D., et al. dMyc functions downstream of yorkie to promote the supercompetitive behavior of hippo pathway mutant Cells. PLoS Genetics. 2010;6, article e1001140(9) doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neto-Silva R. M., de Beco S., Johnston L. A. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the drosophila homolog of Yap. Developmental Cell. 2010;19(4):507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodrigues A. B., Zoranovic T., Ayala-Camargo A., et al. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development. 2012;139(21):4051–4061. doi: 10.1242/dev.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincent J.-P., Kolahgar G., Gagliardi M., Piddini E. Steep Differences in Wingless Signaling Trigger Myc-Independent Competitive Cell Interactions. Developmental Cell. 2011;21(2):366–374. doi: 10.1016/j.devcel.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaughen J., Igaki T. Slit-Robo Repulsive Signaling Extrudes Tumorigenic Cells from Epithelia. Developmental Cell. 2016;39(6):683–695. doi: 10.1016/j.devcel.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson H. E., Portela M. Robo-Enabled Tumor Cell Extrusion. Developmental Cell. 2016;39(6):629–631. doi: 10.1016/j.devcel.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Chen C.-L., Schroeder M. C., Kango-Singh M., Tao C., Halder G. Tumor suppression by cell competition through regulation of the Hippo pathway. Proceedings of the National Acadamy of Sciences of the United States of America. 2012;109(2):484–489. doi: 10.1073/pnas.1113882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doggett K., Grusche F. A., Richardson H. E., Brumby A. M. Loss of the Drosophila cell polarity regulator Scribbled promotes epithelial tissue overgrowth and cooperation with oncogenic Ras-Raf through impaired Hippo pathway signaling. BMC Developmental Biology. 2011;11, article 57 doi: 10.1186/1471-213X-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menéndez J., Pérez-Garijo A., Calleja M., Morata G. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proceedings of the National Acadamy of Sciences of the United States of America. 2010;107(33):14651–14656. doi: 10.1073/pnas.1009376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Froldi F., Ziosi M., Garoia F., et al. The lethal giant larvae tumour suppressor mutation requires dMyc oncoprotein to promote clonal malignancy. BMC Biology. 2010;8, article 33 doi: 10.1186/1741-7007-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroeder M. C., Chen C.-L., Gajewski K., Halder G. A non-cell-autonomous tumor suppressor role for Stat in eliminating oncogenic scribble cells. Oncogene. 2013;32(38):4471–4479. doi: 10.1038/onc.2012.476. [DOI] [PubMed] [Google Scholar]
- 52.Herranz H., Weng R., Cohen S. M. Crosstalk between epithelial and mesenchymal tissues in tumorigenesis and imaginal disc development. Current Biology. 2014;24(13):1476–1484. doi: 10.1016/j.cub.2014.05.043. [DOI] [PubMed] [Google Scholar]
- 53.Ohsawa S., Takemoto D., Igaki T. Dissecting tumour heterogeneity in flies: Genetic basis of interclonal oncogenic cooperation. The Journal of Biochemistry. 2014;156(3):129–136. doi: 10.1093/jb/mvu045. [DOI] [PubMed] [Google Scholar]
- 54.Ohsawa S., Sugimura K., Takino K., Xu T., Miyawaki A., Igaki T. Elimination of oncogenic neighbors by JNK-mediated engulfment in Drosophila. Developmental Cell. 2011;20(3):315–328. doi: 10.1016/j.devcel.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Igaki T., Pastor-Pareja J. C., Aonuma H., Miura M., Xu T. Intrinsic Tumor Suppression and Epithelial Maintenance by Endocytic Activation of Eiger/TNF Signaling in Drosophila. Developmental Cell. 2009;16(3):458–465. doi: 10.1016/j.devcel.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parisi F., Stefanatos R. K., Strathdee K., Yu Y., Vidal M. Transformed epithelia trigger non-tissue-autonomous tumor suppressor response by adipocytes via activation of toll and eiger/TNF signaling. Cell Reports. 2014;6(5):855–867. doi: 10.1016/j.celrep.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 57.Pérez E., Lindblad J. L., Bergmann A. Tumor-promoting function of apoptotic caspases by an amplification loop involving ROS, macrophages and JNK in Drosophila. ELife. 2017;6, article e26747 doi: 10.7554/eLife.26747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diwanji N., Bergmann A. The beneficial role of extracellular reactive oxygen species in apoptosis-induced compensatory proliferation. Flight Journal. 2017;11(1):46–52. doi: 10.1080/19336934.2016.1222997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fogarty C. E., Diwanji N., Lindblad J. L., et al. Extracellular Reactive Oxygen Species Drive Apoptosis-Induced Proliferation via Drosophila Macrophages. Current Biology. 2016;26(5):575–584. doi: 10.1016/j.cub.2015.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katheder N. S., Khezri R., O'Farrell F., et al. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541(7637):417–420. doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muzzopappa M., Murcia L., Milán M. Feedback amplification loop drives malignant growth in epithelial tissues. Proceedings of the National Acadamy of Sciences of the United States of America. 2017;114(35):E7291–E7300. doi: 10.1073/pnas.1701791114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pastor-Pareja J. C., Wu M., Xu T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Disease Models & Mechanisms. 2008;1(2-3):144–154. doi: 10.1242/dmm.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casas-Tintó S., Lolo F.-N., Moreno E. Active JNK-dependent secretion of Drosophila Tyrosyl-tRNA synthetase by loser cells recruits haemocytes during cell competition. Nature Communications. 2015;6, article 10022 doi: 10.1038/ncomms10022. [DOI] [PubMed] [Google Scholar]
- 64.Wood W., Faria C., Jacinto A. Distinct mechanisms regulate hemocyte chemotaxis during development and wound healing in Drosophila melanogaster. The Journal of Cell Biology. 2006;173(3):405–416. doi: 10.1083/jcb.200508161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pagliarini R. A., Xu T. A Genetic Screen in Drosophila for Metastatic Behavior. Science. 2003;302(5648):1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- 66.Leong G. R., Goulding K. R., Amin N., Richardson H. E., Brumby A. M. Scribble mutants promote aPKC and JNK-dependent epithelial neoplasia independently of Crumbs. BMC Biology. 2009;7, article 1741:p. 62. doi: 10.1186/1741-7007-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willecke M., Toggweiler J., Basler K. Loss of PI3K blocks cell-cycle progression in a Drosophila tumor model. Oncogene. 2011;30(39):4067–4074. doi: 10.1038/onc.2011.125. [DOI] [PubMed] [Google Scholar]
- 68.Willoughby L. F., Schlosser T., Manning S. A., et al. An in vivo large-scale chemical screening platform using Drosophila for anti-cancer drug discovery. Disease Models & Mechanisms. 2013;6(2):521–529. doi: 10.1242/dmm.009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doggett K., Turkel N., Willoughby L. F., et al. BTB-zinc finger oncogenes are required for ras and notch-driven tumorigenesis in drosophila. PLoS ONE. 2015;10, article 132987(7) doi: 10.1371/journal.pone.0132987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turkel N., Sahota V. K., Bolden J. E., et al. The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in Drosophila melanogaster through Maintaining a Progenitor-like Cell State. PLoS Genetics. 2013;9, article e1003627(7) doi: 10.1371/journal.pgen.1003627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grzeschik N. A., Parsons L. M., Richardson H. E. Lgl, the SWH pathway and tumorigenesis: It's a matter of context & competition! Cell Cycle. 2010;9(16):3202–3212. doi: 10.4161/cc.9.16.12633. [DOI] [PubMed] [Google Scholar]
- 72.Khan S. J., Bajpai A., Alam M. A., et al. Epithelial neoplasia in Drosophila entails switch to primitive cell states. Proceedings of the National Acadamy of Sciences of the United States of America. 2013;110(24):E2163–E2172. doi: 10.1073/pnas.1212513110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rives-Quinto N., Franco M., de Torres-Jurado A., Carmena A. Synergism between canoe and scribble mutations causes tumor-like overgrowth via Ras activation in neural stem cells and epithelia. Development. 2017;144(14):2570–2583. doi: 10.1242/dev.148171. [DOI] [PubMed] [Google Scholar]
- 74.Huang H.-L., Wang S., Yin M.-X., et al. Par-1 Regulates Tissue Growth by Influencing Hippo Phosphorylation Status and Hippo-Salvador Association. PLoS Biology. 2013;11, aticle e1001620(8) doi: 10.1371/journal.pbio.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bayraktar J., Zygmunt D., Carthew R. W. Par-1 kinase establishes cell polarity and functions in Notch signaling in the Drosophila embryo. Journal of Cell Science. 2006;119(4):711–721. doi: 10.1242/jcs.02789. [DOI] [PubMed] [Google Scholar]
- 76.Brumby A. M., Goulding K. R., Schlosser T., et al. Identification of novel Ras-cooperating oncogenes in Drosophila melanogaster: a RhoGEF/Rho-family/JNK pathway is a central driver of tumorigenesis. Genetics. 2011;188(1):105–125. doi: 10.1534/genetics.111.127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khoo P., Allan K., Willoughby L., Brumby A. M., Richardson H. E. In Drosophila, RhoGEF2 cooperates with activated Ras in tumorigenesis through a pathway involving Rho1-Rok-Myosin-II and JNK signalling. Disease ModelS & Mechanisms. 2013;6(3):661–678. doi: 10.1242/dmm.010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turkel N., Portela M., Poon C., Li J., Brumby A. M., Richardson H. E. Cooperation of the BTB-Zinc finger protein, Abrupt, with cytoskeletal regulators in Drosophila epithelial tumorigenesis. Biology Open. 2015;4(8):1024–1039. doi: 10.1242/bio.012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Enomoto M., Igaki T. Src controls tumorigenesis via JNK-dependent regulation of the Hippo pathway in Drosophila. EMBO Reports. 2013;14(1):65–72. doi: 10.1038/embor.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vidal M., Warner S., Read R., Cagan R. L. Differing Src signaling levels have distinct outcomes in Drosophila. Cancer Research. 2007;67(21):10278–10285. doi: 10.1158/0008-5472.CAN-07-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hirabayashi S., Cagan R. L. Salt-inducible kinases mediate nutrient- sensing to link dietary sugar and tumorigenesis in Drosophila. eLife. 2015;4, aticle e08501(2015) doi: 10.7554/eLife.08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vidal M., Larson D. E., Cagan R. L. Csk-deficient boundary cells are eliminated from normal drosophila epithelia by exclusion, migration, and apoptosis. Developmental Cell. 2006;10(1):33–44. doi: 10.1016/j.devcel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 83.Read R. D., Bach E. A., Cagan R. L. Drosophila C-terminal Src kinase negatively regulates organ growth and cell proliferation through inhibition of the Src, Jun N-terminal kinase, and STAT pathways. Molecular and Cellular Biology. 2004;24(15):6676–6689. doi: 10.1128/MCB.24.15.6676-6689.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirabayashi S., Baranski T. J., Cagan R. L. Transformed drosophila cells evade diet-mediated insulin resistance through wingless signaling. Cell. 2013;154(3):664–675. doi: 10.1016/j.cell.2013.06.030Article. doi: 10.1016/j.cell.2013.06.030Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rudrapatna V. A., Bangi E., Cagan R. L. A Jnk-Rho-Actin remodeling positive feedback network directs Src-driven invasion. Oncogene. 2014;33(21):2801–2806. doi: 10.1038/onc.2013.232. [DOI] [PubMed] [Google Scholar]
- 86.Ho D. M., Pallavi S. K., Artavanis-Tsakonas S. The notch-mediated hyperplasia circuitry in drosophila reveals a Src-JNK signaling axis. eLife. 2015;4, aticle e05996(2015) doi: 10.7554/eLife.05996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Casas-Tintó S., Maraver A., Serrano M., Ferrús A. Troponin-I enhances and is required for oncogenic overgrowth. Oncotarget . 2016;7(33):52631–52642. doi: 10.18632/oncotarget.10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chabu C., Li D.-M., Xu T. EGFR/ARF6 regulation of Hh signalling stimulates oncogenic Ras tumour overgrowth. Nature Communications. 2017;8, article 14688 doi: 10.1038/ncomms14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cordero J. B., Macagno J. P., Stefanatos R. K., Strathdee K. E., Cagan R. L., Vidal M. Oncogenic ras diverts a host TNF tumor suppressor activity into tumor promoter. Developmental Cell. 2010;18(6):999–1011. doi: 10.1016/j.devcel.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uhlirova M., Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO Journal. 2006;25(22):5294–5304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Igaki T., Pagliarini R. A., Xu T. Loss of Cell Polarity Drives Tumor Growth and Invasion through JNK Activation in Drosophila. Current Biology. 2006;16(11):1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 92.Bangi E., Pitsouli C., Rahme L. G., Cagan R., Apidianakis Y. Immune response to bacteria induces dissemination of Ras-activated Drosophila hindgut cells. EMBO Reports. 2012;13(6):569–576. doi: 10.1038/embor.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma X., Yang L., Yang Y., Li M., Li W., Xue L. dUev1a modulates TNF-JNK mediated tumor progression and cell death in Drosophila. Developmental Biology. 2013;380(2):211–221. doi: 10.1016/j.ydbio.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 94.Ma X., Shao Y., Zheng H., Li M., Li W., Xue L. Src42A modulates tumor invasion and cell death via Ben/dUev1a-mediated JNK activation in Drosophila. Cell Death & Disease. 2013;4(10, article e864) doi: 10.1038/cddis.2013.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma X., Li W., Yu H., et al. Bendless modulates JNK-mediated cell death and migration in Drosophila. Cell Death & Differentiation. 2014;21(3):407–415. doi: 10.1038/cdd.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang Y., Scott K. L., Kwak S.-J., Chen R., Mardon G. Sds22/PP1 links epithelial integrity and tumor suppression via regulation of myosin II and JNK signaling. Oncogene. 2011;30(29):3248–3260. doi: 10.1038/onc.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma X., Lu J.-Y., Dong Y., Li D., Malagon J. N., Xu T. PP6 Disruption Synergizes with Oncogenic Ras to Promote JNK-Dependent Tumor Growth and Invasion. Cell Reports. 2017;19(13):2657–2664. doi: 10.1016/j.celrep.2017.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Christofi T., Apidianakis Y. Ras-oncogenic Drosophila hindgut but not midgut cells use an inflammation-like program to disseminate to distant sites. Gut Microbes. 2013;4(1):54–59. doi: 10.4161/gmic.22429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pascual J., Jacobs J., Sansores-Garcia L., et al. Hippo Reprograms the Transcriptional Response to Ras Signaling. Developmental Cell. 2017;42(6):667–680.e4. doi: 10.1016/j.devcel.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 100.Chi C., Zhu H., Han M., Zhuang Y., Wu X., Xu T. Disruption of lysosome function promotes tumor growth and metastasis in Drosophila. The Journal of Biological Chemistry. 2010;285(28):21817–21823. doi: 10.1074/jbc.M110.131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Manent J., Banerjee S., De Matos Simoes R., et al. Autophagy suppresses Ras-driven epithelial tumourigenesis by limiting the accumulation of reactive oxygen species. Oncogene. 2017;36(40):5576–5592. doi: 10.1038/onc.2017.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martinez A.-M., Schuettengruber B., Sakr S., Janic A., Gonzalez C., Cavalli G. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nature Genetics. 2009;41(10):1076–1082. doi: 10.1038/ng.414. [DOI] [PubMed] [Google Scholar]
- 103.Levine B. D., Cagan R. L. Drosophila Lung Cancer Models Identify Trametinib plus Statin as Candidate Therapeutic. Cell Reports. 2016;14(6):1477–1487. doi: 10.1016/j.celrep.2015.12.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martorell Ò., Merlos-Suárez A., Campbell K., et al. Conserved mechanisms of tumorigenesis in the Drosophila adult midgut. PLoS ONE. 2014;9, article e88413(2) doi: 10.1371/journal.pone.0088413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang C., Zhao R., Huang P., et al. APC loss-induced intestinal tumorigenesis in Drosophila: Roles of Ras in Wnt signaling activation and tumor progression. Developmental Biology. 2013;378(2):122–140. doi: 10.1016/j.ydbio.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 106.Bangi E., Murgia C., Teague A. G. S., Sansom O. J., Cagan R. L. Functional exploration of colorectal cancer genomes using Drosophila. Nature Communications. 2016;7, article 13615 doi: 10.1038/ncomms13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Taylor E., Alqadri N., Dodgson L., et al. MRL proteins cooperate with activated Ras in glia to drive distinct oncogenic outcomes. Oncogene. 2017;36(30):4311–4322. doi: 10.1038/onc.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garoia F., Grifoni D., Trotta V., Guerra D., Pezzoli M. C., Cavicchi S. The tumor suppressor gene fat modulates the EGFR-mediated proliferation control in the imaginal tissues of Drosophila melanogaster. Mechanisms of Development. 2005;122(2):175–187. doi: 10.1016/j.mod.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 109.Herranz H., Hong X., Hung N. T., Mathijs Voorhoeve P., Cohen S. M. Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes & Development. 2012;26(14):1602–1611. doi: 10.1101/gad.192021.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eichenlaub T., Cohen S. M., Herranz H. Cell competition drives the formation of metastatic tumors in a drosophila model of epithelial tumor formation. Current Biology. 2016;26(4):419–427. doi: 10.1016/j.cub.2015.12.042. [DOI] [PubMed] [Google Scholar]
- 111.Read R. D., Cavenee W. K., Furnari F. B., Thomas J. B. A Drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genetics. 2009;5, article e1000374(2) doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Witte H. T., Jeibmann A., Klämbt C., Paulus W. Modeling glioma growth and invasion in Drosophila melanogaster. Neoplasia. 2009;11(9):882–888. doi: 10.1593/neo.09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Read R. D., Fenton T. R., Gomez G. G., et al. A Kinome-Wide RNAi Screen in Drosophila Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma. PLoS Genetics. 2013;9, article e1003253(2) doi: 10.1371/journal.pgen.1003253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ho D. M., Artavanis-Tsakonas S. The Notch-Mediated Proliferation Circuitry. Current Topics in Developmental Biology. 2016;116:17–33. doi: 10.1016/bs.ctdb.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 115.Pallavi S. K., Ho D. M., Hicks C., Miele L., Artavanis-Tsakonas S. Notch and Mef2 synergize to promote proliferation and metastasis through JNK signal activation in Drosophila. EMBO Journal. 2012;31(13):2895–2907. doi: 10.1038/emboj.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ferres-Marco D., Gutierrez-Garcia I., Vallejo D. M., Bolivar J., Gutierrez-Aviño F. J., Dominguez M. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439(7075):430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 117.Palomero T., Sulis M. L., Cortina M. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nature Medicine. 2007;13(10):1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Da Ros V. G., Gutierrez-Perez I., Ferres-Marco D., Dominguez M. Dampening the signals transduced through hedgehog via microRNA miR-7 facilitates notch-induced tumourigenesis. PLoS Biology. 2013;11, article e1001554(5) doi: 10.1371/journal.pbio.1001554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vallejo D. M., Caparros E., Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO Journal. 2011;30(4):756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bossuyt W., De Geest N., Aerts S., Leenaerts I., Marynen P., Hassan B. A. The atonal proneural transcription factor links differentiation and tumor formation in Drosophila. PLoS Biology. 2009;7(2, article no. e40) doi: 10.1371/journal.pbio.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhai Z., Ha N., Papagiannouli F., et al. Antagonistic regulation of apoptosis and differentiation by the cut transcription factor represents a tumor-suppressing mechanism in drosophila. PLoS Genetics. 2012;8, article e1002582(3) doi: 10.1371/journal.pgen.1002582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sun S. G., Wu S., Zhang L. The discovery and expansion of Hippo signaling pathway in Drosophila model. Yi Chuan. 2017;39:537–545. doi: 10.16288/j.yczz.17-051. [DOI] [PubMed] [Google Scholar]
- 123.Song S., Herranz H., Cohen S. M. The chromatin remodeling BAP complex limits tumor-promoting activity of the Hippo pathway effector Yki to prevent neoplastic transformation in. Disease Models & Mechanisms. 2017;10(10):1201–1209. doi: 10.1242/dmm.030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang C., Robinson B. S., Xu W., et al. The Ecdysone Receptor Coactivator Taiman Links Yorkie to Transcriptional Control of Germline Stem Cell Factors in Somatic Tissue. Developmental Cell. 2015;34(2):168–180. doi: 10.1016/j.devcel.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vanzomeren-Dohm A., Sarro J., Flannery E., Duman-Scheel M. The drosophila netrin receptor frazzled/DCC functions as an invasive tumor suppressor. BMC Developmental Biology. 2011;11, article 41 doi: 10.1186/1471-213X-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Das T. K., Dana D., Paroly S. S., et al. Centrosomal kinase Nek2 cooperates with oncogenic pathways to promote metastasis. Oncogenesis. 2013;2, article e69 doi: 10.1038/oncsis.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Da Silva S. M., Moutinho-Santos T., Sunkel C. E. A tumor suppressor role of the Bub3 spindle checkpoint protein after apoptosis inhibition. The Journal of Cell Biology. 2013;201(3):385–393. doi: 10.1083/jcb.201210018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dekanty A., Barrio L., Milán M. Contributions of DNA repair, cell cycle checkpoints and cell death to suppressing the DNA damage-induced tumorigenic behavior of Drosophila epithelial cells. Oncogene. 2015;34(8):978–985. doi: 10.1038/onc.2014.42. [DOI] [PubMed] [Google Scholar]
- 129.Pérez-Garijo A., Shlevkov E., Morata G. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development. 2009;136(7):1169–1177. doi: 10.1242/dev.034017. [DOI] [PubMed] [Google Scholar]
- 130.Pérez-Garijo A., Martín F. A., Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131(22):5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- 131.Pérez-Garijo A., Martín F. A., Struhl G., Morata G. Dpp signaling and the induction of neoplastic tumors by caspase-inhibited apoptotic cells in Drosophila. Proceedings of the National Acadamy of Sciences of the United States of America. 2005;102(49):17664–17669. doi: 10.1073/pnas.0508966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ryoo H. D., Gorenc T., Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the wingless signaling pathways. Developmental Cell. 2004;7(4):491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 133.Kondo S., Senoo-Matsuda N., Hiromi Y., Miura M. DRONC coordinates cell death and compensatory proliferation. Molecular and Cellular Biology. 2006;26(19):7258–7268. doi: 10.1128/MCB.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huh J. R., Guo M., Hay B. A. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase dronc in a nonapoptotic role. Current Biology. 2004;14(14):1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 135.Kagey J. D., Brown J. A., Moberg K. H. Regulation of Yorkie activity in Drosophila imaginal discs by the Hedgehog receptor gene patched. Mechanisms of Development. 2012;129(9-12):339–349. doi: 10.1016/j.mod.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Uhlirova M., Jasper H., Bohmann D. Non-cell-autonomous induction of tissue overgrowth by JNK/Ras cooperation in a Drosophila tumor model. Proceedings of the National Acadamy of Sciences of the United States of America. 2005;102(37):13123–13128. doi: 10.1073/pnas.0504170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu M., Pastor-Pareja J. C., Xu T. Interaction between RasV12 and scribbled clones induces tumour growth and invasion. Nature. 2010;463(7280):545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ohsawa S., Sato Y., Enomoto M., Nakamura M., Betsumiya A., Igaki T. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature. 2012;490(7421):547–551. doi: 10.1038/nature11452. [DOI] [PubMed] [Google Scholar]
- 139.Nakamura M., Ohsawa S., Igaki T. Mitochondrial defects trigger proliferation of neighbouring cells via a senescence-associated secretory phenotype in Drosophila. Nature Communications. 2014;5, article 5264 doi: 10.1038/ncomms6264. [DOI] [PubMed] [Google Scholar]
- 140.Dekanty A., Barrio L., Muzzopappa M., Auer H., Milán M. Aneuploidy-induced delaminating cells drive tumorigenesis in Drosophila epithelia. Proceedings of the National Acadamy of Sciences of the United States of America. 2012;109(50):20549–20554. doi: 10.1073/pnas.1206675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nakajima Y.-I., Meyer E. J., Kroesen A., McKinney S. A., Gibson M. C. Epithelial junctions maintain tissue architecture by directing planar spindle orientation. Nature. 2013;500(7462):359–362. doi: 10.1038/nature12335. [DOI] [PubMed] [Google Scholar]
- 142.Clemente-Ruiz M., Murillo-Maldonado J. M., Benhra N., et al. Gene Dosage Imbalance Contributes to Chromosomal Instability-Induced Tumorigenesis. Developmental Cell. 2016;36(3):290–302. doi: 10.1016/j.devcel.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 143.Vidal M. The dark side of fly TNF: An ancient developmental proof reading mechanism turned into tumor promoter. Cell Cycle. 2010;9(19):3851–3856. doi: 10.4161/cc.9.19.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Andersen D. S., Colombani J., Palmerini V., et al. The Drosophila TNF receptor Grindelwald couples loss of cell polarity and neoplastic growth. Nature. 2015;522(7557):482–486. doi: 10.1038/nature14298. [DOI] [PubMed] [Google Scholar]
- 145.Lolo F.-N., Casas-Tintó S., Moreno E. Cell Competition Time Line: Winners Kill Losers, which Are Extruded and Engulfed by Hemocytes. Cell Reports. 2012;2(3):526–539. doi: 10.1016/j.celrep.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 146.Lolo F.-N., Tintó S. C., Moreno E. How winner cells cause the demise of loser cells: Cell competition causes apoptosis of suboptimal cells: Their dregs are removed by hemocytes, thus preserving tissue homeostasis. BioEssays. 2013;35(4):348–353. doi: 10.1002/bies.201200156. [DOI] [PubMed] [Google Scholar]
- 147.Tamori Y., Suzuki E., Deng W.-M. Epithelial Tumors Originate in Tumor Hotspots, a Tissue-Intrinsic Microenvironment. PLoS Biology. 2016;14, article e1002537(9) doi: 10.1371/journal.pbio.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bunker B. D., Nellimoottil T. T., Boileau R. M., Classen A. K., Bilder D. The transcriptional response to tumorigenic polarity loss in Drosophila. eLife. 2015;2015(4) doi: 10.7554/eLife.03189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma X., Chen Y., Zhang S., et al. Rho1-Wnd signaling regulates loss-of-cell polarity-induced cell invasion in Drosophila. Oncogene. 2016;35(7):846–855. doi: 10.1038/onc.2015.137. [DOI] [PubMed] [Google Scholar]
- 150.Wu C., Chen C., Dai J., et al. Toll pathway modulates TNF-induced JNK-dependent cell death in Drosophila. Open Biology. 2015;5, article 140171(7) doi: 10.1098/rsob.140171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kajita M., Fujita Y. EDAC: Epithelial defence against cancer - Cell competition between normal and transformed epithelial cells in mammals. The Journal of Biochemistry. 2015;158(1):15–23. doi: 10.1093/jb/mvv050. [DOI] [PubMed] [Google Scholar]
- 152.Maruyama T., Fujita Y. Cell competition in mammals — novel homeostatic machinery for embryonic development and cancer prevention. Current Opinion in Cell Biology. 2017;48:106–112. doi: 10.1016/j.ceb.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 153.Anastasiou D. Tumour microenvironment factors shaping the cancer metabolism landscape. British Journal of Cancer. 2017;116(3):277–286. doi: 10.1038/bjc.2016.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wong G. S., Rustgi A. K. Matricellular proteins: priming the tumour microenvironment for cancer development and metastasis. British Journal of Cancer. 2013;108(4):755–761. doi: 10.1038/bjc.2012.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kenny P. A., Bissell M. J. Tumor reversion: Correction of malignant behavior by microenvironmental cues. International Journal of Cancer. 2003;107(5):688–695. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liotta L. A., Kohn E. C. The microenvironment of the tumour—host interface. Nature. 2001;411(6835):375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 157.Hanahan D., Weinberg R. A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 158.Hirabayashi S. The interplay between obesity and cancer: A fly view. Disease Models & Mechanisms. 2016;9(9):917–926. doi: 10.1242/dmm.025320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hariharan I. K., Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annual Review of Genetics. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- 160.Grifoni D., Sollazzo M., Fontana E., Froldi F., Pession A. Multiple strategies of oxygen supply in Drosophila malignancies identify tracheogenesis as a novel cancer hallmark. Scientific Reports. 2015;5, article 9061 doi: 10.1038/srep09061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Calleja M., Morata G., Casanova J. Tumorigenic Properties of Drosophila Epithelial Cells Mutant for lethal giant larvae. Developmental Dynamics. 2016;245(8):834–843. doi: 10.1002/dvdy.24420. [DOI] [PubMed] [Google Scholar]
- 162.Wang C.-W., Purkayastha A., Jones K. T., Thaker S. K., Banerjee U. In vivo genetic dissection of tumor growth and the Warburg effect. eLife. 2016;5, article e18126 doi: 10.7554/eLife.18126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rosin D., Schejter E., Volk T., Shilo B.-Z. Apical accumulation of the Drosophila PDGF/VEGF receptor ligands provides a mechanism for triggering localized actin polymerization. Development. 2004;131(9):1939–1948. doi: 10.1242/dev.01101. [DOI] [PubMed] [Google Scholar]
- 164.Das T. K., Cagan R. L. KIF5B-RET Oncoprotein Signals through a Multi-kinase Signaling Hub. Cell Reports. 2017;20(10):2368–2383. doi: 10.1016/j.celrep.2017.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ballesteros-Arias L., Saavedra V., Morata G. Cell competition may function either as tumour-suppressing or as tumour-stimulating factor in Drosophila. Oncogene. 2014;33(35):4377–4384. doi: 10.1038/onc.2013.407. [DOI] [PubMed] [Google Scholar]
- 166.Das T. K., Sangodkar J., Negre N., Narla G., Cagan R. L. Sin3a acts through a multi-gene module to regulate invasion in Drosophila and human tumors. Oncogene. 2013;32(26):3184–3197. doi: 10.1038/onc.2012.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xie G., Chen H., Jia D., et al. The SWI/SNF complex protein Snr1 is a tumor suppressor in drosophila imaginal tissues. Cancer Research. 2017;77(4):862–873. doi: 10.1158/0008-5472.CAN-16-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rudrapatna V. A., Bangi E., Cagan R. L. Caspase signalling in the absence of apoptosis drives Jnk-dependent invasion. EMBO Reports. 2013;14(2):172–177. doi: 10.1038/embor.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Petzoldt A. G., Gleixner E. M., Fumagalli A., Vaccari T., Simons M. Elevated expression of the V-ATPase C subunit triggers JNK-dependent cell invasion and overgrowth in a Drosophila epithelium. Disease Models & Mechanisms. 2013;6(3):689–700. doi: 10.1242/dmm.010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Portela M., Richardson H. E. Death takes a holiday-non-apoptotic role for caspases in cell migration and invasion. EMBO Reports. 2013;14(2):107–108. doi: 10.1038/embor.2012.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang X., Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharmacologica Sinica. 2008;29(11):1275–1288. doi: 10.1111/j.1745-7254.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bangi E. Drosophila at the intersection of infection, inflammation, and cancer. Frontiers in Cellular and Infection Microbiology. 2013;3, article 103 doi: 10.3389/fcimb.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wagner E. F., Nebreda Á. R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nature Reviews Cancer. 2009;9(8):537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 174.Heasley L. E., Han S.-Y. JNK regulation of oncogenesis. Molecules and Cells. 2006;21(2):167–173. [PubMed] [Google Scholar]
- 175.Rodahl L. M., Haglund K., Sem-Jacobsen C., et al. Disruption of Vps4 and JNK Function in Drosophila Causes Tumour Growth. PLoS ONE. 2009;4(2):p. e4354. doi: 10.1371/journal.pone.0004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bossuyt W., Kazanjian A., De Geest N., et al. Atonal homolog 1 is a tumor suppressor gene. PLoS Biology. 2009;7(2):0311–0326. doi: 10.1371/journal.pbio.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jansen S., Gosens R., Wieland T., Schmidt M. Paving the Rho in cancer metastasis: Rho GTPases and beyond. Pharmacology & Therapeutics. 2017 doi: 10.1016/j.pharmthera.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 178.Zhu M., Xin T., Weng S., et al. Activation of JNK signaling links lgl mutations to disruption of the cell polarity and epithelial organization in Drosophila imaginal discs. Cell Research. 2010;20(2):242–245. doi: 10.1038/cr.2010.2. [DOI] [PubMed] [Google Scholar]
- 179.Sun G., Irvine K. D. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Developmental Biology. 2011;350(1):139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Warner S. J., Yashiro H., Longmore G. D. The Cdc42/Par6/aPKC Polarity Complex Regulates Apoptosis-Induced Compensatory Proliferation in Epithelia. Current Biology. 2010;20(8):677–686. doi: 10.1016/j.cub.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Humbert P. O., Russell S. M., Smith L., Richardson H. E. The Scribble-Dlg-Lgl Module in Cell Polarity Regulation. In: Ebnet K., editor. Cell Polarity. Vol. 1. Switzerland: Springer International Publishing; 2015. pp. 65–111. [Google Scholar]
- 182.Igaki T. Correcting developmental errors by apoptosis: Lessons from Drosophila JNK signaling. Apoptosis. 2009;14(8):1021–1028. doi: 10.1007/s10495-009-0361-7. [DOI] [PubMed] [Google Scholar]
- 183.Richardson H. E., Portela M. Tissue growth and tumorigenesis in Drosophila: cell polarity and the Hippo pathway. Current Opinion in Cell Biology. 2017;48:1–9. doi: 10.1016/j.ceb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 184.Külshammer E., Uhlirova M. The actin cross-linker filamin/cheerio mediates tumor malignancy downstream of JNK signaling. Journal of Cell Science. 2013;126(4):927–938. doi: 10.1242/jcs.114462. [DOI] [PubMed] [Google Scholar]
- 185.Atkins M., Potier D., Romanelli L., et al. An Ectopic Network of Transcription Factors Regulated by Hippo Signaling Drives Growth and Invasion of a Malignant Tumor Model. Current Biology. 2016;26(16):2101–2113. doi: 10.1016/j.cub.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 186.Davie K., Jacobs J., Atkins M., et al. Discovery of transcription factors and regulatory regions driving in vivo tumor development by ATAC-seq and FAIRE-seq open chromatin profiling. PLoS Genetics. 2015;11(2):p. e1004994. doi: 10.1371/journal.pgen.1004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kulshammer E., Mundorf J., Kilinc M., Frommolt P., Wagle P., Uhlirova M. Interplay among Drosophila transcription factors Ets21c, Fos and Ftz-F1 drives JNK-mediated tumor malignancy. Disease Models & Mechanisms. 2015;8(10):1279–1293. doi: 10.1242/dmm.020719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hariharan I. K. How growth abnormalities delay "puberty" in Drosophila. Science Signaling. 2012;5(229, article 2003238) doi: 10.1126/scisignal.2003238. [DOI] [PubMed] [Google Scholar]
- 189.Garelli A., Gontijo A. M., Miguela V., Caparros E., Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. 2012;336(6081):579–582. doi: 10.1126/science.1216735. [DOI] [PubMed] [Google Scholar]
- 190.Colombani J., Andersen D. S., Léopol P. Secreted peptide dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336(6081):582–585. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- 191.Figueroa-Clarevega A., Bilder D. Malignant drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Developmental Cell. 2015;33(1):47–56. doi: 10.1016/j.devcel.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Daniel S. G., Russ A. D., Guthridge K. M., et al. miR-9a mediates the role of Lethal giant larvae as an epithelial growth inhibitor in Drosophila. Biology Open. 2017 doi: 10.1242/bio.027391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Grzeschik N. A., Parsons L. M., Allott M. L., Harvey K. F., Richardson H. E. Lgl, aPKC, and Crumbs Regulate the Salvador/Warts/Hippo Pathway through Two Distinct Mechanisms. Current Biology. 2010;20(7):573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 194.Parsons L. M., Grzeschik N. A., Amaratunga K., Burke P., Quinn L. M., Richardson H. E. A kinome RNAi screen in Drosophila identifies novel genes interacting with Lgl, aPKC, and Crb cell polarity genes in epithelial tissues. G3: Genes, Genomes, Genetics. 2017;7(8):2497–2509. doi: 10.1534/g3.117.043513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Portela M., Parsons L. M., Grzeschik N. A., Richardson H. E. Regulation of Notch signaling and endocytosis by the Lgl neoplastic tumor suppressor. Cell Cycle. 2015;14(10):1496–1506. doi: 10.1080/15384101.2015.1026515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Parsons L. M., Portela M., Grzeschik N. A., Richardson H. E. Lgl regulates notch signaling via endocytosis, independently of the apical aPKC-Par6-Baz polarity complex. Current Biology. 2014;24(18):2073–2084. doi: 10.1016/j.cub.2014.07.075. [DOI] [PubMed] [Google Scholar]
- 197.Dow L. E., Elsum I. A., King C. L., Kinross K. M., Richardson H. E., Humbert P. O. Loss of human Scribble cooperates with H-Ras to promote cell invasion through deregulation of MAPK signalling. Oncogene. 2008;27(46):5988–6001. doi: 10.1038/onc.2008.219. [DOI] [PubMed] [Google Scholar]
- 198.Pearson H. B., Perez-Mancera P. A., Dow L. E., et al. SCRIB expression is deregulated in human prostate cancer, and its deficiency in mice promotes prostate neoplasia. The Journal of Clinical Investigation. 2011;121(11):4257–4267. doi: 10.1172/JCI58509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pearson H. B., McGlinn E., Phesse T. J., et al. The polarity protein Scrib mediates epidermal development and exerts a tumor suppressive function during skin carcinogenesis. Molecular Cancer. 2015;14(1, article 169) doi: 10.1186/s12943-015-0440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Godde N. J., Sheridan J. M., Smith L. K., et al. Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland. PLoS Genetics. 2014;10, article e1004323(5) doi: 10.1371/journal.pgen.1004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Elsum I. A., Yates L. L., Pearson H. B., et al. Scrib heterozygosity predisposes to lung cancer and cooperates with KRas hyperactivation to accelerate lung cancer progression in vivo. Oncogene. 2014;33(48):5523–5533. doi: 10.1038/onc.2013.498. [DOI] [PubMed] [Google Scholar]
- 202.Cordenonsi M., Zanconato F., Azzolin L., et al. The hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 203.Archibald A., Al-Masri M., Liew-Spilger A., McCaffrey L. Atypical protein kinase C induces cell transformation by disrupting Hippo/Yap signaling. Molecular Biology of the Cell (MBoC) 2015;26(20):3578–3595. doi: 10.1091/mbc.E15-05-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhan L., Rosenberg A., Bergami K. C., et al. Deregulation of Scribble Promotes Mammary Tumorigenesis and Reveals a Role for Cell Polarity in Carcinoma. Cell. 2008;135(5):865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jang A. C.-C., Chang Y.-C., Bai J., Montell D. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nature Cell Biology. 2009;11(5):569–579. doi: 10.1038/ncb1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gibson M. C., Lehman D. A., Schubiger G. Lumenal transmission of decapentaplegic in Drosophila imaginal discs. Developmental Cell. 2002;3(3):451–460. doi: 10.1016/S1534-5807(02)00264-2. [DOI] [PubMed] [Google Scholar]
- 207.Speicher S., Fischer A., Knoblich J., Carmena A. The PDZ Protein Canoe Regulates the Asymmetric Division of Drosophila Neuroblasts and Muscle Progenitors. Current Biology. 2008;18(11):831–837. doi: 10.1016/j.cub.2008.04.072. [DOI] [PubMed] [Google Scholar]
- 208.Radziwill G., Erdmann R. A., Margelisch U., Moelling K. The Bcr kinase downregulates Ras signaling by phosphorylating AF-6 and binding to its PDZ domain. Molecular and Cellular Biology. 2003;23(13):4663–4672. doi: 10.1128/MCB.23.13.4663-4672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fernández B. G., Gaspar P., Brás-Pereira C., Jezowska B., Rebelo S. R., Janody F. Actin-capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 2011;138(11):2337–2346. doi: 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- 210.Sansores-Garcia L., Bossuyt W., Wada K.-I., et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO Journal. 2011;30(12):2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Richardson H. E. Actin up for Hippo. EMBO Journal. 2011;30(12):2307–2309. doi: 10.1038/emboj.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ann Mack N., Georgiou M. The interdependence of the Rho GTPases and apicobasal cell polarity. Small GTPases. 2014;5(2) doi: 10.4161/21541248.2014.973768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schroeder M. C., Halder G. Regulation of the Hippo pathway by cell architecture and mechanical signals. Seminars in Cell & Developmental Biology. 2012;23(7):803–811. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 214.Guarino M. Src signaling in cancer invasion. Journal of Cellular Physiology. 2010;223(1):14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 215.Ashton-Beaucage D., Therrien M. How genetics has helped piece together the MAPK signaling pathway. Methods in Molecular Biology. 2017;1487:1–21. doi: 10.1007/978-1-4939-6424-6_1. [DOI] [PubMed] [Google Scholar]
- 216.Malartre M. Regulatory mechanisms of EGFR signalling during Drosophila eye development. Cellular and Molecular Life Sciences. 2016;73(9):1825–1843. doi: 10.1007/s00018-016-2153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Simanshu D. K., Nissley D. V., McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell. 2017;170(1):17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mitsudomi T., Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS Journal. 2010;277(2):301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 219.Karnoub A. E., Weinberg R. A. Ras oncogenes: split personalities. Nature Reviews Molecular Cell Biology. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Nakamura M., Igaki T. Induction and detection of oncogene-induced cellular senescence in Drosophila. Methods in Molecular Biology. 2017;1534:211–218. doi: 10.1007/978-1-4939-6670-7_20. [DOI] [PubMed] [Google Scholar]
- 221.DeNicola G. M., Tuveson D. A. RAS in cellular transformation and senescence. European Journal of Cancer. 2009;45(1):211–216. doi: 10.1016/S0959-8049(09)70036-X. [DOI] [PubMed] [Google Scholar]
- 222.Sumabat T. M., Hariharan I. K. Ras brakes for hippo. Developmental Cell. 2017;42(6):561–562. doi: 10.1016/j.devcel.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 223.Classen A.-K., Bunker B. D., Harvey K. F., Vaccari T., Bilder D. A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nature Genetics. 2009;41(10):1150–1155. doi: 10.1038/ng.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kimmelman A. C., White E. Autophagy and Tumor Metabolism. Cell Metabolism. 2017;25(5):1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Thompson B. J., Cohen S. M. The Hippo Pathway Regulates the bantam microRNA to Control Cell Proliferation and Apoptosis in Drosophila. Cell. 2006;126(4):767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 226.Nolo R., Morrison C. M., Tao C., Zhang X., Halder G. The bantam MicroRNA is a target of the hippo tumor-suppressor pathway. Current Biology. 2006;16(19):1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 227.Herranz H., Hong X., Cohen S. M. Mutual repression by bantam miRNA and capicua links the EGFR/MAPK and hippo pathways in growth control. Current Biology. 2012;22(8):651–657. doi: 10.1016/j.cub.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 228.Milán M. Tumor models: Tumor-stroma interactions drive neoplastic transformation in Drosophila. Current Biology. 2014;24(14):R658–R659. doi: 10.1016/j.cub.2014.05.074. [DOI] [PubMed] [Google Scholar]
- 229.Gregory P. A., Bert A. G., Paterson E. L., et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 230.Niu N., Mercado-Uribe I., Liu J. Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene. 2017;36(34):4887–4900. doi: 10.1038/onc.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lv H., Shi Y., Zhang L., et al. Polyploid giant cancer cells with budding and the expression of cyclin E, S-phase kinase-associated protein 2, stathmin associated with the grading and metastasis in serous ovarian tumor. BMC Cancer. 2014;14(1, article 576) doi: 10.1186/1471-2407-14-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhang S., Mercado-Uribe I., Hanash S., Liu J. iTRAQ-based proteomic analysis of polyploid giant cancer cells and budding progeny cells reveals several distinct pathways for ovarian cancer development. PLoS ONE. 2013;8, article e80120(11) doi: 10.1371/journal.pone.0080120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Davoli T., De Lange T. The causes and consequences of polyploidy in normal development and cancer. Annual Review of Cell and Developmental Biology. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 234.Read R. D. Drosophila melanogaster as a model system for human brain cancers. Glia. 2011;59(9):1364–1376. doi: 10.1002/glia.21148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ntziachristos P., Lim J. S., Sage J., Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25(3):318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dominguez M. Oncogenic programmes and Notch activity: An 'organized crime'? Seminars in Cell & Developmental Biology. 2014;28:78–85. doi: 10.1016/j.semcdb.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 237.Palomero T., Dominguez M., Ferrando A. A. The role of the PTEN/AKT pathway in NOTCH1-induced leukemia. Cell Cycle. 2008;7(8):965–970. doi: 10.4161/cc.7.8.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Chen Y., Zhang L. Members of the microRNA-200 family are promising therapeutic targets in cancer (Review) Experimental and Therapeutic Medicine. 2017;14(1):10–17. doi: 10.3892/etm.2017.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Brabletz S., Bajdak K., Meidhof S., et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO Journal. 2011;30(4):770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Meza-Sosa K. F., Pérez-García E. I., Camacho-Concha N., López-Gutiérrez O., Pedraza-Alva G., Pérez-Martínez L. MiR-7 promotes epithelial cell transformation by targeting the tumor suppressor KLF4. PLoS ONE. 2014;9, article e103987(9) doi: 10.1371/journal.pone.0103987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jones S., Zhang X., Parsons D. W., et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wegorzewska M., Krauss R. S., Kang J.-S. Overexpression of the immunoglobulin superfamily members CDO and BOC enhances differentiation of the human rhabdomyosarcoma cell line RD. Molecular Carcinogenesis. 2003;37(1):1–4. doi: 10.1002/mc.10121. [DOI] [PubMed] [Google Scholar]
- 243.Oh H., Irvine K. D. Yorkie: the final destination of Hippo signaling. Trends in Cell Biology. 2010;20(7):410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Grusche F. A., Richardson H. E., Harvey K. F. Upstream regulation of the Hippo size control pathway. Current Biology. 2010;20(13):574–582. doi: 10.1016/j.cub.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 245.Pfleger C. M. The Hippo Pathway: A Master Regulatory Network Important in Development and Dysregulated in Disease. Current Topics in Developmental Biology. 2017;123:181–228. doi: 10.1016/bs.ctdb.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 246.Zygulska A. L., Krzemieniecki K., Pierzchalski P. Hippo pathway - brief overview of its relevance in cancer. Journal of Physiology and Pharmacology. 2017;68(3):311–335. [PubMed] [Google Scholar]
- 247.Marenda D. R., Zraly C. B., Dingwall A. K. The Drosophila Brahma (SWI/SNF) chromatin remodeling complex exhibits cell-type specific activation and repression functions. Developmental Biology. 2004;267(2):279–293. doi: 10.1016/j.ydbio.2003.10.040. [DOI] [PubMed] [Google Scholar]
- 248.Brumby A. M., Zraly C. B., Horsfield J. A., et al. Drosophila cyclin E interacts with components of the brahma complex. EMBO Journal. 2002;21(13):3377–3389. doi: 10.1093/emboj/cdf334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Armstrong J. A., Sperling A. S., Deuring R., et al. Genetic screens for enhancers of brahma reveal functional interactions between the BRM chromatin-remodeling complex and the Delta-Notch signal transduction pathway in Drosophila. Genetics. 2005;170(4):1761–1774. doi: 10.1534/genetics.105.041327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Staehling-Hampton K., Ciampa P. J., Brook A., Dyson N. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics. 1999;153(1):275–287. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Herr A., Mckenzie L., Suryadinata R., et al. Geminin and Brahma act antagonistically to regulate EGFR-Ras-MAPK signaling in Drosophila. Developmental Biology. 2010;344(1):36–51. doi: 10.1016/j.ydbio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 252.Zraly C. B., Marenda D. R., Dingwall A. K. SNR1 (INI1/SNF5) mediates important cell growth functions of the Drosophila Brahma (SWI/SNF) chromatin remodeling complex. Genetics. 2004;168(1):199–214. doi: 10.1534/genetics.104.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zraly C. B., Middleton F. A., Dingwall A. K. Hormone-response genes are direct in vivo regulatory targets of Brahma (SWI/SNF) complex function. The Journal of Biological Chemistry. 2006;281(46):35305–35315. doi: 10.1074/jbc.M607806200. [DOI] [PubMed] [Google Scholar]
- 254.Terriente-Félix A., de Celis J. F. Osa, a subunit of the BAP chromatin-remodelling complex, participates in the regulation of gene expression in response to EGFR signalling in the Drosophila wing. Developmental Biology. 2009;329(2):350–361. doi: 10.1016/j.ydbio.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 255.Zhu Y., Li D., Wang Y., et al. Brahma regulates the Hippo pathway activity through forming complex with Yki-Sd and regulating the transcription of Crumbs. Cellular Signalling. 2015;27(3):606–613. doi: 10.1016/j.cellsig.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 256.Jin Y., Xu J., Yin M., et al. Brahma is essential for Drosophila intestinal stem cell proliferation and regulated by Hippo signaling. eLife. 2013;2 doi: 10.7554/eLife.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Fang Y., Zhang X. Targeting NEK2 as a promising therapeutic approach for cancer treatment. Cell Cycle. 2016;15(7):895–907. doi: 10.1080/15384101.2016.1152430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Dar A. C., Das T. K., Shokat K. M., Cagan R. L. Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature. 2012;486(7401):80–84. doi: 10.1038/nature11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Vidal M., Wells S., Ryan A., Cagan R. ZD6474 suppresses oncogenic RET isoforms in a Drosophila model for type 2 multiple endocrine neoplasia syndromes and papillary thyroid carcinoma. Cancer Research. 2005;65(9):3538–3541. doi: 10.1158/0008-5472.CAN-04-4561. [DOI] [PubMed] [Google Scholar]
- 260.Read R. D., Goodfellow P. J., Mardis E. R., Novak N., Armstrong J. R., Cagan R. L. A drosophila model of multiple endocrine neoplasia type 2. Genetics. 2005;171(3):1057–1081. doi: 10.1534/genetics.104.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bonavia R., Inda M.-D., Cavenee W. K., Furnari F. B. Heterogeneity maintenance in glioblastoma: A social network. Cancer Research. 2011;71(12):4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zhou H., Neelakantan D., Ford H. L. Clonal cooperativity in heterogenous cancers. Seminars in Cell & Developmental Biology. 2017;64:79–89. doi: 10.1016/j.semcdb.2016.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tissot T., Thomas F., Roche B. Non-cell-autonomous effects yield lower clonal diversity in expanding tumors. Scientific Reports. 2017;7, article 11157(1) doi: 10.1038/s41598-017-11562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tissot T., Ujvari B., Solary E., Lassus P., Roche B., Thomas F. Do cell-autonomous and non-cell-autonomous effects drive the structure of tumor ecosystems? Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2016;1865(2):147–154. doi: 10.1016/j.bbcan.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 265.Tin Su T. Non-autonomous consequences of cell death and other perks of being metazoan. AIMS Genetics. 2015;2(1):54–69. doi: 10.3934/genet.2015.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Fogarty C. E., Bergmann A. The sound of silence: Signaling by apoptotic cells. Current Topics in Developmental Biology. 2015;114:241–265. doi: 10.1016/bs.ctdb.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Fan Y., Bergmann A. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends in Cell Biology. 2008;18(10):467–473. doi: 10.1016/j.tcb.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Martín F. A., Peréz-Garijo A., Morata G. Apoptosis in Drosophila: compensatory proliferation and undead cells. The International Journal of Developmental Biology. 2009;53(8–10):1341–1347. doi: 10.1387/ijdb.072447fm. [DOI] [PubMed] [Google Scholar]
- 269.Fogarty C. E., Bergmann A. Killers creating new life: Caspases drive apoptosis-induced proliferation in tissue repair and disease. Cell Death & Differentiation. 2017;24(8):1390–1400. doi: 10.1038/cdd.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Grusche F. A., Degoutin J. L., Richardson H. E., Harvey K. F. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Developmental Biology. 2011;350(2):255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 271.Graves H. K., Woodfield S. E., Yang C.-C., Halder G., Bergmann A. Notch signaling activates yorkie non-cell autonomously in drosophila. PLoS ONE. 2012;7, article e37615(6) doi: 10.1371/journal.pone.0037615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Moberg K. H., Schelble S., Burdick S. K., Hariharan I. K. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Developmental Cell. 2005;9(5):699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 273.Reynolds-Kenneally J., Mlodzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Developmental Biology. 2005;285(1):38–48. doi: 10.1016/j.ydbio.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 274.Christiansen A. E., Ding T., Bergmann A. Ligand-independent activation of the Hedgehog pathway displays non-cell autonomous proliferation during eye development in Drosophila. Mechanisms of Development. 2012;129(5-8):98–108. doi: 10.1016/j.mod.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Christiansen A. E., Ding T., Fan Y., et al. Non-cell autonomous control of apoptosis by ligand-independent Hedgehog signaling in Drosophila. Cell Death & Differentiation. 2013;20(2):302–311. doi: 10.1038/cdd.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Recasens-Alvarez C., Ferreira A., Milán M. JAK/STAT controls organ size and fate specification by regulating morphogen production and signalling. Nature Communications. 2017;8, article 13815 doi: 10.1038/ncomms13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Huang J., Wu S., Barrera J., Matthews K., Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122(3):421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 278.Dekanty A., Milán M. Aneuploidy, cell delamination and tumorigenesis in Drosophila epithelia. Cell Cycle. 2013;12(5):728–731. doi: 10.4161/cc.23949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Milán M., Clemente-Ruiz M., Dekanty A., Muzzopappa M. Aneuploidy and tumorigenesis in Drosophila. Seminars in Cell & Developmental Biology. 2014;28:110–115. doi: 10.1016/j.semcdb.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 280.Shlevkov E., Morata G. A dp53/JNK-dependant feedback amplification loop is essential for the apoptotic response to stress in Drosophila. Cell Death & Differentiation. 2012;19(3):451–460. doi: 10.1038/cdd.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hariharan I. K., Serras F. Imaginal disc regeneration takes flight. Current Opinion in Cell Biology. 2017;48:10–16. doi: 10.1016/j.ceb.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Markstein M. Modeling colorectal cancer as a 3-dimensional disease in a dish: The case for drug screening using organoids, zebrafish, and fruit flies. Drug Discovery Today: Technologies. 2013;10(1):e73–e81. doi: 10.1016/j.ddtec.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 283.Fernández-Hernández I., Scheenaard E., Pollarolo G., Gonzalez C. The translational relevance of Drosophila in drug discovery. EMBO Reports. 2016;17(4):471–472. doi: 10.15252/embr.201642080. [DOI] [PMC free article] [PubMed] [Google Scholar]