Abstract
The AP-1 transcription factor is activated by oncogenic signal transduction cascades and its function is critical for both mitogenesis and carcinogenesis. To define the role of AP-1 in the context of a human fibrosarcoma cell line, HT1080, we expressed a dominant negative c-jun mutant fused to the green fluorescent protein in an ecdysone-inducible system. We demonstrated that high levels of this mutant, GFP-TAM67, inhibit AP-1 activity and arrest cells predominately in the G1 phase of the cell cycle. This arrest is reversible and occurs only above a threshold concentration; low to moderate levels of GFP-TAM67 are insufficient for growth arrest. Contrary to expectations based on the literature, GFP-TAM67 does not inhibit expression of cyclin D1, cyclin E, or their respective cyclin-dependent kinases. However, pRB is hypophosphorylated in GFP-TAM67-arrested cells and the activity of both the cyclin D1:cdk and the cyclin E:cdk complexes are impaired. Both of these complexes show an increased association with p21CIP1/WAF1, concomitantly with induction of the p21 mRNA by GFP-TAM67. These results suggest a novel function of AP-1 in the activation of the G1 cyclin:cdk complexes in human tumor cells by regulating the expression of the p21CIP1/WAF1 gene.
INTRODUCTION
AP-1 is a dimeric transcription factor that is composed of members of the Jun and Fos proto-oncogene families. AP-1 both activates and represses transcription through a cis-acting element in the promoter of target genes and has long been associated with proliferation. Both c-fos and c-jun are immediate early genes that are rapidly and transiently induced by a large variety of mitogens via the ras-mitogen–activated protein (MAP) kinase pathway (Karin et al., 1997). Microinjection of antibodies to AP-1 family members inhibits entry into S phase, indicating that general AP-1 activity is necessary for cell cycle progression (Kovary and Bravo, 1991). Despite the necessity of AP-1 activity for proliferation, the specific function of AP-1 in the cell cycle is not well understood. Given that it exerts its biological effects by regulating the transcription of target genes, AP-1 presumably directs the expression of a critical target gene, or genes, in response to mitogenic signals. The cyclin D1 gene has emerged as one such possible target (Brown et al., 1998). The promoter for cyclin D1 contains an AP-1 site and ectopic expression of either c-fos or c-jun induces cyclin D1 mRNA expression (Miao and Curran, 1994; Albanese et al., 1995). Fibroblasts derived from mice that are null for both c-fos and FosB have an impaired proliferation and reduced levels of cyclin D1 (Brown et al., 1998), as do fibroblasts that are null for c-jun (Schreiber et al., 1999). Re-expression of cyclin D1 in these cells partially restores the proliferative phenotype (Brown et al., 1998). These results suggest that cyclin D1 is a critical target of AP-1 during mitogenesis. However, transcriptional activation of the cyclin D1 gene might not be the sole function of AP-1 in regulating the cell cycle. It has been recently demonstrated that v-jun activates the cyclin E:cdk2 complex in chick embryo fibroblasts without affecting the level of expression of either cyclin E or cyclin D1 (Clark et al., 2000). This result argues against the activation of the cyclin D1 gene expression being a critical function for AP-1 in all cell types. c-jun has also been demonstrated to positively regulate mouse embryo fibroblast proliferation in a p53-dependent manner (Schreiber et al., 1999). However, there is evidence that AP-1 regulates the cell cycle in a p53-independent manner as well. Dominant negative c-jun constructs inhibit colony formation of transformed cells that have mutant p53 (Rapp et al., 1994). c-jun has been reported to directly regulate the p21CIP1/WAF1 promoter, both positively and negatively, via an SP-1 site, also suggesting a p53-independent mechanism (Kardassis et al., 1999;Wang et al., 2000). These somewhat contradictory findings were derived from several different experimental systems and are not necessarily mutually exclusive. It is possible that AP-1 regulates proliferation by a variety of mechanisms that are cell type and growth context specific. Given this specificity, it is important to evaluate the function of AP-1 in a cell type that more closely models human disease than the mouse embryo fibroblast cells used in the above studies.
To investigate the role of AP-1 in the context of a human tumor cell line, HT1080, we have utilized the dominant negative mutant of c-jun, TAM67, fused to the green fluorescent protein (GFP) under the control of an ecdysone-inducible expression system. We demonstrate that high levels of GFP-TAM67 lead to a reversible G1 arrest in HT1080 cells; whereas low or moderate levels of GFP-TAM67 allow cells to proliferate normally. GFP-TAM67–mediated G1 arrest does not significantly alter the expression of cyclin D1 or cyclin E complex components. However, GFP-TAM67 arrest leads to hypophosphorylation of pRB and loss of activity of the cyclin D1:cdk4/6 and cyclin E:cdk2 complexes. The loss of cyclin E- and cyclin D1-dependent kinase activity correlates with an increase in the association of p21CIP1/WAF1 with these complexes and an increase in p21CIP1/WAF1 gene expression. These data suggest that the biochemical activation of existing cyclin complexes is a novel function of AP-1 in the regulation of human tumor cell proliferation and that this function is distinct from that previously described for AP-1 in untransformed cells.
MATERIALS AND METHODS
Plasmids
pCMV-TAM67 was a gift of Dr. Bradford W. Ozanne (Beatson Institute, Glasgow, Scotland) and the insert was fully sequenced at the University of Cincinnati DNA Core facility (Cincinnati, OH). Standard cloning techniques were utilized (Sambrook et al., 1989). DNA fragments were excised from agarose gels and purified with kits from Qiagen (Chatsworth, CA). Plasmids were purified with Concert Maxiprep columns (GIBCO, Grand Island, NY). TAM67 was excised from pCMV-TAM67 as a RsaI-BamHI fragment and cloned into the EcoRV-BamHI site of the pIRESpuro vector (Clontech, Palo Alto, CA). To construct a GFP fusion, a BamHI-HindIII fragment was cut from the pIRES-TAM67puro plasmid and then ligated into the BamHI-HindIII sites of pEGFP-C3 (Clontech). To shift the TAM67 insert into frame with the GFP, this plasmid was linearized with HindIII, and the recessed ends were filled with T4 polymerase (GIBCO) and religated to generate an in-frame fusion between GFP and TAM67, designated pGFP-TAM67. To express GFP-TAM67 from a bicistronic vector the chimeric DNA encoding GFP-TAM67 was cut out of pGFP-TAM67 with the use of Eco47III and BamHI and cloned into the EcoRV and BamHI sites of pIRESpuro to generate pGFP-TAM67puro. To construct ecdysone-inducible GFP-TAM67, the insert was excised from pGFP-TAM67 with NheI and BamHI and cloned into the NheI and BamHI sites of pIND (Clontech) to generate pIND-GFP-TAM67.
Cell Culture
HT-1080 fibrosarcoma cells were originally obtained from the American Type Tissue Collection (Rockville, MD). Cells were grown in high glucose DMEM supplemented with 10% fetal calf serum, penicillin, and streptomycin (GIBCO Life Technologies, Gaithersburg, MD) at 37°C, 5% CO2, and 90% humidity. Transfections were routinely with the use of Fugene-6 (Roche Biochemicals, Indianapolis, IN). For selection of ecdysone-inducible clones, 1 μg each of pIND-GFP-TAM67 and the regulatory plasmid pVgRXR were transfected as above selected in 400 μg/ml Zeocin (Invitrogen, San Diego, CA) and 800 μg/ml G418 (GIBCO Life Technologies). Transfections with an empty pIND vector and pVgRXR were performed to generate control cell lines. Medium was changed every 3 d and resistant colonies were isolated, expanded, and screened for induction of green fluorescence by flow cytometry.
Confocal Microscopy
GFP was visualized after fixation in 4% paraformaldehyde with the use of a LSM510 laser scanning confocal microscope (Zeiss, Oberkochen, Germany). For ponasterone induction of GFP-TAM67, iGT1a cells were grown in four-well chamber slides with and without 10 μM ponasterone A for 24 h. The samples were fixed as above permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 5 min. Cells were counterstained with 13.2 nM Alexafluor568 phalloidin (Molecular Probes, Eugene, OR) in PBS for 20 min at room temperature, washed three times with PBS, and mounted with Gelmount (Fisher Scientific, Pittsburgh, PA).
Reporter Gene Assays
The reporter plasmids used were pAP1-SEAP (Clontech), containing four tandem copies of the AP-1 consensus sequence, TGA(G/C)TCA, fused upstream of a minimal TATA-like promoter from the Herpes simplex thymidine kinase gene controlling the expression a heat-stable secreted alkaline phosphatase (SEAP) reporter gene. The pTAL-SEAP (Clontech), an identical plasmid lacking enhancer elements, served as a negative control. pSEAP2 (Clontech) expressing SEAP under the control of the SV40 early region promoter and enhancer elements served as a positive control. iGT1a and iC1 cells were each plated in two six-well plates at 1 × 105 cells per plate, and ponasterone A was added to 10 μM to one plate. Cells were transfected with 0.5 μg pCMV-β-galactosidase (Clontech) and 1.5 μg of the SEAP reporter plasmid and incubated for 48 h. Conditioned medium was collected and serum-derived alkaline phosphatases were heat inactivated at 65°C for 30 min. Chemiluminescent assays for SEAP activity were performed in triplicate in opaque microtiter plates (Fisher, Pittsburgh, PA) with the use of CSPD (Boehringer Mannheim, Indianapolis, IN) as a substrate. Unconditioned heat-inactivated medium was used a negative control and unheated medium served as a positive control for the CSPD assay. For each reaction 20 μl of conditioned medium were mixed with 200 μl of CSPD reaction buffer (11 mM NaCl, 11 mM Tris, pH 9.5, 16.66 μM CSPD), sealed, and incubated for 15 min at 37°C. Chemiluminescence was then measured with a Topcount microplate scintillation counter (Packard, Meriden, CT). β-Galactosidase activity was determined from transfected cell lysates by a microtiter ONPG assay, and 20 μl of the protein lysate were mixed with 200 μl of ONPG reaction buffer (1 mg/ml ONPG (Sigma, St. Louis, MO), 50 mM β-mercaptoethanol, 0.1 M sodium phosphate buffer, pH 7.5). Plates were sealed and incubated at 37°C for 30–90 min and the adsorbance at 410 nm was read with a MR700 microtiter plate reader (Dynatech, Chantilly, VA). Secreted alkaline phosphatase activity was the expressed as counts per second (cps) normalized for β-galactosidase activity per milligram of protein.
Cell Cycle Analysis
Control and GFP-TAM67 cell lines were plated to ∼80% confluence, 3 × 105 cells/35-mm dish, and treated with the indicated doses of ponasterone A from a 2 mM stock in ethanol. After 24 h cells were trypsinized, washed three times in cold PBS, and fixed in cold 70% ethanol for 15 min at −20°C. Cells were then stained with 10 μg/ml propidium iodide (Molecular Probes) and 40 μg/ml RNase A (Sigma) in PBS for 20 min at room temperature. Cells were then washed and resuspended in 1 ml of PBS and analyzed on a Epics XL flow cytometer (Coulter, Palo Alto, CA). Figures were prepared from listmode data with the use of WinMIDI software and quantitation of the cell cycle profile was performed by curve fitting with the use of the ModFitLT program (Becton Dickinson, Franklin Lakes, NJ).
Growth Curves
iC1 and iGT1a cells were each seeded into two 10-cm dishes at 1 × 106 per plate and treated with or without ponasterone A as described above for 24 h. The plates were then washed three times with PBS and trypsinized, and each was seeded into four six-well plates at 1 × 105 cells/plate. Ponasterone was added to 10 μM to three wells per plate. Cell counts were determined every day for 4 d with a Z1 Coulter counter.
Immunoprecipitation
Immunoprecipitations were performed as previously described (Rauscher et al., 1988a). Briefly, iGT1a cells were plated in 35-mm dishes with 10 μM ponasterone A or vehicle for 24 h. Cells were then washed and grown in 0.5 ml of free DMEM (GIBCO). After addition of 0.5 mCi 35S-labeled cysteine and methionine, cells were incubated for 30 min. Samples were then rinsed twice with cold PBS and lysates were made with 1 ml of RIPA (0.15 M NaCl, 10 mM Tris-Cl, pH 7.4, 1 mM EDTA, 1% Triton X-100, 0.5% desoxycholate, 0.1% SDS). Boiled lysates were made with 0.2 ml of denaturing lysis buffer (2% SDS, 50 mM Tris-Cl, pH 7.4, 1 mM EDTA), boiled for 5 min, and then added to 0.8 ml of RIPA. Both types of lysate were cleared by centrifugation and immunoprecipitated with 0.4 μg of anti-GFP or control immunoglobulin G and protein A/G agarose. Immunoprecipitations were run on a 10% PAGE and autoradiography was performed after soaking in Enhance (Amersham, Piscataway, NJ).
Immunoblotting
Cell lysates were prepared with RIPA buffer containing cocktail of protease inhibitors (Sigma). Protein concentrations were determined with the BCA kit (Pierce, Rockford, IL). GFP fluorescence was visualized from nondenaturing SDS-PAGE gels with the use of a Molecular Dynamics STORM phosphoimager in blue fluorescence mode. For immunoblotting, 20 μg of cell lysates were electrophoresed on SDS-polyacrylamide gels, transferred to polyvinylidene difluoride (PVDF) membranes (Amersham) and processed as previously described. (Sambrook et al., 1989).
Kinase Assays
Cyclin E and cyclin D1 kinase assays were performed as previously described (Guadagno and Assoian, 1991; Saha et al., 1997). Briefly, 1 × 106 iGT1a cells were plated in a 10-cm dish and treated with 10 μM ponasterone A or vehicle control for 24 h, lysed with the appropriate buffers, precleared, and then immunoprecipitated with either anti-cyclin E (antibody-1; Labvision, Fremont, CA) or an anti-cyclin D1 antibody (DCS-11; LabVision). Immunoprecipitates were then split, and one aliquot was used in the kinase assay with either histone H1 or pRB-GST as a substrate. The other aliquot was used in immunoblot analysis to confirm immunoprecipitation.
Northern Blots
Total RNA was prepared from iGT1a cells with and without ponasterone A with Trizol (GIBCO). Five micrograms of each was run on a 1% agarose-formaldehyde gel, transferred to Hybond-N (Pharmacia), and probed with randomly primed 33P-labeled probe in Rapid-Hyb solution (Amersham, Piscataway, NJ). Blots were washed in 2× SSC, 1% SDS at 65°C for 2 × 10 min and twice in 0.5× SSC, 1% SDS at 65°C for 20 min.
Antibodies
In this study we used antibodies to c-jun (antibody-1; Oncogene Research, San Diego, CA), GFP, (7.1 and 13.1; Boehringer Mannheim), cyclin B1 (GNS-1; Santa Cruz), cyclin A (H432; Santa Cruz, a gift from Dr. Kenji Fukasawa), RB (1F8; Lab Vision), cyclin E (HE12 and antibody-1; Lab Vision), cyclin D1 (DCS-11 and antibody-3; Lab Vision), CDK2 (antibody-3; Lab Vision), CDK4 (antibody-4; Lab Vision), p21WAF1/CIP1 (antibody-7; Lab Vision), p27Kip1(sc-776; Santa Cruz, a gift from Dr. Larry Sherman), and p53 (Pab1801; a gift from Dr. Kenji Fukasawa).
RESULTS
Construction of a GFP-TAM67 Fusion Protein
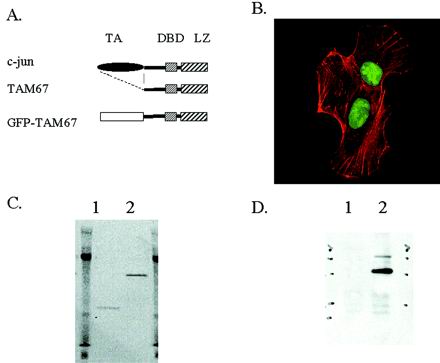
As a means of monitoring TAM67 expression, we fused it to the C terminus of GFP, which allowed easy visualization by fluorescence microscopy (Figure 1A). When transfected into HT1080 cells, GFP-TAM67 is localized to the nucleus (Figure 1B). To confirm the identity of this nuclear localizing protein, HT1080 cells were transiently transfected with pCMV-GFP-TAM67 or control pEGFP-C3 plasmids. Lysates from these transfections were immunoprecipitated with an anti-GFP monoclonal antibody and fractionated on an SDS-polyacrylamide gel under nonreducing conditions. The gel was then scanned with a Molecular Dynamics phosphoimager in blue fluorescence mode revealing an ∼53-kDa green fluorescent band in the GFP-TAM67-transfected lane (Figure 1C, lane 2) and a 26-kDa fluorescent band in the control GFP-transfected lane (Figure 1C, lane 1). After transfer to a PVDF membrane, the 53-kDa band reacted with anti-jun antibodies, confirming its identity as a GFP-TAM67 fusion protein (Figure 1D).
Figure 1.
The GFP-TAM67 mutant localizes to the nucleus. (A) A schematic diagram depicting c-jun (top) with its constituent transactivating domain (TA), DNA-binding domain (DBD), and leucine zipper domain (LZ). The TAM67 dominant negative mutant has a deletion of amino acids 3–122, removing the transactivating domain. GFP was fused to the N terminus of TAM67 to generate GFP-TAM67. (B) pCMV-GFP-TAM67 transiently transfected into HT1080 cells is localized to the nucleus (green). F-actin is stained with phalloidin (red). (C) HT1080 cells transfected transiently with pEGFP (lane 1) or with pCMV-GFP-TAM67 (lane 2) were immunoprecipitated with anti-GFP antibodies and then run on a 10% polyacrylamide gel under nonreducing conditions. A fluorescent image of the gel acquired with a Molecular Dynamics phosphoimager in blue fluorescence mode shows the 26-kDa GFP protein and the 54-kDa GFP-TAM67 fusion. (D) This gel was transferred to a PVDF membrane and probed with anti-c-jun antibodies.
GFP-TAM67 Inhibits HT1080 Colony Formation
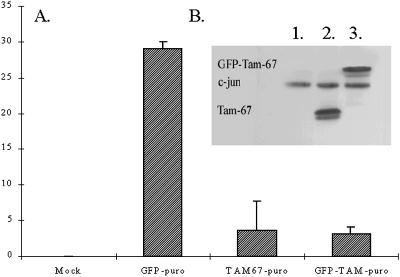
To more effectively express the GFP-TAM67 fusion protein we subcloned it into the bicistronic expression plasmid pIRESpuro. This vector expresses the gene of interest and the gene encoding resistance to puromycin as a single transcription unit separated by a viral internal ribosome entry site. Because of this physical linkage, selection for puromycin ensures the expression of GFP-TAM67 in resistant cells. To determine whether the GFP fusion interferes with TAM67 function, colony formation assays were performed. Bicistronic plasmids expressing TAM67, GFP-TAM67, or GFP were transfected into HT1080 cells, in duplicate. After 2 d, one set of transfections was harvested for Western blot analysis and the other set was plated in selective medium containing 0.5 μg/ml puromycin. After 2 weeks colonies were counted. Immunoblot analysis of the transient transfections showed equivalent expression of TAM67 and GFP-TAM67 (Figure 2B). Both TAM67- and GFP-TAM67–transfected cells yielded a small number of colonies, 3.7 ± 4 and 3.0 ± 1/plate, respectively. In contrast, the GFP-transfected cells developed 29 ± 1 colonies/plate, indicating that overexpression of GFP alone is not growth inhibitory. No colonies grew in the mock-transfected controls (Figure 2A). This 10-fold reduction in colony formation relative to GFP shows that the GFP-TAM67 construct has the same biological activity as TAM67. Although colony formation was significantly inhibited, viable colonies expressing GFP-TAM67 could be isolated. Further experiments revealed that the number of GFP-TAM67–expressing colonies was dependent on the concentration of puromycin. Fewer colonies were seen at high levels of puromycin and more colonies were seen at lower doses (Hennigan and Stambrook, unpublished results). High concentrations of puromycin select for cells expressing high levels of the puromycin resistance gene. The concurrent high level expression of TAM67 or GFP-TAM67 resulted in inhibition of growth. At lower concentrations of puromycin, lower levels of the puromycin resistance gene are sufficient for drug resistance and the lower levels TAM67 and GFP-TAM67 do not inhibit growth. These results suggest that there is a threshold above which expression of GFP-TAM67 and TAM67 are growth inhibitory in HT1080 cells.
Figure 2.
GFP-TAM67 inhibits HT1080 colony formation. GFP, TAM67, and GFP-TAM67 were cloned into the bicistronic vector pIRESpuro. HT1080 cells were then transfected with these constructs in duplicate. (A) One set of transfections was plated in triplicate in DMEM, 10% fetal calf serum, and 0.5 μg/ml puromycin. Colonies were counted after 14 d. The figure is representative of three independent experiments. (B) Lysates were made from the other set of transfections and probed with anti-c-jun antibodies; lane 1, GFP; lane 2, TAM67; lane 3, GFP-TAM67.
Ecdysone-inducible GFP-TAM67
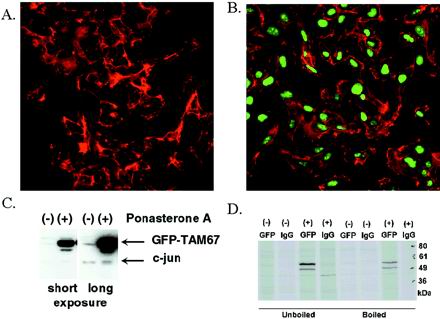
To define the growth inhibitory activity of GFP-TAM67 more fully, we generated cell lines that express GFP-TAM67 conditionally by cotransfecting HT1080 with a regulatory vector pVgRXR and the ecdysone-inducible expression vector pIND containing GFP-TAM67 or with empty vector as a control. After coselection in G-418 and Zeocin, single colonies were isolated and the induction of GFP-TAM67 by the ecdysone analogue ponasterone A was measured by flow cytometry. To confirm inducible expression one clone, iGT1a, was treated with 10 μM ponasterone A (Figure 3B) or with vehicle (Figure 3A) for 16 h and observed by confocal microscopy. A weak green fluorescence, localized to the nucleus, was apparent in the untreated cells indicating a low level expression of GFP-TAM67. Addition of 10 μM ponasterone induced a significant increase of green nuclear fluorescence in all cells in the field. Immunoblots of iGT1a lysates probed with an anti-c-jun antibody confirmed that this induced protein is GFP-TAM67 (Figure 3C). Longer exposure of this blot revealed that the levels of GFP-TAM67 in the uninduced cells were roughly equivalent to the endogenous c-jun, indicating that induction by ponasterone A results in overexpression of GFP-TAM67 without affecting the amount of endogenous c-jun. To determine whether heterodimerization occurs between GFP-TAM67 and other leucine zipper proteins, uninduced and induced iGT1a cells were metabolically labeled and lysates were either prepared under nondenaturing conditions that allow Fos and Jun heterodimerization (Rauscher et al., 1988a) or denatured by boiling. Immunoprecipitation with anti-GFP antibodies revealed GFP-TAM67 as two major bands from 50–60 kDa that are induced by ponasterone. Autoradiography revealed an identical pattern between the boiled and unboiled lysates (Figure 3D), suggesting that GFP-TAM67 does not interact with other leucine zipper proteins.
Figure 3.
Ponasterone A induces GFP-TAM67. The iGT1a clone was treated with either vehicle (A) or 10 μM ponasterone A (B) for 24 h. Cells were fixed, stained with Alexafluor568-conjugated phalloidin and imaged by laser scanning confocal microscopy. (C) Immunoblot analysis of lysates from clone iGT1a treated with (+) and without (−) ponasterone A probed with antibodies to c-jun. A short and a long exposure of this blot is presented. (D) Immunoprecipitation of lysates isolated under denaturing or nondenaturing conditions from [35S]methionine-labeled, uninduced or induced iGT1a cells with anti-GFP or control immunoglobulin G.
GFP-TAM67 Expression Inhibits AP-1 Activity
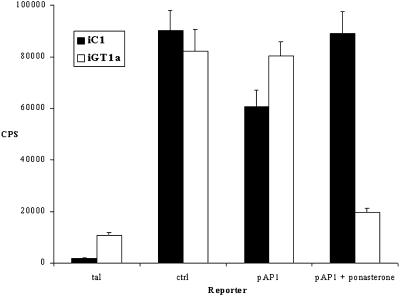
To determine the effect of GFP-TAM67 on AP-1 activity, iGT1a or control iC1 cells were plated in the presence or absence of 10 μM ponasterone A and then cotransfected with a plasmid containing a minimal promoter with four copies of an AP-1 consensus site driving a SEAP reporter gene. SEAP activity was normalized for transfection efficiency with the use of a cytomegalovirus-driven immediate early promotor β-galactosidase reporter construct. Induction of GFP-TAM67 by ponasterone A inhibited AP-1 activity by fivefold in iGT1a cells but not in control iC1 cells (Figure 4). The magnitude of the reduction of AP-1 activity did not correlate with the large increase in GFP-TAM67 concentration. This might be a result of with the use of a transient transfection system in which the reporter gene is expressed from many copies of the reporter plasmid in an episomal state, requiring a higher concentration of GFP-TAM67 for inhibition. These results demonstrate a clear negative effect of GFP-TAM67 expression on AP-1 activity and show that it is specific for GFP-TAM67 expression and is not an artifact of the inducible expression system.
Figure 4.
Ponasterone A induction of GFP-TAM67 inhibits AP-1 reporter gene activity. Control iC1 cells and iGT1a cells were treated with 10 μM ponasterone A or vehicle for 24 h and then transiently transfected with constructs driving a heat-stable SEAP reporter gene. Alkaline phosphatase activity from heat–inactivated conditioned media normalized to β-galactosidase activity is presented. The promoter constructs used were: tal, an enhancerless minimal promoter; ctrl, a full SV40 early promoter; pAP-1, a minimal promoter with four tandem repeats of the consensus AP-1 site, TGA(G/C)TCA.
GFP-TAM67 Arrests Cells in G1
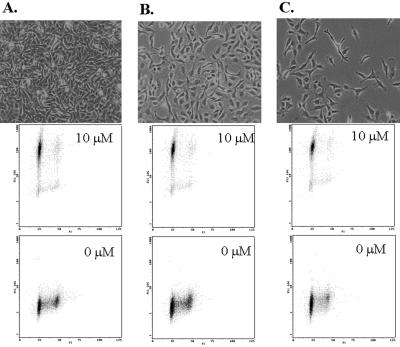
To determine whether GFP-TAM67 expression affected a specific stage of the cell cycle, dual parameter flow cytometry experiments were performed. The control iC1 cell line and the inducible GFP-TAM67 cell line, iGT1a, were treated with 10 μM ponasterone A for 16 h. Samples were then fixed, stained with propidium iodide, and analyzed by flow cytometry. In the absence of ponasterone A, the cell cycle profile of iGT1a cells was normal (Figure 5). Uninduced iGT1a cells had a slightly elevated green fluorescence relative to uninduced iC1 cells, indicative of the basal expression of GFP-TAM67 that was apparent by confocal microscopy and Western blot. As expected ponasterone A had no affect on the cell cycle profile of control iC1 cells and did not increase their green fluorescence (Figure 5, A and B). Administration of ponasterone to iGT1a cells caused a 100-fold increase in green fluorescence in 90% of the cells. Of this induced population, 95% were arrested in G1, none were arrested in S, and 5% were arrested in G2 (Figure 5, C and D). The remaining 10% were moderately induced, if at all, and the cell cycle profiles of this population were identical to uninduced controls. Identical ponasterone A-induced cell cycle arrest was seen with two other GFP-TAM67–inducible cell lines and transient transfection studies have demonstrated arrest in other human tumor-derived cell lines, i.e., HeLa, TSU pr1, and A431 (Hennigan and Stambrook, unpublished results). These results demonstrate that a high concentration of GFP-TAM67 expression in HT1080 cells caused a cell cycle arrest predominantly in G1 within 24 h.
Figure 5.
GFP-TAM67 reversibly arrests HT1080 cells in G1. Control iC1 Cells (A and B) and iGT1a cells (C and D) were treated with 0 μM ponasterone A (A and C) or 10 μM ponasterone A (B and D) for 24 h. GFP fluorescence is depicted on the Y-axis and DNA content is depicted on the X-axis. (E) iGT1a cells were treated with 10 μM ponasterone A (squares) or vehicle (triangles) for 24 h and trypsinized, and then 5 × 104 cells from each treatment were cultured in the presence (filled) or absence (open) of ponasterone A. Cell counts were determined daily for 4 d.
GFP-TAM67–induced Arrest Is Reversible
To determine whether the G1 arrest induced by GFP-TAM67 is reversible, iGT1a cells were treated with ponasterone A for 24 h and cultured in the presence or absence of the inducer; then the growth kinetics of these populations were measured. As a control the growth kinetics of uninduced cells in the presence and absence of ponasterone A were also measured. iGT1a cells that had been arrested and then washed free of ponasterone displayed a lag phase in the first 24 h period relative to the asynchronously growing uninduced cells (Figure 5E). However, after 48 h, the previously arrested population grew at nearly the same rate as the asynchronous population (Figure 5E). Adding ponasterone to the asynchronous population or maintaining ponasterone in the arrested population dramatically slowed, but did not stop, growth in both populations. This slow increase in cell number was due to the expansion of the uninduced population seen in flow cytometry; the induced population remained viable and arrested in G1 for days after ponasterone A treatment (Hennigan and Stambrook, unpublished results). No sub-G1 cell populations were apparent in GFP-TAM67–positive cells and growth resumed upon removal of ponasterone, indicating that GFP-TAM67 did not induce apoptosis or terminal differentiation.
GFP-TAM67 Does Not Restore Contact Inhibition of Growth to HT1080 Cells
HT1080 cells have an activated N-ras mutation that is responsible for their transformed phenotype (Paterson et al., 1987). Because ras activation induces expression of the AP-1 components c-fos (Stacey et al., 1987) and c-jun and this expression is necessary for oncogenic ras function (Ledwith et al., 1990; Lloyd et al., 1991), it is possible that the G1 arrest caused by GFP-TAM67 is an indirect consequence of reversion of the transformed phenotype. Specifically, GFP-TAM67 might restore contact inhibition of proliferation caused by activated N-ras in HT1080 cells, a condition that would also engage a late G1 checkpoint (Guadagno and Assoian, 1991). To test this possibility, iGT1a cells were plated at low, medium, and high density and then treated with 10 μM ponasterone A, and 24 h later cell cycle profiles were determined. Ponasterone A–induced cell cycle arrest occurred at all three densities, (Figure 6), indicating that GFP-TAM67 did not restore contact inhibition of growth and suggesting that the role of AP-1 activity in regulating progression through the G1 phase of the cell cycle is direct.
Figure 6.
Cell cycle arrest is independent of cell density. iGT1a cells were plated at high (A), medium (B), and low density (C) in the presence (top dot plot) or absence (bottom dot plot) of 10 μM ponasterone A. GFP fluorescence is depicted on the Y-axis and DNA content is depicted on the X-axis.
Threshold Effect of GFP-TAM67 Expression on G1 Arrest
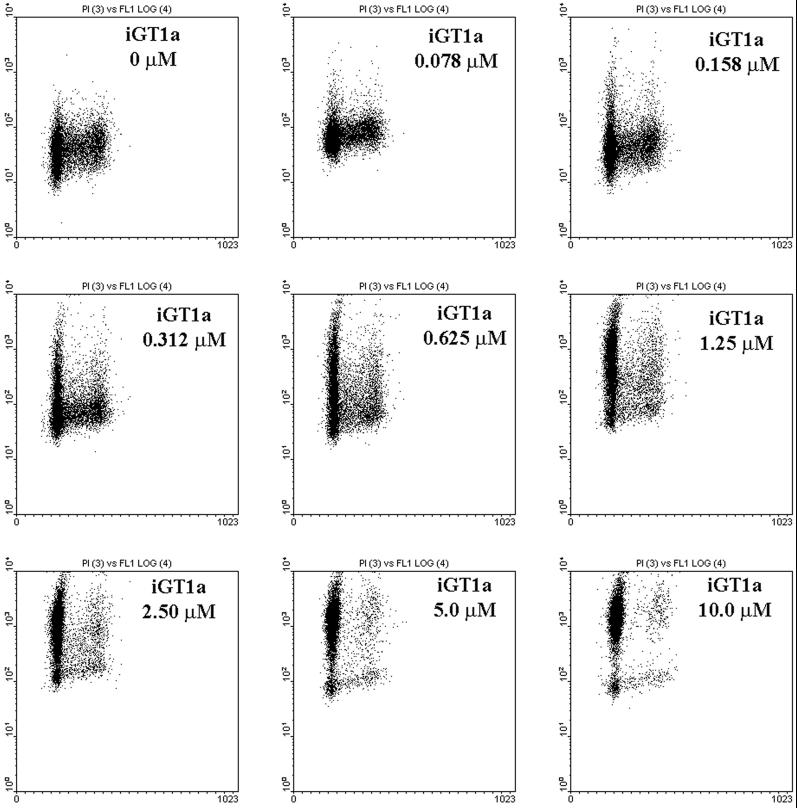
Previous experiments with the bicistronic vectors suggested that cell cycle arrest occurred only in cells expressing high levels of GFP-TAM67. To confirm this, the dose response of ponasterone A on iGT1a cell cycle progression was measured by flow cytometry for a range of ponasterone A concentrations from 0.078 to 10 μM. The green fluorescence intensity of uninduced cells was between 1 and 10 U in an arbitrary 4-log scale and these cells had a G1:S:G2/M ratio of 53.13, 32.02, and 14.86%, respectively (Figure 7; Table 1). Induction of GFP-TAM67 expression was measurable at the lowest doses of ponasterone A, at 0.078 and 0.158 μM, with the proportion of G1 cells increasing in 2% increments with successive doses of ponasterone (ΔG1%, Table 1). Greater concentrations of inducer, from 0.312 to 1.25 μM, resulted in heterogeneous expression of GFP-TAM67, as indicated by the broad distribution of cells in the vertical axis of the dot plots. Cells expressing moderate levels of GFP-TAM67, with green fluorescence intensities between 10 and 100 U, still proliferated but the proportion of cells in G1 increased more dramatically, now by 10% increments with successive doses of ponasterone (ΔG1%, Table 1). Cell cycle arrest was seen only in those cells expressing the most GFP-TAM67, at fluorescence intensities of 100 U or higher. This trend continued; at the highest doses used, 2.5–10 μM, the proportion of cells in G1 approached 90%. At 10 μM ponasterone, most of the cells had a green fluorescence intensity of >100 U and this population had a G1:S:G2/M ratio of 92.59, 3.57, and 3.84%, respectively. These data indicate that high levels of GFP-TAM67 are required to arrest cells in G1.
Figure 7.
G1 arrest by GFP-TAM67 is dose dependent. Cells were treated with the indicated doses of ponasterone A for 24 h and analyzed by flow cytometry. GFP fluorescence is depicted on the Y-axis and DNA content is depicted on the X-axis. The proportion of cells in each the three phases of the cell cycle and increase in G1 phase (Δ%G1) with successive doses of ponasterone A is presented in Table 1.
Table 1.
Changes in cell cycle profile with increasing ponasterone
| Ponasterone A (μM) | G1 (%) | ΔG1 (%) | S (%) | G2/M (%) |
|---|---|---|---|---|
| 0.000 | 53.13 | – | 32.02 | 14.86 |
| 0.078 | 55.21 | 2.08 | 30.80 | 13.99 |
| 0.156 | 56.25 | 1.04 | 28.11 | 15.64 |
| 0.313 | 58.05 | 1.80 | 28.23 | 13.72 |
| 0.625 | 64.61 | 6.56 | 23.13 | 12.26 |
| 1.250 | 74.40 | 9.79 | 14.85 | 9.75 |
| 2.500 | 84.30 | 9.90 | 10.05 | 5.65 |
| 5.000 | 90.41 | 6.11 | 5.50 | 4.08 |
| 10.000 | 92.59 | 2.18 | 3.57 | 3.84 |
The proportion of cells in G1, S, or G2/M phase of the cell cycle was derived from the data presented in figure 7. The second column (ΔG1%) represents the increase in the proportion of cells in G1 compared to the previous dose.
GFP-TAM67 Does Not Affect the Expression of Components of G1 Cyclin-CDK Complexes
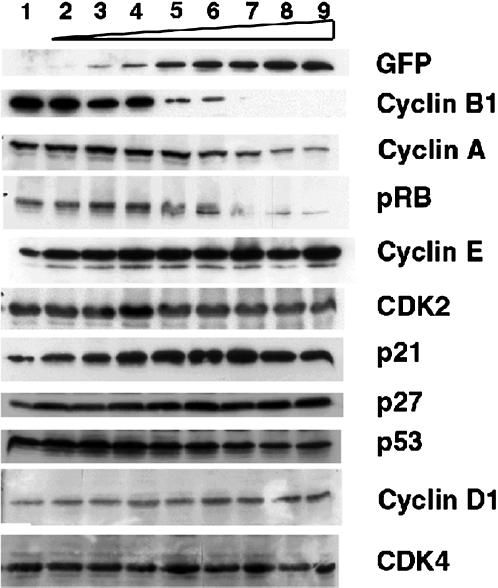
It has been suggested that AP-1 family members regulate cell cycle by inducing the expression of cyclin D1 via AP-1 sites in its promoter region (Brown et al., 1998). It is therefore possible that GFP-TAM67 arrests cells in G1 by inhibiting the expression of cyclin D1 or of another component of the cyclin D1-cdk4/6 or cyclin E-cdk2 complexes. To test this, immunoblot analysis was performed on lysates from iGT1a cells exposed to a range of ponasterone A concentrations ranging from 0.078 to 10 μM (Figure 7). As a control for GFP-TAM67 induction, samples were probed with an anti-GFP monoclonal antibody. Increasing amounts of the 53 kDa GFP-TAM67 mirrored the induction seen by the same concentrations of ponasterone A by flow cytometry in Figure 7. Note the steady rise in GFP-TAM67 expression in contrast to the sharp increases in the proportion of cells in G1 presented in Table 1. Expression of cyclin B1 was inversely proportional to GFP-TAM67 expression. Cyclin A levels also decreased with increasing ponasterone A concentration but less dramatically than seen for cyclin B1. This reduction in cyclin B1 and cyclin A expression is consistent with a late G1 arrest. The retinoblastoma gene product showed a progressive hypophosphorylation with increasing ponasterone as indicated by the appearance of a higher mobility band, accompanied by a reduction in the overall levels of this protein. This suggests that the function of the G1 cyclin-kinase complexes are impaired by GFP-TAM67. Immunoblot analysis showed that the amounts of cyclin E and cyclin D1 were essentially unchanged in response to ponasterone, as were the levels of cdk2 and cdk4. Cyclin D3 and cdk6 levels were equivalent between treated and untreated cells, whereas cyclin D2 could not be detected by Western blot (Hennigan and Stambrook, unpublished results). The results of this experiment rule out the possibility that GFP-TAM67 inhibits the expression of any component of the cyclin D1 or cyclin E complexes in this cell type.
We also tested whether GFP-TAM67 increased the expression of a variety of cyclin D1-cdk inhibitors. Immunoblot analysis failed to show expression of either p16ink4a or any of the other cyclin D1 inhibitors, p15INK4b, p18INK4c, or p19INK4d (Hennigan and Stambrook, unpublished results). Levels of p27KIP1 were constitutively high and modestly elevated upon ponasterone administration (Figure 8).
Figure 8.
G1 arrest by GFP-TAM67 does not affect the levels of G1 cyclin-cdk complex components. The iGT1a cells were treated with ponasterone A in the following doses: 1, 0 μM; 2, 0.078 μM; 3, 0.158 μM; 4, 0.313 μM; 5, 0.624 μM; 6, 1.25 μM; 7, 2.5 μM; 8, 5 μM; 9, 10 μM. Cell lysates were electrophoresed, transferred to PVDF membranes, and probed with the indicated antibodies.
It has been shown that c-jun represses p53 induction in mouse embryo fibroblasts, causing increased amounts of p21CIP1/WAF1 to inhibit growth (Schreiber et al., 1999). Ponasterone A significantly increased p21CIP1/WAF1 expression. Basal expression of p53 were high and ponasterone caused a reduction of p53 levels. The increase in p21CIP1/WAF1 expression in response to GFP-TAM67, coupled with the reduction levels of p53 caused by ponasterone, rules out a mechanism for AP-1 function similar to that described in mouse embryo fibroblasts, in which GFP-TAM67 relieves an AP-1–mediated repression of the p53 gene. It is possible that this increase in p21CIP1/WAF1 expression is responsible for GFP-TAM67–induced G1 arrest. To determine this a more careful analysis of the activity and constitution of the cyclin D1 and the cyclin E complexes was necessary.
GFP-TAM67 Expression Inhibits Cyclin D1- and Cyclin E-dependent Kinase Activities and Induces p21CIP1/WAF1
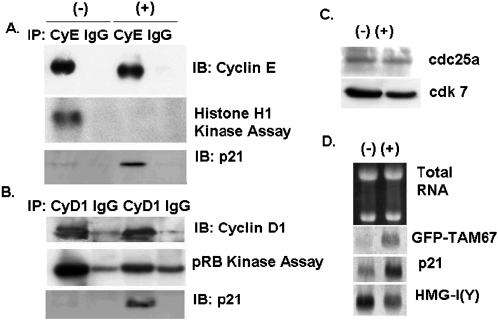
Because the protein levels of the cyclin D1 and cyclin E complex were unchanged, an alternate explanation of the G1 arrest is that GFP-TAM67 inhibits phosphorylation of pRB by preventing the activation of these complexes rather than the expression of their component genes. To confirm this in vitro kinase assays were performed. Cyclin E was immunoprecipitated from lysates of iGT1a cells treated with vehicle or 10 μM ponasterone A for 24 h. Kinase assays were then performed with histone H1 as a substrate. This experiment showed that the cyclin E immunoprecipitated from induced cells was unable to phosphorylate histone H1 (Figure 9A). Uninduced cells had a robust histone H1 kinase activity, and immunoblot analysis of these immunoprecipitates showed that the amount of cyclin E in the reactions was equivalent between induced and uninduced cells. Cyclin D1 immunoprecipitates also had a reduced kinase activity to a recombinant pRB-GST fusion protein (Figure 9B). Blots of these immunoprecipitates also showed equal levels of cyclin D1. Although the reduction in cyclin D1 kinase activity was not as large as that seen in the cyclin E immunoprecipitates, the degree of inhibition of cyclin D1 kinase activity was significant, from 5- to 10-fold. Significantly, immunoblots of both cyclin E and cyclin D1 immunoprecipitates revealed a dramatic increase in the amount of p21CIP1/WAF1 that associated with these complexes (Figure 9, A and B). Northern blot analysis confirmed that the mRNA for p21CIP1/WAF1 was induced by ponasterone A by sixfold, comparable to the induction of GFP-TAM67. In contrast mRNA for an unrelated gene, HMG-I(Y), was reduced by twofold (Figure 9D). No change in the levels of two proteins that activate cdks, cdc25a and cdk7, were seen in response to GFP-TAM67 (Figure 9C.). Also, overexpression of cdc25a failed to rescue the growth inhibitory function of GFP-TAM67 in a colony formation assay (Hennigan and Stambrook, unpublished results). This experiment confirmed that the activity of the cyclin D1- and cyclin E-dependent kinase complexes was inhibited in cells expressing GFP-TAM67 and suggests that the induction of the p21CIP1/WAF1 gene by GFP-TAM67 is responsible for the inhibition of cyclin E and cyclin D1 kinase activity.
Figure 9.
GFP-TAM67 expression inhibits activation of both cyclin D1 and cyclin E kinase complexes and induces p21CIP1/WAF1. The iGT1a cells were treated with and without 10 μM ponasterone A for 24 h. (A) Cyclin E immunoprecipitates were assayed for histone H1 kinase activity, blotted, and probed with antibodies to cyclin E and p21CIP1/WAF1. (B) Cyclin D1 immunoprecipitates were assayed for retinoblastoma protein kinase activity, blotted, and probed with antibodies to cyclin D1 and p21CIP1/WAF1. (C) Immunoblots of cell lysates from iGT1a cells treated with 0 or 10 μM ponasterone A and probed with antibodies to cdc25a and cdk7. (D) Northern blot analysis of 5 μg of total RNA isolated from iGT1a cells treated with 0 or 10 μM ponasterone A and probed with the indicated cDNAs.
DISCUSSION
We have previously demonstrated that expression of TAM67 in FBR v-fos-transformed rat fibroblasts inhibits invasion in vitro (Lamb et al., 1997). Others have shown a similar inhibition by TAM67 of mouse keratinocyte invasion in response to 12-O-tetradecanoylphorbol 13-acetate (Dong et al., 1997). When expressed in human A431 carcinoma cells, TAM67 inhibits motility and cytoskeletal rearrangements response to epithelial growth factor by preventing activation of rac and rho (Malliri et al., 1998). These publications demonstrate an important role for AP-1 in mediating biological functions that are critical for motility and metastasis, aspects of tumor progression that do not directly involve regulation of proliferation. However, the existence of tumor cell lines constitutively expressing TAM67 is inconsistent with previous work showing that overexpression of this mutant inhibits proliferation of transformed cells (Brown et al., 1993; Rapp et al., 1994). We sought to resolve this inconsistency by fusing TAM67 to GFP and expressing it from both the bicistronic and ecdysone-inducible expression vectors. These experiments show that a high level of GFP-TAM67 expression in HT1080 inactivates the cyclin D1:cdk4/6 and cyclin E:cdk2 complexes and arrests in the cells in G1. The p21CIP1/WAF1 mRNA is induced by GFP-TAM67, and the p21CIP1/WAF1 protein associates with inactive cyclin D1 and cyclin E complexes in arrested cells.
Cell Cycle Control
The mechanism by which the ras-MAP kinase signal transduction cascade interacts with the RB pathway during cell cycle progression is poorly understood. The induction of AP-1 activity in response to upstream signaling and the identification of cyclin D1 as a potential target of AP-1 indicates that the ras-MAP kinase and RB pathways are connected through AP-1 via cyclin D1 regulation (Miao and Curran, 1994; Bakiri et al., 2000). However, our data argue against cyclin D1 as a critical target of AP-1 in HT1080 cells and suggest an alternative role for AP-1 in controlling the activity of G1 cyclin:cdk complexes by regulating the expression of p21CIP1/WAF1. Infection of chick embryo fibroblasts with v-jun induces proliferation by activating the cyclin E:cdk2 complex without increasing the expression of its component genes or cyclin D1 (Clark et al., 2000), an observation that is complimentary to the results described here. Induction of p21CIP1/WAF1 mRNA by GFP-TAM67 is p53 independent, as indicated by the reduction of p53 levels that is coincident with the increase in GFP-TAM67 and p21CIP1/WAF1 levels. This is in contrast to a report describing c-jun as participating in a p53-dependent regulation of p21CIP1/WAF1 in c-jun null MEFs (Schreiber et al., 1999). The fact that GFP-TAM67 expression did not reduce cyclin D1 levels is probably due to a combination of the complexity of the endogenous cyclin D1 promoter region and the cell background used. The cyclin D1 promoter region has elements that are responsive to a variety of stimuli (Albanese et al., 1995; Lee et al., 1999; Matsumura et al., 1999), indicating that the cyclin D1 promoter is regulated by multiple, redundant signaling pathways. Many of the experiments identifying cyclin D1 as a biologically significant target of AP-1 were preformed in primary mouse fibroblast cells derived from nullizygous embryos and represent the behavior of AP-1 in normal, untransformed cells (Brown et al., 1998; Wisdom et al., 2000). In contrast, HT-1080 is a fully transformed human tumor cell that is known to have impaired p53 function, activated N-ras, and to express autocrine motility and growth factors (Paterson et al., 1987; Silletti and Raz, 1993; Paulson et al., 1998). The finding that AP-1 has different targets in HT1080 cells and mouse embryo fibroblasts is significant in terms of understanding the role of AP-1 in malignant disease. HT1080 is derived from a tumor cell population that has undergone successive rounds of selection in vivo for aggressive growth and metastasis and is a better model for the type of cell that is the ultimate target of antitumor therapies.
Mechanism of GFP-TAM67 Action
The TAM67 mutant is particularly well suited as a tool to investigate AP-1 function in transformed cells. TAM67 fails to induce transcription via AP-1 elements because of a deletion, from amino acids 3–122, that removes the major transactivating domain but retains the DNA-binding region and the leucine zipper dimerization domain (Alani et al., 1991). As a result TAM67 has the ability to interact with the same range of AP-1 proteins as c-jun, thereby functioning as a global inhibitor of the AP-1 transcription factor complex (Curran and Franza, 1988; Benbrook and Jones, 1990). In addition, TAM67 has the ability to interfere with interactions on composite elements between AP-1 complexes and other transcription factors such as the glucocorticoid response factor and NF-AT (Petark et al., 1994; Cippitelli et al., 1995). The dominant negative activity of TAM67 is thought to be the result of two possible mechanisms. The first mechanism is “quenching,” the formation of inactive heterodimers between TAM67 and AP-1 proteins (Brown et al., 1994). A second mechanism is “blocking,” TAM67 dimers preventing functional AP-1 complexes from binding to target DNA, thus inhibiting AP-1–mediated gene expression (Brown et al., 1994). Immunoprecipitation experiments with radiolabeled lysates prepared under denaturing and nondenaturing conditions suggest that at high concentrations, GFP-TAM67 predominately forms homodimers. The absence of heterodimers between GFP-TAM67 and endogenous leucine zipper proteins is unexpected. It is possible that GFP-TAM67 interacts with less abundant binding partners that are not visible in the radioimmunoprecipitation assay. However, this preponderance of GFP-TAM67 homodimers might account for the requirement of high levels of GFP-TAM67 to induce G1 arrest. Because Jun-Jun homodimers have a 30-fold lower affinity for DNA than Fos-Jun heterodimers (Rauscher et al., 1988b), higher concentrations of GFP-TAM67 homodimers might be necessary to block endogenous AP-1 DNA-binding activity in vivo.
The induction of p21CIP1/WAF1 by GFP-TAM67 suggests that it counteracts an AP-1–mediated repression of the p21CIP1/WAF1 promoter. One report suggests that p21CIP1/WAF1 is repressed via an AP-1–like site at −1510 in the promoter (Crowe et al., 2000). Intriguingly, c-Jun has been implicated in separate reports as either inducing or repressing p21CIP1/WAF1 expression by an unconventional mechanism (Kardassis et al., 1999; Wang et al., 2000). Although these reports demonstrate opposing effects of c-jun in different cell backgrounds, they both map the c-jun interacting site as a proximal Sp-1 element located from −122 to −64 of the promoter. Both papers describe an atypical interaction between c-jun and Sp-1 that does not involve the binding of c-jun to DNA. Interestingly, it was found that the leucine zipper domain alone mediated this “superactivator” function of c-jun. This suggests that rather than inhibiting an AP-1–mediated repression by the blocking mechanism described above, GFP-TAM67 might directly activate the p21CIP1/WAF1 promoter by acting as a superactivator. Further experiments are required to determine whether interactions between Sp-1 and either endogenous c-jun or GFP-TAM67 directly regulate p21CIP1/WAF1 expression in HT1080 cells. However, this kind of c-jun–mediated activation, one that is independent of the transactivating domain, could explain the previously reported behavior of a fusion between TAM67 and the TAF2 ligand-dependent transactivating domain of the estrogen receptor. This construct displayed a restored AP-1–transactivating activity but still inhibited transformation by activated c-Ha-ras or c-raf, thus separating the AP-1 transactivation from the dominant negative function of TAM67 (Kim et al., 1996). This also is interesting in light of experiments with a temperature-sensitive mutant of v-fos, which suggest that the ability to activate AP-1–dependent transcription is dispensable for transforming activity (Joos and Muller, 1992).
Many studies utilizing antisense and dominant negative strategies have demonstrated that AP-1 is a critical downstream target of the ras-MAP kinase pathway (Mercola et al., 1987; Ledwith et al., 1990; Roux et al., 1990; Rapp et al., 1994; Suzuki et al., 1994; Johnson et al., 1996; Kralova et al., 1998). It is a transcription factor that translates short-term biochemical signals generated by mitogenic and oncogenic signal transduction cascades into long-term changes in gene expression that constitute the neoplastic phenotype. The work described here establishes a novel, AP-1–dependent, link between oncogene-mediated signal transduction cascades and cell cycle control by regulating the activation of cyclin-dependent kinase activity via the p21CIP1/WAF1 gene. Ultimately, elucidation of the mechanism by which GFP-TAM67 regulates p21CIP1/WAF1 will provide valuable insight into the role of AP-1 in the regulation of tumor cell proliferation.
ACKNOWLEDGMENTS
We thank Dr. Michael Kaminsky for providing GFP plasmids and the ecdysone-inducible system, Drs. Erik Knudsen, Kenji Fukasawa, and Larry Sherman for providing antibodies, and Dr. Karen Knudesn for help with the cyclin D1 kinase assay. Flow cytometry was performed by Jim Cornealius at the Cincinnati Shriners Hospital. We would also like to thank Dr. Sherman and Dr. Robert Brackenbury for critical reading of the manuscript. This work was supported by American Cancer Society institutional grant IRG-92-026-06.
Footnotes
Corresponding author. E-mail address: Robert.Hennigan@uc.edu.
REFERENCES
- Alani R, Brown P, Binetruy B, Dosaka H, Rosenberg RK, Angel P, Karin M, Birrer MJ. The transactivating domain of c-Jun is required for cotransformation of rat embryo cells. Mol Cell Biol. 1991;11:6286–6295. doi: 10.1128/mcb.11.12.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-ETS-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M. Cell cycle-dependent variations in c-Jun, and JunB phosphorylation: a direct role in the control of cyclin D1 expression. EMBO J. 2000;19:2056–2068. doi: 10.1093/emboj/19.9.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrook DM, Jones NC. Heterodimer formation between CREB and Jun proteins. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- Brown JR, Nigh E, Lee RJ, Ye H, Thompson MA, Saudou F, Pestell RG, Greenberg ME. Fos family members induce cell cycle entry by activating cyclin D1. Mol Cell Biol. 1998;18:5606–5619. doi: 10.1128/mcb.18.9.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PH, Alani R, Preis LH, Szabo E, Birrer MJ. Suppression of oncogene induced transformation by a deletion mutant of c-Jun. Oncogene. 1993;8:877–886. [PubMed] [Google Scholar]
- Brown PH, Chen TK, Birrer MJ. Mechanism of action of a dominant negative mutant of c-Jun. Oncogene. 1994;9:791–799. [PubMed] [Google Scholar]
- Cippitelli M, Sica A, Viggiano V, Ye J, Ghosh P, Birrer MJ, Young S. Negative transcriptional regulation of the interferon-gamma promoter by glucocorticoids and dominant negative mutants of c-Jun. J Biol Chem. 1995;270:12548–12556. doi: 10.1074/jbc.270.21.12548. [DOI] [PubMed] [Google Scholar]
- Clark W, Black EJ, MacLaren A, Kruse U, LaThange N, Vogt PK, Gillespie DAF. v-Jun overrides the mitogen dependence of S-phase entry by deregulating retinoblastoma protein phosphorylation, and E2F-pocket protein interactions as a consequence of enhanced cyclin E-cdk2 catalytic activity. Mol Cell Biol. 2000;20:2529–2542. doi: 10.1128/mcb.20.7.2529-2542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe DL, Brown TN, Kim R, Smith SM, Lee MK. A c-fos/estrogen receptor fusion protein promotes cell cycle progression and proliferation of human cancer cell lines. Mol Cell Biol Res Commun. 2000;4:243–248. doi: 10.1006/mcbr.2000.0221. [DOI] [PubMed] [Google Scholar]
- Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55:395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- Dong Z, Crawford HC, Lavrovsky V, Taub D, Watts R, Matrisian LM, Colburn NH. A dominant negative mutant of jun blocking 12-O-tetradecanoylphorbol-13-acetate induced invasion in mouse keratinocytes. Mol Carcinog. 1997;19:204–212. doi: 10.1002/(sici)1098-2744(199707)19:3<204::aid-mc8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Guadagno TM, Assoian RK. G1/S control of anchorage independent growth in the fibroblast cell cycle. J Cell Biol. 1991;115:1419–1425. doi: 10.1083/jcb.115.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Spiegelman BM, Hanahan D, Wisdom R. Cellular transcription and malignancy induced by ras require c-jun. Mol Cell Biol. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos K, Muller R. Analysis of temperature sensitive functions of Fos: lack of a correlation between transformation and TRE dependant trans activation. Oncogene. 1992;7:1933–1939. [PubMed] [Google Scholar]
- Kardassis D, Papakosta P, Pardali K, Moustakas A. c-Jun transactivates the promoter of the human p21WAF1/Cip1 gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J Biol Chem. 1999;274:29572–29581. doi: 10.1074/jbc.274.41.29572. [DOI] [PubMed] [Google Scholar]
- Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- Kim S, Brown PH, Birrer MJ. The inhibitory activity of a transdominant c-Jun mutant fused to the ligand binding domain of the estrogen receptor. Oncogene. 1996;12:1043–1053. [PubMed] [Google Scholar]
- Kovary K, Bravo R. The Jun and Fos families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralova J, Liss AS, Bargmann W, Bose HR. AP-1 factors play an important role in transformation induced by the v-rel oncogene. Mol Cell Biol. 1998;18:2997–3009. doi: 10.1128/mcb.18.5.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RF, Hennigan RF, Turnbull K, Katsanakis KD, MacKenzie ED, Birnie GD, Ozanne BW. AP-1 mediated invasion requires increased expression of the hyaluronan receptor CD44. Mol Cell Biol. 1997;17:963–976. doi: 10.1128/mcb.17.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledwith BJ, Manam S, Kraynak AR, Nichols WW, Bradley MO. Antisense-fos RNA causes partial reversion of the transformed phenotypes induced by the c-Ha-Ras oncogene. Mol Cell Biol. 1990;10:1545–1555. doi: 10.1128/mcb.10.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RJ, Albanese C, Stenger RJ, Watanabe G, Inghirami G, Haines GK, Webster M, Muller WJ, Brugge JS, Davis RJ, Pestell RG. pp60v-src induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. J Biol Chem. 1999;274:7341–7350. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- Lloyd A, Yancheva N, Wasylyk B. Transformation supressor activity of a Jun transcription factor lacking its activation domain. Nature. 1991;352:635–638. doi: 10.1038/352635a0. [DOI] [PubMed] [Google Scholar]
- Malliri A, Symons M, Hennigan RF, Hurlstone AFL, Lamb RF, Wheeler T, Ozanne BW. The transcription factor AP-1 is required for EGF-induced activation of Rho-like GTPases, cytoskeletal rearrangements, motility and in vitro invasion of A431 cells. J Cell Biol. 1998;143:1087–1099. doi: 10.1083/jcb.143.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura I, Kitamura T, Wakao H, Tanaka H, Hashimoto K, Albanese C, Downward J, Pestell RG, Kane S. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999;18:1367–1377. doi: 10.1093/emboj/18.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercola D, Rundell A, Westwick J, Edwards SA. Antisense RNA to the c-fos Gene: restoration of density-dependant growth arrest in a transformed cell line. Biochem Biophys Res Commun. 1987;147:288–294. doi: 10.1016/s0006-291x(87)80119-5. [DOI] [PubMed] [Google Scholar]
- Miao GG, Curran T. Cell transformation by c-fos requires an extended period of expression and is independent of the cell cycle. Mol Cell Biol. 1994;14:4295–4310. doi: 10.1128/mcb.14.6.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson H, Reeves R, Brown R, Hall A, Furth M, Bos J, Jones P, Marshall C. Activated N-ras controls the transformed phenotype of HT1080 human fibrosarcoma cells. Cell. 1987;51:803–812. doi: 10.1016/0092-8674(87)90103-6. [DOI] [PubMed] [Google Scholar]
- Paulson TG, Almasan A, Brody LL, Whal GM. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petark D, Sarfaz SA, Birrer MJ, Ashwell JJ, Zacharchuk CM. Dominant negative mutant of c-Jun inhibits NF-AT transcriptional activity and prevents Il-2 gene transcription. J Immunol. 1994;153:2046–2051. [PubMed] [Google Scholar]
- Rapp UR, Troppmair J, Beck T, Birrer MJ. Transformation by Raf and other oncogenes renders cells differentially sensitive to growth inhibition by a dominant negative c-Jun mutant. Oncogene. 1994;9:3493–3498. [PubMed] [Google Scholar]
- Rauscher FJ, Cohen DR, Curran T, Bos TJ, Vogt PK, Bohmann D, Tjian R, Franza BR., Jr Fos associated protein p39 is the product of the Jun proto-oncogene. Science. 1988a;240:1010–1016. doi: 10.1126/science.3130660. [DOI] [PubMed] [Google Scholar]
- Rauscher FJ, Voulalas PJ, Franza BR, Jr, Curran T. Fos and Jun bind cooperatively to the AP-1 site: reconstitution in vitro. Growth Dev. 1988b;2:1687–1699. doi: 10.1101/gad.2.12b.1687. [DOI] [PubMed] [Google Scholar]
- Roux P, Blanchard J-M, Fernandez A, Lamb N, Jeanteur P, Piechaczyk M. Nuclear localization of c-Fos, but not v-Fos proteins, is controlled by extracellular signals. Cell. 1990;341:351. doi: 10.1016/0092-8674(90)90167-d. [DOI] [PubMed] [Google Scholar]
- Saha P, Eichbaum Q, Silberman ED, Mayer B, Dutta A. p21CIP1 and Cdc25A: competetion between an inhibitor and an activator of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4338–4345. doi: 10.1128/mcb.17.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silletti S, Raz A. Autocrine motility factor is a growth factor. Biochem Biophys Res Commun. 1993;194:446–457. doi: 10.1006/bbrc.1993.1840. [DOI] [PubMed] [Google Scholar]
- Stacey DW, Watson T, Kung H-F, Curran T. Microinjection of transforming ras protein induces c-fos expression. Mol Cell Biol. 1987;7:523–537. doi: 10.1128/mcb.7.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Murakami M, Onai N, Fukuda E, Hashimoto Y, Sonobe MH, Kameda T, Ichinose M, Miki K, Iba H. Analysis of AP-1 function in cellular transformation pathways. J Virol. 1994;68:3527–3535. doi: 10.1128/jvi.68.6.3527-3535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-H, Tsao Y-P, Chen H-J, Wang H-W, Chen S-L. Transcriptional repression of p21(Waf1/CIP1/Sdi1) gene by c-jun through Sp1 site. Biochem Biophys Res Commun. 2000;270:303–310. doi: 10.1006/bbrc.2000.2422. [DOI] [PubMed] [Google Scholar]
- Wisdom R, Johnson RS, Moore C. c-Jun regulates cell cycle progression, and apoptosis by distinct mechanisms. EMBO J. 2000;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]