De Muyt et al. identified the ZZS (Zip2–Zip4–Spo16) complex, required for crossover formation, which carries two distinct activities: one provided by Zip4, which acts as hub through physical interactions with components of the chromosome axis and the crossover machinery, and the other carried by Zip2 and Spo16, which preferentially bind branched DNA molecules in vitro.
Keywords: homologous recombination, crossover, meiosis, Zip2–Zip4–Spo16 complex, XPF–ERCC1, synaptonemal complex
Abstract
Meiotic crossover formation requires the stabilization of early recombination intermediates by a set of proteins and occurs within the environment of the chromosome axis, a structure important for the regulation of meiotic recombination events. The molecular mechanisms underlying and connecting crossover recombination and axis localization are elusive. Here, we identified the ZZS (Zip2–Zip4–Spo16) complex, required for crossover formation, which carries two distinct activities: one provided by Zip4, which acts as hub through physical interactions with components of the chromosome axis and the crossover machinery, and the other carried by Zip2 and Spo16, which preferentially bind branched DNA molecules in vitro. We found that Zip2 and Spo16 share structural similarities to the structure-specific XPF–ERCC1 nuclease, although it lacks endonuclease activity. The XPF domain of Zip2 is required for crossover formation, suggesting that, together with Spo16, it has a noncatalytic DNA recognition function. Our results suggest that the ZZS complex shepherds recombination intermediates toward crossovers as a dynamic structural module that connects recombination events to the chromosome axis. The identification of the ZZS complex improves our understanding of the various activities required for crossover implementation and is likely applicable to other organisms, including mammals.
During meiosis, cells undergo DNA recombination to form at least one crossover (CO) between each pair of homologous chromosomes, which ensures their proper segregation during meiosis I and thus avoids aneuploidy. Meiotic recombination is initiated by the induction of DNA double-strand breaks (DSBs) catalyzed by the topoisomerase relative Spo11. DSBs are subsequently resected to generate 3′ single-stranded tails that invade a homologous intact DNA duplex, preferentially on the homologous chromosome, forming a D-loop structure to yield either CO or non-CO (NCO) recombinant products (Bishop and Zickler 2004). In all studied species, the number of DSBs exceeds the final number of COs, indicating that meiotic cells designate a subpopulation of DSBs to specifically become a CO. Two antagonistic activities play an important role in controlling the CO/NCO balance. Sgs1, the Bloom (BLM) syndrome RecQ family helicase ortholog in budding yeast, functions together with Top3 and Rmi1 to promote the disassembly of strand invasion intermediates (Oh et al. 2007; De Muyt et al. 2012; Zakharyevich et al. 2012; Kaur et al. 2015; Tang et al. 2015). Such an activity favors NCO production by synthesis-dependent strand annealing (SDSA), in which the extended invading strand is displaced and annealed to the other side of the break. In parallel, a meiosis-specific pathway ensures that a subset of DSBs is repaired into COs. For this, the single-end invasion (SEI) recombination intermediate, a metastable D-loop structure, is formed, which is subsequently transformed via capture of the second break end into a double Holliday junction (dHJ) (Hunter and Kleckner 2001). The dHJ is then resolved into a CO product. A group of proteins collectively termed “ZMM” (for Zip, Msh, and Mer) has been proposed to protect recombination intermediates from the anti-recombination activity of Sgs1 (Jessop et al. 2006; Oh et al. 2007). The ZMM proteins are important to stabilize nascent joint molecules in vivo and are responsible for the formation of ∼85% of all COs while being dispensable for NCOs (Börner et al. 2004; Lynn et al. 2007). The ZMM group includes proteins that act directly on recombination intermediates in vitro, such as the Mer3 helicase, which preferentially recognizes and migrates D loops, as well as the MutS homolog heterodimer Msh4–Msh5, which stabilizes dHJs (Mazina et al. 2004; Snowden et al. 2004; Duroc et al. 2017). The ZMM group also includes Zip1 (the central element of the synaptonemal complex [SC]) and Zip3 (an E3 ligase) as well as Zip2, Zip4, and Spo16 with less well-defined functions.
In budding yeast, the ZMM proteins also participate in SC assembly, which starts with the formation of the axial element (or chromosome axis) along each pair of sister chromatids. Axial elements tether loops of chromatin at their bases by meiosis-specific axis proteins, which include Hop1, Red1, and the cohesin subunit Rec8 (Smith and Roeder 1997; Klein et al. 1999; Panizza et al. 2011). Next, Zip1 progressively polymerizes between each pair of homologous axes. The ZMM proteins play an important role to coordinately promote SC polymerization (Chua and Roeder 1998; Agarwal and Roeder 2000; Tsubouchi et al. 2006; Shinohara et al. 2008). However, it is not clear how ZMM-dependent CO formation and SC assembly are functionally linked (Lynn et al. 2007).
Intriguingly, Zip2 shows predicted structural similarity to SHOC1 and C9orf84 of plants and humans, respectively, and possesses a predicted XPF domain (Macaisne et al. 2008). XPF-related proteins are conserved endonucleases involved in branched DNA structure recognition and processing (Ciccia et al. 2008). During evolution, ancestors of the XPF family diverged from a common homodimeric complex into several types of heterodimers (Dehé and Gaillard 2017). Each heterodimer is composed of an XPF subunit (which may contain a catalytic domain) and a noncatalytic subunit that belongs to the ERCC1 protein family (Ciccia et al. 2008). The XPF–ERCC1-like heterodimers MUS81–EME1 (Mus81–Mms4 in budding yeast), MUS81–EME2, and XPF–ERCC1 (Rad1–Rad10 in budding yeast) have key roles in DNA replication, recombination, and repair and share a conserved catalytic domain that specifically cleaves secondary DNA structures (Dehé and Gaillard 2017). Surprisingly, the FANCM–FAAP24 complex, another member of this family, does not show any endonuclease activity (Ciccia et al. 2007, 2008). Instead, it is suggested that FANCM–FAAP24 works as a recognition module that binds to a DNA structure caused by a stalled replication fork at an interstrand cross-link and recruits other factors for subsequent repair (Xue et al. 2015). Therefore, two types of XPF–ERCC1-like heterodimer might coexist: “nuclease-active” and “nuclease-inactive” variants.
In this study, we provide evidence that two ZMM proteins, Zip2 and Spo16, form a meiosis-specific XPF–ERCC1-like complex. The recombinant Zip2 XPF domain together with Spo16 preferentially binds branched DNA structures, such as D loops and HJs. We suggest that Zip2–Spo16 forms a “nuclease-inactive” heterodimer involved in the recognition of DNA junctions formed during meiotic homologous recombination. In vivo, the Zip2 XPF domain is important to promote wild-type levels of COs. In addition, we show that the Zip2–Spo16 heterodimer forms a stable complex with a third ZMM protein, Zip4. We found that Zip4 physically interacts with other ZMM proteins and with the axial element of the SC, providing a direct physical link between CO-designated recombination intermediates and SC assembly.
Results
Zip2 forms a complex with Zip4 and Spo16 in meiotic cells
Despite exhibiting a well-defined predicted XPF domain (Macaisne et al. 2008), the molecular role of Zip2 is unknown. To gain insights into its function during meiosis, we conducted a tandem affinity purification (TAP) tag pull-down of the Zip2 complexes in meiotic cell extract at 4.25 h after meiosis induction, the expected time when recombination occurs. The relative abundance of Zip2 partners from three biological replicates was determined by label-free quantitative mass spectrometry-based proteomics (Fig. 1A; Supplemental Table S1). Interestingly, Spo16 and Zip4 were the most abundant identified partners among the Zip2 copurified proteins. Estimation of the relative abundance of Zip2 partners suggested that, on average, about one copy of Zip4 and Spo16 is associated with one Zip2 protein, which would be compatible with Zip2, Zip4, and Spo16 forming a stoichiometric complex of a 1:1:1 ratio (Fig. 1B; Supplemental Table S1). Reciprocally, Zip2- and Spo16-derived peptides were found as top candidates in the Zip4-TAP precipitates (Fig. 1C). Likewise, Zip2 and Zip4 were the highest-ranked candidates identified in the Spo16-TAP eluates (Fig. 1C). In addition, coimmunoprecipitation assays confirmed that Zip2 is physically associated with Zip4 and Spo16 in meiotic cells (Fig. 1D). Furthermore, yeast two-hybrid analysis revealed that Zip2, Zip4, and Spo16 interact with each other (Fig. 1F; Supplemental Table S2). These results are consistent with previous studies that showed that Zip2 colocalizes with Zip4 on chromosome spreads and that Spo16 coimmunoprecipitates with Zip4 (Tsubouchi et al. 2006; Shinohara et al. 2008). Altogether, we conclude that Zip2, Zip4, and Spo16 form a stable complex, referred to here as the “ZZS” complex.
Figure 1.

Zip2, Zip4, and Spo16 form a stable complex and are physically connected to the chromosome axis and post-translational modifiers. (A, left) Representative silver-stained gel of TAP eluates from meiotic cell extracts of Zip2-TAP. (Right) Mass spectrometry analysis of Zip2-TAP partners during meiosis. t = 4.25 h. The volcano plot indicates in red the proteins significantly copurified with Zip2. Log2(fold change) > 3; log10(P-value) < 2. Selected top candidates are indicated. Msh5 protein is also shown despite being just below the fixed cutoff. The entire list is in Supplemental Table S1. The experiment was done in triplicate. (B) Determination of relative abundance by mass spectrometry of Spo16, Zip4, Red1, Hop1, and Zip3 relative to the Zip2 protein. Values are the mean ± SD. of three biological replicates. (C, left) Representative silver-stained gels of TAP eluates from meiotic cell extracts of Zip4-TAP and Spo16-TAP. (Right) Mass spectrometry analysis of Zip4-TAP and Spo16-TAP partners during meiosis. t = 4.25 h. The corresponding volcano plots are shown using the same parameters as for the Zip2-TAP experiment. The entire list is in Supplemental Table S1. The experiments were done in triplicate. (D) Coimmunoprecipitation between Spo16-TAP, Zip4-Flag, and Zip2-Myc from meiotic cells at 4 h in meiosis, analyzed by Western blot. The asterisks indicated a nonspecific cross-hybridizing band. (E) Coimmunoprecipitation between Zip4-Flag, Zip2-Myc, and V5-Red1 from meiotic cells at 4 h in meiosis, analyzed by Western blot. (F) Yeast two-hybrid assays. Interaction between the prey fused with the GAL4 activation domain (Gal4AD) and the bait fused with the GAL4 DNA-binding domain (Gal4BD). Growth on −His/Ade medium indicates an interaction. A cartoon summarizing the network for protein–protein interactions identified by TAP and supported by yeast two-hybrid assays is indicated. See also Supplemental Tables S1 and S2.
To obtain more information about the size and composition of the ZZS complex, we performed gel filtration analysis of the Spo16-TAP-associated complexes purified from meiotic cell extract at 4 h of meiosis (Fig. 2A). Western blot analysis revealed that a large fraction of the Spo16 complex eluted between 500 and 600 kDa (around fraction 11). Importantly, this fraction of Spo16 coelutes perfectly with Zip4 and Zip2, confirming the presence of a stable ZZS complex. The theoretical size of a 1:1:1 ratio complex is only 263 kDa, suggesting that the ZZS complex may have a different composition. The higher apparent molecular weight may result from multimerization of the ZZS complex or from other proteins associated with the ZZS complex. Interestingly, a second type of complex (fraction 13) contained Zip2 and Spo16 but not Zip4. Importantly, the full-length Zip2 was absent, and Zip2 was strongly degraded in this fraction. This suggests that Zip2 protein stability may require Zip4. Consistently, semiquantitative Western blot analyses indicated that Zip2 protein levels are strongly decreased in the zip4Δ mutant (Fig. 2B). Moreover, the absence of Spo16 also affected Zip2 protein levels, suggesting that Zip2 requires being in complex with both Spo16 and Zip4 to be stable. In contrast, Spo16 protein levels were reduced by only ∼30% in the absence of either Zip4 or Zip2 (Fig. 2C). Finally, we did not observe any reduction of Zip4 protein levels in zip2Δ and spo16Δ mutants, where it even accumulated at higher levels, possibly due to impaired meiotic progression (Fig. 2D). Altogether, these data suggest that the stability of Zip2 and Spo16 is mutually dependent, and both require Zip4 to prevent their degradation.
Figure 2.
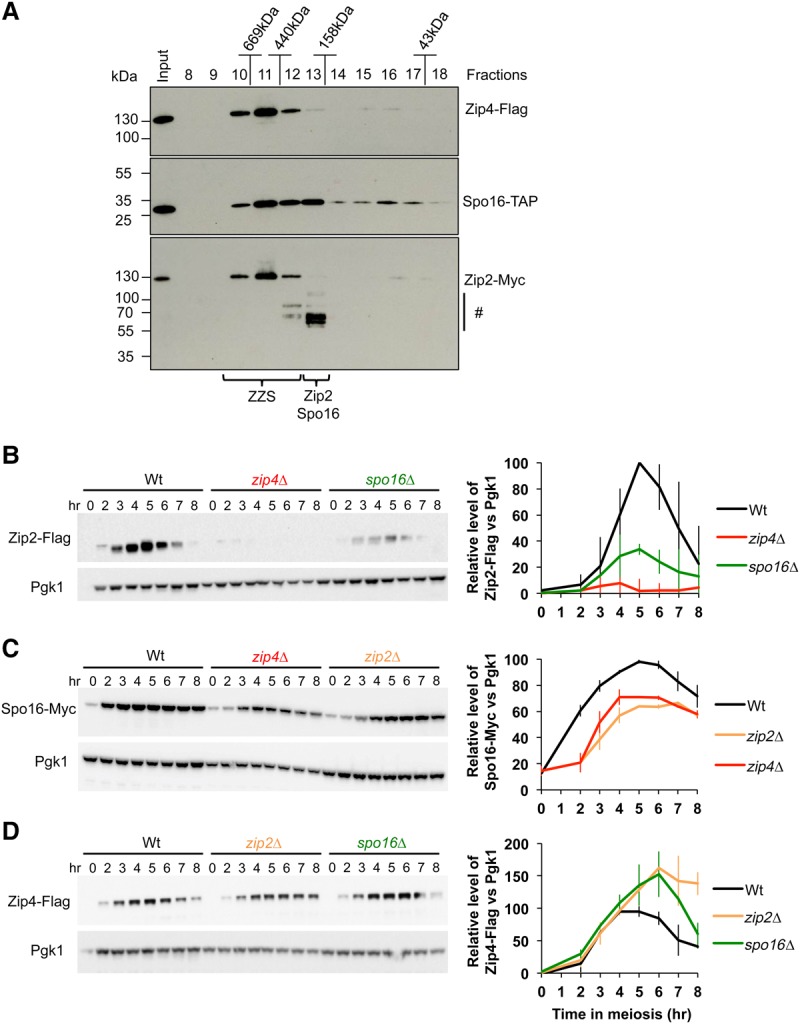
Analysis of ZZS complex formation during meiosis. (A) Western blot of Superdex-200 gel filtration column fractions. The positions of molecular weight markers are indicated. Braces show the migration positions of the ZZS complex (fractions 10–12) and the Zip4-free Zip2–Spo16 complex (fraction 13), respectively. (#) Possible degradation products of Zip2-Myc. (B–D) Western blot time course analysis of Zip2-Flag (B), Spo16-Myc (C), and Zip4-Flag (D) in wild-type cells or in the absence of one ZZS member, as indicated. Graphs of the quantification of Zip2-Flag, Spo16-Myc, and Zip4-Flag relative levels are at the right. Values are the mean ± SD of two independent experiments.
The ZZS complex interacts physically with components of the chromosome axis and with factors involved in post-translational modification
Besides Zip2, Zip4, and Spo16 forming a complex, we identified two components of the SC axial element, Red1 and Hop1, in all three purifications (Zip2-TAP, Zip4-TAP, and Spo16-TAP), suggesting that the entire ZZS complex interacts physically with the chromosome axis (Fig. 1A,C; Supplemental Table S1). This was confirmed by coimmunoprecipitation of Red1 with Zip4 (Fig. 1E). We also observed an interaction between Zip4, but not the other ZZS members, and Red1 in a yeast two-hybrid assay, suggesting that Zip4 makes the connection between the ZZS complex and the chromosome axis (Fig. 1F; Supplemental Table S2). However, we note that two-hybrid assays are not an absolute indicator of the absence of an interaction, and current data do not exclude other possible interactions. In addition, we identified Pch2 in our Spo16-TAP eluates (Fig. 1C; Supplemental Table S1). Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1 (Chen et al. 2014), suggesting that axis remodeling may occur in the vicinity of the ZZS complex.
We detected the E3 ligase Zip3 as a Zip2-TAP-interacting partner (Fig. 1A; Supplemental Table S1), which is in accordance with a previous report (Agarwal and Roeder 2000). Interestingly, Zip4, but not the other ZZS members, interacted in a two-hybrid test with Zip3 (Fig. 1F), suggesting that in vivo, Zip4 mediates the interaction between Zip2 and Zip3. Finally, we identified the Cdc7 kinase in our Zip4-TAP pull-down (Fig. 1C; Supplemental Table S1). Cdc7 is a Dbf4-dependent protein kinase important for DNA replication but also numerous meiotic recombination processes, including CO formation (Masai and Arai 2002; Sasanuma et al. 2008; Wan et al. 2008; Chen et al. 2015). In particular, Cdc7-Dbf4 is important for the CO/NCO decision between homologs by phosphorylating the Zip1 protein (Chen et al. 2015). Further investigation will be required to determine the role of this interaction. Altogether, these interactions suggest that several types of post-translational modifications occur near the ZZS complex.
Zip2, Zip4, and Spo16 preferentially bind DSB hot spots and are mutually dependent for their association with chromosomes
Since the ZZS complex interacts physically with components of the chromosome axis and promotes CO formation, we asked how the ZZS distributes between axis attachment sites and DSB sites. For this, we mapped Zip2-, Zip4-, and Spo16-binding sites by ChIP (chromatin immunoprecipitation) sequencing (ChIP-seq) experiment at 4 h of meiosis. We also mapped Zip3, which had been localized previously at lower resolution (Serrentino et al. 2013). As seen in Figure 3, A and B, all four proteins showed highly similar profiles and associated strongly with DSB hot spots but more weakly with Red1 axis sites. This was confirmed by Spearman correlation analyses that indicated that the 1000 highest Zip2, Zip3, Zip4, Spo16 peaks show a better correlation with the DSB peaks than with the Red1 peaks (Supplemental Fig. S1). This is consistent with our stoichiometry estimation, where only a fraction of the ZZS complex interacts with Red1 and Hop1 (Fig. 1B). Interestingly, we found that Zip2, Zip4, and, to a lesser extent, Spo16 were also located at centromeres, like Zip3 (Supplemental Fig. S2), which is consistent with previous cytological data (Tsubouchi et al. 2008).
Figure 3.
Zip2, Zip4, and Spo16 preferentially bind DSB hot spots and are mutually dependent on each other for chromosome binding. (A) ChIP-seq DNA-binding profiles for Zip3-Flag, Zip2-Flag, Zip4-Flag, and Spo16-Myc plotted for 110 kb of chromosome VI. DSB sites mapped by Spo11 oligos (Zhu and Keeney 2015) and Red1-binding sites (Sun et al. 2015) are indicated. All experiments were carried out at 4 h in meiosis. (*) The GAT1 DSB hot spot. (B) Heat map of Zip3-Flag, Zip2-Flag, Zip4-Flag, and Spo16-Myc signals within 4-kb regions centered around Spo11 oligo peaks (Zhu and Keeney 2015) (top panel) and at Red1-binding sites (Sun et al. 2015) (bottom panel), both arranged to their peak strength. (C) Mutant analysis of the genetic requirements for Zip2, Zip4, and Spo16 association with different chromosomal regions, assessed by ChIP. Maximum Zip2-Flag, Zip4-Flag, and Spo16-Myc levels observed by quantitative PCR (qPCR) using primers that cover the indicated regions are shown. Values are the mean ± SD from two independent experiments. The full corresponding time courses are in Supplemental Figure S3.
Because Zip2, Zip4, and Spo16 form a complex and associate with DSB hot spots genome-wide, we thought that they may function as a single unit to bind meiotic chromosomes. We examined their binding to DSB hot spots and chromosome axes by quantitative PCR (qPCR)-based ChIP experiments (Fig. 3C). Zip2, Zip4, and Spo16 showed a similar and reproducible dynamic localization during meiosis. They bound to DSB sites and axis sites from 3 h after meiosis induction to reach a maximum enrichment at 4 h and almost completely disappeared at 8 h (Supplemental Fig. S3). Their association with DSB sites was completely abolished in the spo11Δ mutant, indicating that it depends on DSB formation. Moreover, Zip2 and Zip4 recruitment to DSB sites was delayed and reduced in the dmc1Δ mutant, indicating that ZZS binding to chromatin is dependent on Dmc1-mediated strand exchange activity (Supplemental Fig. S3). Interestingly, the absence of any member of the complex strongly affected the binding of the other two members, revealing that Zip2, Zip4, and Spo16 are mutually dependent for their optimal association with DSB hot spots and chromosome axes (Fig. 3C; Supplemental Fig. S4). This supports the hypothesis that a tight complex formed by these three proteins binds to chromosomes.
Biochemical studies suggested that the Msh4–Msh5 heterodimer forms a clamp on strand exchange products, such as HJs, facilitating their maturation into COs (Snowden et al. 2004). Interestingly, we found that Zip2 and Zip4 binding to DSB hot spots and chromosome axes was not affected in a msh4Δ mutant (Fig. 3C; Supplemental Fig. S4). These data are consistent with cytological analyses showing that Zip4 still localizes on chromosomes when Msh4 is missing (Shinohara et al. 2008). Taken together, these results suggest that the ZZS complex preferentially binds DSB hot spots genome-wide and is still recruited even if the stabilization of recombination intermediates by Msh4–Msh5 is compromised.
The XPF domain is important for Zip2 function during meiotic recombination
Previous reports identified two domains within the Zip2 protein: a WD40 repeat motif (Perry et al. 2005) and a XPF domain (Macaisne et al. 2008), respectively. However, these two domains overlap, suggesting that one of the domain predictions might not be completely accurate. After a careful computational analysis of the Zip2 structure (see the Supplemental Material), we identified only the XPF domain, suggesting that the WD40 repeat domain is likely not present in this protein.
The reported XPF domain structure can be divided into two subdomains: the ERCC4 domain that typically carries the nuclease activity and a dimerization motif as well as the (HhH)2 domain that is a major determinant for binding to DNA junctions with a precise polarity (Ciccia et al. 2008). A previous study identified similarities between the C-terminal domain of Zip2 and the XPF domain (Macaisne et al. 2008). Consistent with this finding, we could predict with 97% confidence (probability HHpred) the whole three-dimensional (3D) structure of the XPF domain of Zip2 by using the solved structure of the XPF domain of human FANCM as a template (Protein Data Bank 4bxo) (Fig. 4A; Coulthard et al. 2013).
Figure 4.

The XPF domain of Zip2 is important for its pro-CO activity. (A) Schematic representation of the Zip2 protein and its XPF domain and the predicted 3D structure of the XPF domain of Zip2. The ERCC4 domain and HhH2 motif are indicated. (B) Meiotic progression as assessed by DAPI staining of strains with the indicated genotype. (C) Indicated strains were sporulated for 24 h at 30°C. The average sporulation of two independent colonies was graphed. Errors bars represent SD of two independent experiments. (D) Spore viability assays of strains with the indicated genotype. Numbers of dissected tetrads are indicated. (E) Representative Southern blot images of one-dimensional (1D) gel CO analysis at the HIS4–LEU2 hot spot. (F) Quantitative analysis of CO products after 8 h. Errors bars represent SD of two independent experiments. (G) ChIP monitoring of Zip2-Flag and Zip2ΔXPF-Flag association with different chromosomal regions, measured by qPCR using primers that cover the indicated regions. Values are the mean ± SD of two independent experiments.
To investigate the role of the Zip2 XPF domain during meiotic recombination, we generated a truncated form of Zip2, Zip2ΔXPF, lacking the ERCC4-(HhH)2 structure. Strains expressing Zip2ΔXPF showed a delay in meiotic progression and reduced sporulation efficiency and spore viability (Fig. 4B–D). Western blot analysis indicated that the meiotic phenotypes observed were not due to reduced levels of Zip2ΔXPF (Supplemental Fig. S5A). Moreover, CO frequency of the zip2ΔXPF mutant was reduced compared with the wild type, indicating that the XPF domain is important for the pro-CO function of Zip2 (Fig. 4E,F). We next studied the role of the XPF domain in Zip2 localization on meiotic chromosomes by ChIP and qPCR. Zip2ΔXPF binding was strongly impaired compared with the full-length protein, indicating that the XPF domain is critical for Zip2 binding to chromosomes (Fig. 4G).
Interestingly, despite a minor difference between CO levels, sporulation efficiency and spore viability of zip2ΔXPF mutant were significantly higher than for the zip2-null mutant (zip2Δ), suggesting that the N-terminal part of Zip2 also participates in its pro-CO activity (Fig. 4D–F). Consistently, residual binding of Zip2ΔXPF was still observed at DSB hot spots and axes, suggesting that Zip2 protein is still able to weakly bind chromosomes even in the absence of its XPF domain (Fig. 4G).
Spo16 specifically binds to the XPF domain of Zip2 in vivo
Many XPF domain proteins assemble into a heterodimeric complex with an ERCC1 domain protein to bind DNA (Ciccia et al. 2008). To identify factors that may interact specifically with the Zip2 XPF domain, we analyzed Zip2ΔXPF-TAP-binding partners by quantitative proteomics in synchronous cell extract at 4.25 h after induction of meiosis and compared the obtained results with the set of identified Zip2-TAP-interacting proteins. Zip2ΔXPF-TAP still pulled down Zip4 and the axial components of the SC (Red1 and Hop1) in amounts and stoichiometry comparable with those of the full-length Zip2-TAP construct (Fig. 5A; Supplemental Fig. S5B,C; Supplemental Table S1). Likewise, although the enrichment was less significant, we still recovered peptides from the Zip3 E3 ligase. In contrast, one member of the ZZS, Spo16, was no longer copurified with Zip2ΔXPF-TAP (Fig. 5A; Supplemental Table S1), suggesting that Spo16 interacts specifically with the Zip2 XPF domain. Therefore, we assessed the specificity of the interaction between the two parts of Zip2—the N terminus and the XPF domain—with Zip4 and Spo16 by yeast two-hybrid assay. Consistent with our TAP tag experiment, we found that the N-terminal part of Zip2 interacted with Zip4, while the XPF domain interacted with Spo16 (Fig. 5B).
Figure 5.

Zip2 and Spo16 form a XPF–ERCC1-like heterodimer that recognizes branched DNA structures. (A) Representative silver-stained gel of TAP eluates from meiotic cell extracts of Zip2-TAP and Zip2ΔXPF-TAP. The experimental design and analysis are same as in Figure 1, A and C. Only selected top candidates are represented. The entire list is in Supplemental Table S1. The dash indicates that Spo16 was not identified. Although both were highly enriched, Msh5 was above our enrichment threshold only in the Zip2ΔXPF-TAP experiment, whereas Zip3 was above the threshold only for Zip2-TAP. (B) Yeast two-hybrid interaction analysis between Zip2 domains and Zip4 and Spo16. The two-hybrid protein–protein interactions identified are diagrammed below. (C) A structural model of the Zip2 (green) XPF domain/Spo16 (cyan and gray) complex and its representative diagram. The gray region in Spo16 indicates poor quality of the model due to unreliable alignment with the reference structure 4bxo (FAAP24).
We also identified the MutS homolog Msh5 as a significantly enriched partner of Zip2ΔXPF-TAP (Fig. 5A; Supplemental Fig. S5B). Interestingly, Msh5 was also enriched in full-length Zip2-TAP experiments, although its false discovery rate had a P-value of >1% (Figs. 1A, 5A; Supplemental Table S1). Therefore, we tested whether Msh5 interacted with ZZS members in a yeast two-hybrid assay. Indeed, we could detect a physical interaction between Msh5 and Zip4 (Supplemental Fig. S5D; Supplemental Table S2), suggesting that the in vivo interaction between the Msh4–Msh5 heterodimer and the ZZS complex is mediated through Zip4 and Msh5.
Taken together, our data indicate that the N-terminal part of Zip2 is linked to the chromosome axis (Red1 and Hop1) and the other ZMM components of CO machinery (Zip3 and Msh4–Msh5) through its interaction with Zip4, while the XPF domain interacts exclusively with Spo16.
Spo16 shows a highly diverged ERCC1-like domain
The specificity of interaction between the XPF domain of Zip2 and Spo16 was reminiscent of an XPF–ERCC1-like heterodimer. The sequence of Zip2 diverged rapidly during the evolution of yeasts, with Saccharomyces cerevisiae Zip2 sharing only 26% and 23% sequence identities with its orthologous proteins in the closely related species Zygosaccharomyces rouxii (XP_002499352.1) and Kluyveromyces lactis (XP_453570.1), respectively. Despite this divergence, using remote homology detection tools, significant evolutionary relationships were detected previously between Zip2 in S. cerevisiae, SHOC1 in Arabidopsis thaliana, and C9orf84 in Homo sapiens (Macaisne et al. 2008). However, analysis of the Spo16 primary amino acid sequence failed to detect any similarity to known functional domains, including ERCC1-like structures. The S. cerevisiae Spo16 sequence shares only 20% and 11% sequence identities with its orthologous proteins in Z. rouxii (XP_002495594.1) and K. lactis (XP_454656.1), respectively, indicating that it diverged rapidly during the evolution of yeasts, significantly more so than Zip2. Besides, no homologs in species more distantly related than K. lactis could be identified even when using sensitive tools for remote homology detection such as HHblits (Remmert et al. 2011). We also searched for Spo16 homologs in several fully sequenced species of the Ascomycete clade. When the profile of S. cerevisiae Spo16 was queried with HHsearch (Söding 2005) against the database of profiles built for all protein sequences in each of these Ascomycete species, no match had a probability >32%, and no candidate Spo16 ortholog could be reliably identified (Supplemental Table S3). Nevertheless, in each species, at least one of the 10 best matches could be assigned to some members of the XPF–ERCC1 superfamily, supporting the possibility that Spo16 might be a highly diverged version of the ERCC1-like domain (Supplemental Table S4). We therefore explored whether the Spo16 sequence could be aligned with a structure of known members of the XPF–ERCC1 superfamily. Through weak profile–profile alignment relationships and secondary structure similarity analysis, the Spo16 sequence could be aligned to a reference structure belonging to the ERCC1 family (Supplemental Fig. S6). The structure of the FANCM–FAAP24 complex was used as a template to create a model of Spo16 in complex with the Zip2 XPF domain using the Rosetta CM program (Song et al. 2013). Interestingly, the regions of Spo16 that could be matched with the best agreement of secondary structure prediction from the profile–profile alignment correspond to those interacting with the XPF-like model of Zip2 (Fig. 5C; Supplemental Fig. S6A,B). Upstream of these regions, the sequence alignment and the secondary structure prediction do not ideally match with the structural template and may adopt a different conformation. Intriguingly, the Spo16 predicted structure shows only one HhH motif, while the majority of ERCC1 domain proteins carries a tandem of HhH motifs, reflecting the high divergence of Spo16 compared with other ERCC1 domain members. Nevertheless, the likelihood of the model in the region of the interface between Zip2 and Spo16 supports the hypothesis that the two proteins form a XPF–ERCC1-like assembly (Fig. 5C).
The XPF domain of the Zip2 and Spo16 heterodimer shows a strong affinity for branched DNA structures in vitro and likely lacks endonuclease activity
Members of the XPF family recognize various types of branched ssDNA/dsDNA molecules (Ciccia et al. 2008; Dehé and Gaillard 2017). Therefore, we investigated whether the complex formed by the Zip2 XPF domain and Spo16 [XPF(Zip2)–Spo16] could bind DNA. We copurified recombinant Spo16 with XPF(Zip2)-10xHis, confirming that Zip2 and Spo16 interact directly with each other (Fig. 6A). During size exclusion chromatography, the XPF(Zip2)–Spo16 behaved as a complex with 1:1 stoichiometry (Supplemental Fig. S7A). DNA-binding analysis of the XPF(Zip2)–Spo16 complex by electrophoretic mobility shift assays indicated that it binds weakly to ssDNA or dsDNA (Fig. 6B,C). In contrast, the XPF(Zip2)–Spo16 complex showed a marked binding preference for branched DNA structures, particularly D loops and HJs. The preferential binding to branched DNA molecules was confirmed by competition assays in which the XPF(Zip2)–Spo16 complex was prebound to a HJ substrate and then challenged with unlabeled HJs or dsDNA (Fig. 6D–F). Moreover, this analysis also indicated that the binding of the complex to DNA is dynamic and that the protein can relocate to other DNA molecules. Therefore, the preference of the XPF(Zip2)–Spo16 complex for binding branched DNA molecules is similar to that of the XPF–ERCC1 complexes (Ciccia et al. 2008), indicating that the Zip2–Spo16 heterodimer recognizes DNA substrates like a bona fide XPF–ERCC1 complex in vitro.
Figure 6.

The recombinant XPF domain of Zip2, XPF(Zip2), and Spo16 preferentially bind branched DNA structures but do not show any endonuclease activity in vitro. (A, left) XPF(Zip2)-10xHis–Spo16 purification protocol. (Right) Coomassie-stained SDS-PAGE showing samples from a representative purification of XPF(Zip2)-10xHis–Spo16. We observed a single band of XPF(Zip2)-10xHis, which migrated at a position corresponding to its molecular weight of 26 kDa. The 23-kDa Spo16 polypeptide comigrated with XPF(Zip2)-10xHis. Fractions 7 and 8, obtained by size exclusion chromatography, show 95% purity and were used for the electrophoretic mobility shift assays. (B) DNA-binding specificities of XPF(Zip2)–Spo16. (*) 32P label. The smeared migration profile is likely due to partial dissociation of the protein–DNA complex during electrophoresis (see Supplemental Fig. S7B). (C) Quantitation of the electrophoretic mobility shift assays with XPF(Zip2)–Spo16 and various oligonucleotide-based DNA substrates such as those shown in B. Values are the mean ± SD from three to six independent experiments. (D) A scheme of the competition assay to test the specificity of XPF(Zip2)–Spo16 binding to branched molecules as HJs. (E) The XPF(Zip2)–Spo16 complex was prebound to 1 nM 32P-labeled HJs and challenged with the indicated concentrations of unlabeled HJs or dsDNA. (F) Quantitation of the experiments such as that shown in E. Values are the mean ± SD from two independent experiments. (G) Nuclease assay with XPF(Zip2)–Spo16 and Dna2 on a 3′ end-labeled Y structure.
The catalytic subunits of XPF and MUS81 display a GDXnERKX3D active site motif that is required for metal-dependent endonuclease activity but not DNA junction binding (Enzlin and Schärer 2002; Nishino et al. 2003). In contrast, Zip2, like the XPF domain protein FANCM, lacks the conserved endonuclease motif that is characteristic of the eukaryotic XPF family (Macaisne et al. 2008, 2011). Moreover, unlike in the case of the Dna2 nuclease, in our conditions, we did not observe any nuclease activity of the XPF(Zip2)–Spo16 complex on the Y structure (Fig. 6G), a branched DNA molecule that is processed by members of the XPF–ERCC1 family (Ciccia et al. 2008). Altogether, these results strongly suggest that within the ZZS complex, the Zip2 XPF domain in association with Spo16 represents a DNA recognition module with a structural rather than catalytic function.
Discussion
Zip2 and Spo16 likely form a catalytically inactive XPF–ERCC1-like complex that recognizes DNA structures and promotes CO formation
The XPF–ERCC1 complex is known to act on various branched DNA structures, such as Y structures, stem–loop DNA structures, or HJs (de Laat et al. 1998; Enzlin and Schärer 2002). Although Zip2 shares primary sequence similarities to the XPF endonuclease family, there was no evidence about the requirement of the XPF domain of Zip2 in the formation of interfering COs. Our studies showed that the XPF domain is important not only for Zip2 binding to the DSB hot spot and axis but also to promote CO formation. Interestingly, Zip2 homologs in plants and mammals, SHOC1 and C9orf84, respectively, also possess a predicted XPF domain, and the SHOC1 protein is critical for the production of interfering COs in Arabidopsis (Macaisne et al. 2008). Therefore, the XPF domain may also be important for CO formation in higher eukaryotes such as plants and mammals.
In their functionally competent state, XPF domain-containing proteins form a heterodimer with their ERCC1 domain partners. However, it was unclear whether Zip2 is active as a heterodimer or homodimerizes similarly to XPF of archaea (Dehé and Gaillard 2017). We did not identify any self-interactions between individual Zip2 proteins by yeast two-hybrid assay, suggesting that Zip2 does not homodimerize. In contrast, the Zip2 XPF domain interacts directly with Spo16 in vitro and in vivo. Moreover, despite being highly divergent, we found that Spo16 protein shows structural similarities to ERCC1 domain proteins. Together, this suggests that Spo16 is a new member of the ERCC1 family and is the missing partner to form with Zip2 a bona fide XPF–ERCC1-like complex. Therefore, we propose that a third member of the XPF family exists alongside the Mus81/Mms4 and Rad1/Rad10 complexes in yeast and is likely conserved in mammals (see below).
Our data provide evidence suggesting that despite having a XPF domain, Zip2 does not have an intrinsic nuclease activity on meiotic recombination intermediates. First, the in vitro purified XPF(Zip2)–Spo16 complex is not able to cleave Y structures, while members of the XPF family that have nuclease activity are able to cleave such branched DNA substrates (Ciccia et al. 2008). Second, the conserved sequence GDXnERKX3D active site motif of the XPF domain is highly diverged in Zip2 compared with canonical XPF endonucleases (Macaisne et al. 2008). Besides, the catalytic site for DNA cleavage is also missing in plants and mammalian Zip2 orthologs SHOC1 and C9orf84, respectively, suggesting that nuclease activity is also likely missing in these species (Macaisne et al. 2008). Because the formation of the single end invasion recombination intermediate (SEI) is affected in zip2 and spo16 mutants (Börner et al. 2004; Shinohara et al. 2008), we favor the hypothesis that the Zip2–Spo16 heterodimer functions as a DNA recognition module that binds and stabilizes D loops rather than being a DNA endonuclease or resolvase. Nevertheless, the biochemical activity of the entire complex, obtained from recombinant proteins or purified from meiotic cells, will be needed before totally excluding a cleavage activity of the ZZS complex.
The ZZS complex shepherds joint molecules toward a pro-CO state
In budding yeast, the CO/NCO decision is made soon after DSB formation and is subjected to the perpetual antagonistic roles of pro-CO versus anti-CO activities (Fig. 7). The Sgs1–Top3–Rmi1 (STR) complex orchestrates this CO/NCO decision by preventing the accumulation of unregulated joint molecules through disassembling unprotected branched DNA structures, thus driving events toward both NCOs and SC-associated ZMM proteins (De Muyt et al. 2012; Zakharyevich et al. 2012; Kaur et al. 2015; Tang et al. 2015). In budding yeast, the earliest CO-specific DNA intermediates, SEIs and dHJs, are reduced in the zip2, zip4, and spo16 mutants (Börner et al. 2004; Shinohara et al. 2008). Interestingly, genetic and molecular analyses suggest that ZMM proteins prevent D-loop and HJ disassembly by Sgs1 (Jessop et al. 2006; Oh et al. 2007). Together with these data, our results suggest that the ZZS complex acts early in the CO differentiation pathway, in which the XPF domain of Zip2 and Spo16 recognize and stabilize the D-loop early recombination intermediates, protecting them from being dismantled by the anti-recombination STR complex (Fig. 7, step 1). Such an activity would consolidate D loops to become a metastable CO-correlated joint molecule, the SEI. During the SEI-to-dHJ transition, the displaced 3′ strand of an SEI could undergo annealing to the second DSB end (Lao et al. 2008). In theory, this type of DNA junction can still be sensitive to anti-CO activities until it is fully ligated and destabilized to form a NCO. Therefore, we suggest that the ZZS complex also recognizes and protects these DNA junctions that are formed during the post-invasion step of the second end capture, thus stabilizing the second HJ (Fig. 7, step 2). Interestingly, a zip3Δ mutant shows longer gene conversion tracts associated with CO that are dependent on the presence of Sgs1, suggesting that Zip3 helps to promote the second end capture by reducing the ability of Sgs1 to act on the D-loop intermediate (Oke et al. 2014). Since Zip4 interacts physically with Zip3, it would be interesting to test whether the ZZS complex and Zip3 cooperate to promote the second end capture.
Figure 7.
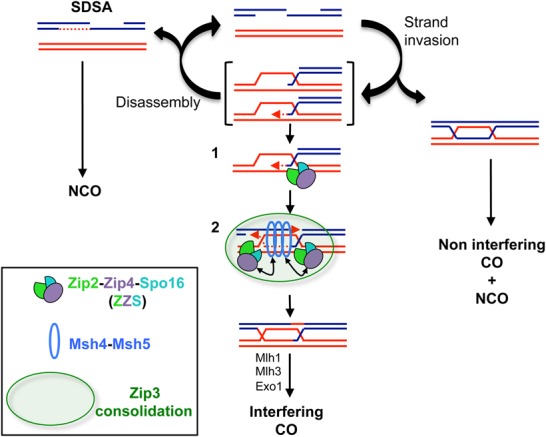
A model of how the ZZS complex regulates meiotic recombination intermediates to promote interfering COs (adapted from Kaur et al. 2015). Arrows indicate the interactions between the ZZS complex and the Msh4–Msh5 complex. The green circle illustrates the proposed consolidation of interactions by the E3 ligase Zip3. Details are in the text.
What could be the roles of Zip4 in the ZZS complex?
Zip4 forms a stable complex with the Zip2–Spo16 heterodimer, but its molecular function during meiotic recombination remains obscure. Zip4 is a giant tetratricopeptide repeat protein (TPR), a domain present in many scaffold proteins (D'Andrea and Regan 2003; Perry et al. 2005; Zeytuni and Zarivach 2012). Consistently, we identified several protein interactions between Zip4 and other ZMM members. In particular, we found an interaction between Zip4 and Msh5. Since mammalian MSH4–MSH5 form a meiosis-specific sliding clamp that recognizes and stabilizes HJs (Snowden et al. 2004), the interaction with Zip4 could locally bridge the ZZS complex and Msh4–Msh5 on DNA intermediates (Fig. 7, step 2). This would coordinate the action of the Zip2–Spo16 and Msh4–Msh5 complexes on the branched DNA structure to convert D loops into highly stable recombination intermediates, the SEIs, and then the dHJs. Another Zip4-interacting partner identified was Zip3. Zip3 is inferred to catalyze SUMO conjugation, and mutations in the SUMO ligase activity abolish CO formation, indicating a predominant role for post-translational modifications in CO formation (Cheng et al. 2006). Interestingly, SUMOylation was proposed to provide glue-like properties to nearby substrates to foster their physical interactions and stabilize protein complexes (Psakhye and Jentsch 2012). Therefore, the SUMO activity of Zip3 might spatially consolidate interactions between several components of the recombination machinery in the vicinity of recombination intermediates, including the ZZS complex (Fig. 7, step 2). Consistent with this, recent data suggest that the mouse Zip3-related proteins RNF212 and HEI10 are important to stabilize recombination factors at designated CO sites (Reynolds et al. 2013; Qiao et al. 2014).
A recent study suggested the importance of the local axis-enriched context to repair DSBs by the interfering CO pathway (Medhi et al. 2016). Consistently, our data identified a link between the ZZS complex and the chromosome axis. However, genome-wide localization data show that Zip2, Zip4, and Spo16 were more often associated with DSB sites than chromosome axis sites. These apparent contradictions can be reconciled by a transient localization of CO factors to the axis sites (Storlazzi et al. 2010). Indeed, the localization of the ZMM proteins Msh4 and Mer3 in Sordaria macrospora suggests that recombination complexes move from chromosome axes to the interaxis region (Storlazzi et al. 2010). Similarly, in budding yeast, the Zip3 protein shows the same dynamic of localization (Serrentino et al. 2013). Therefore, we suggest that the ZZS complex follows the same pattern of localization, in which it is tethered to the chromosome axis, possibly via a Zip4–Red1 interaction. The complex is then progressively delocalized from the axis (but remains on recombination intermediates) to the interaxis region to perform its pro-CO activity. Interestingly, interaction between the Zip4 ortholog TEX11 and the lateral component of the SC, SYCP2, has been reported in mouse testis (Yang et al. 2008). Moreover, TEX11 localizes between chromosome axes during synapsis formation (Yang et al. 2008; Reynolds et al. 2013), suggesting that this dynamic localization is conserved between yeast and mammals.
XPF members are often associated with scaffold proteins that control the optimal repair outcome. For instance, human SLX4 is important to recruit MUS81–EME1 and XPF–ERCC1 to DNA repair sites and stimulate their catalytic activity (Dehé and Gaillard 2017). Interestingly, the TPR domain of the Fanconi anemia protein FANCG binds to the XPF–ERCC1 endonuclease, suggesting that a TPR domain protein can mediate the activity of some XPF family members (Wang and Lambert 2010). The TPR Zip4 is necessary for the recruitment of Zip2 and Spo16 to DSB sites and the chromosome axis. Our data indicate a strong degradation of Zip2 when Zip4 is no longer associated with Zip2 and Spo16. Taken together, these data suggest that Zip4 plays a role in stabilizing the Zip2–Spo16 complex.
Last, the crystal structure of the nuclease of Pseudomonas aeruginosa PaFAN1 showed that the TPR domain binds directly to the 5′ flap DNA structure in vitro (Gwon et al. 2014). Therefore, there is a possibility worth testing that the TPR domain of Zip4 may also participate in DNA binding of the ZZS complex.
Is the ZZS complex evolutionarily conserved?
Presumed ZMM orthologs have also been identified in plants and mammals, suggesting evolutionary conservation of this group (Lynn et al. 2007). In particular in Arabidopsis, SHOC1 and AtZIP4 share domains similar to Zip2 and Zip4, respectively, and both are required for the formation of interfering COs (Chelysheva et al. 2007; Macaisne et al. 2008). Interestingly, parting dancer (PTD), shows sequence similarity to the ERCC1 protein family and is specifically involved in the formation of interfering COs (Wijeratne et al. 2005; Macaisne et al. 2011). Similarly to yeast, a fragment of the SHOC1 protein that contains the XPF domain interacts with PTD in yeast two-hybrid assays, suggesting that SHOC1 and PTD form an XPF–ERCC1-like complex to promote COs (Macaisne et al. 2011). Based on this comparison, we suggest that Spo16 and PTD are functional homologs and propose that the whole ZZS complex is conserved in plants, composed of SHOC1–AtZIP4–PTD.
In mammals, C9orf84 and TEX11 are the closest relatives to Zip2 and Zip4, respectively (Macaisne et al. 2008), suggesting that at least two components of the ZZS complex are also present in mammals. However, putative Spo16 and PTD orthologs were not identified in the mammalian kingdom. Therefore, it is possible that mammalian Zip2 might act alone as a homodimer or, alternatively, interact with an already known mammalian ERCC1 domain-containing protein, such as EME1, EME2, ERCC1, or FAAP24. Alternatively, the example of Spo16 shows how difficult it is to identify an ERCC1 domain protein, and we cannot exclude that C9orf84 may interact with a yet unidentified ERCC1-related Spo16/PTD-like protein.
Concluding remarks
Meiotic roles of the XPF–ERCC1 members have been focused mainly on extensive studies of the MUS81–EME1 nuclease during CO formation. However, the discovery of a new complex of this family, Zip2–Spo16, in budding yeast and its possible conservation among eukaryotes indicate that another set of XPF–ERCC1 actors is important for CO production. Moreover, this study provides evidence that not all XPF–ERCC1 family members are active as nucleases. In contrast, as has been hypothesized for the mammalian FANCM–FAAP24 complex, these proteins may serve as recognition modules of branched DNA structures (Coulthard et al. 2013). In closing, the extreme divergence observed for the Zip2–Spo16 complex among yeast species shows how XPF–ERCC1 family members can be versatile but also calls for future explorations of not yet identified new XPF–ERCC1-like actors.
Materials and methods
Yeast strains, constructions, two-hybrid analysis, TAP, proteomic analyses, coimmunoprecipitation, size exclusion chromatography, Western blotting, Southern blot, ChIP, detection of homology relationship of Spo16, and structural modeling of the XPF(Zip2)–Spo16 are described in the Supplemental Material.
Purification of recombinant proteins
The viruses were produced using a Bac-to-Bac system (Invitrogen) according to manufacturer's recommendations. Spodoptera frugiperda Sf9 cells were then infected with optimal ratios of both viruses, and the cells were harvested 72 h after infection, washed with phosphate-buffered saline (PBS), frozen in liquid nitrogen, and kept at −80°C until use. Purification was performed with cell pellets from 1 L of culture. All subsequent steps were carried out at 4°C. Cells were resuspended in lysis buffer (1× PBS, 150 mM NaCl, 1% Triton X-100, 10% glycerol, Roche protease inhibitory cocktail, 30 mM imidazole) and incubated for 1 h. The cell suspension was centrifuged at 25,000 rpm for 1 h to obtain a soluble extract. The cleared extract was loaded onto a 1-mL Macherey-nagel NiNTA column equilibrated with buffer A (1× PBS, 150 mM NaCl, 10% glycerol, 30 mM imidazole). The resin was washed extensively with buffer A. The XPF(Zip2)-10xHis and Spo16 complex was eluted in 1× PBS containing 150 mM imidazole. The eluted protein peak was concentrated at a 30-kDa cutoff and then loaded onto a Superdex-75 column (GE Healthcare) equilibrated in buffer B (1× PBS, 150 mM NaCl, 10% glycerol). The samples was aliquoted, frozen in liquid nitrogen, and stored at −80°C. The quality of the purified protein was determined with SDS-PAGE gel analysis, and the protein concentration was determined using Bradford protocol with BSA as a standard. Yeast Dna2 was purified as described previously (Levikova et al. 2013). The Ku70–80 complex was purified as described (Reginato et al. 2017).
Biochemical assays
DNA structures—including ssDNA, dsDNA, Y structure, HJs, and 3W junctions—for DNA-binding experiments were prepared as described previously (Ranjha et al. 2014). The D loop was prepared similarly by annealing 5′ end-labeled PC1253 with oligonucleotides 315 (5′-AACGTCATAGACGATGATCCGATGCATATCCGCCTGCCCACGTTGACCC-3′), 320 (5′-GCGATAGTCTCTAGACAGCATGTCCTAGCAATACATTGCTAGGACATCTT-3′) and X12-3SC (5′-TTGCTAGGACATGCTGTCTAGAGACTATCGC-3′). The Y structure used for the nuclease assay was prepared by annealing unlabeled PC1253 with 3′ end-labeled PC1254. The 3′ end labeling was carried out with terminal deoxynucleotidyl transferase (New England Biolabs) and [α-32P]cordycepin 5′ triphosphate according to the manufacturer's instructions. The DNA-binding assays with the XPF(Zip2)–Spo16 complex were carried out in a 15-µL volume in 25 mM Tris-acetate (pH 7.5), 1 mM dithiothreitol, 100 µg/mL bovine serum albumin (BSA) (New England Biolabs), 2 mM magnesium acetate, and 1 nM DNA substrate (in molecules). The reactions were assembled on ice and incubated for 30 min at 30°C. In competition experiments, the XPF(Zip2)–Spo16 complex was prebound to labeled HJs for 15 min at 30°C and then challenged with the indicated concentrations of either unlabeled HJs or dsDNA for another 15 min at 30°C. At the end of the incubation, 5 µL of binding dye (50% glycerol, 0.25% bromophenol blue) was added to each sample. The products were separated on 6% native polyacrylamide gels (acrylamide:bisacrylamide 19:1; Bio-Rad) at 4°C. After separation, the gels were dried on 17 CHR paper (Whatman) and exposed to storage phosphor screens (GE Healthcare). The screens were scanned using a Typhoon FLA 9500 (GE Healthcare) and quantified by ImageQuant software. The Ku70–80 DNA-binding experiments were carried out in an identical buffer under the same conditions as used for the XPF(Zip2)–Spo16 complex.
Illumina sequencing of ChIP DNA and read normalization
Purified DNA was sequenced using an Illumina HiSeq 2500 instrument following the Illumina TruSeq procedure, generating single-end 50-base-pair (bp) reads for Zip2, Zip3, Zip4, and untagged anti-Flag ChIP or paired-end 50-bp reads for Spo16 and untagged anti-Myc ChIP. Each experiment was performed in two independent replicates. Reads were aligned to the Sacccer2 version (SGD June 2008) of the S. cerevisiae S288C genome using Bowtie version 1.0.0 (Langmead et al. 2009), allowing for two mismatches in the 20-bp seed. Reads that matched more than once in the genome or matched to mitochondrial or ribosomal DNA were eliminated from further analysis. Single-end reads were extended to a size of 150 bp (the estimated size of our sonicated chromatin preparations). Aligned and extended reads were then normalized using a custom script as follows: For normalization for the untagged control, the top and bottom outliers of the distribution (Q3 + 1.5IQR and Q1 − 1.5IQR, respectively) were removed. The average read number per position (coverage) of the remaining reads of the untagged control was then normalized to 1 (scaled untagged control). Similarly, top and bottom outliers of the distribution were removed from the average read number in the tagged samples. Average coverage for the remaining reads was then calculated for the tagged and untagged control at the same positions. This was used to compute the immunoprecipitate/untagged control ratio. Next, all reads of the tagged sample were scaled using this ratio (scaled tagged control). Both scaled tagged and untagged samples were converted to bigwig format, and the scaled untagged sample was subtracted from the scaled tagged sample. The obtained normalized tagged sample values were used for Figure 3, A and B, and Supplemental Figure S2. For peak detection and comparison using Spearman correlation in Supplemental Figure S1, we used the bpeaks version 1.2 bioinformatics tool (Merhej et al. 2014). We used the following parameters for thresholds: T1 was the bottom outlier limit of UIP (untagged immunoprecipitate control) coverage (Q3UIP + 1.5IQRUIP), T2 was the top outlier limit of UIP coverage (Q1UIP − 1.5IQRUIP), T3 was 0.3 (immunoprecipitate at least 1.23 times higher than UIP), and T4 was 0. Using these criteria, we obtained for each replicate 2753 and 2466 peaks for Zip2, 2736 and 2545 peaks for Zip3, 3126 and 2853 peaks for Zip4, and 2037 and 2080 peaks for Spo16. Peaks for Red1 ChIP-seq and Spo11 oligonucleotides were from Sun et al. (2015) and Zhu and Keeney (2015) genetics, respectively.
Accession numbers
The ChIP-seq data generated in this study have been deposited at the Gene Expression Omnibus database with accession number GSE103877. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Proteomics Identifications) partner repository with the data set identifier PXD007735.
Supplementary Material
Acknowledgments
We thank Franz Klein for the V5-Red1 strain, and Jean-Baptiste Charbonnier for the pKL and pUCDM plasmids. We thank the Next-Generation Sequencing and Recombinant Proteins platforms and the discovery platform and informatics group at EDyP. We thank Mathilde Grelon and Wayne Crismani for critical reading of the manuscript. We thank Dr. M. Levikova for purified Dna2. This work was supported by Institut Curie, Centre Nationnal de la Recherche Scientifique, Labex DEEP (Development, Epigenesis, Epigenetics, and life-time Potential; ANR-11-LBX-0044; part of the IDEX Idex PSL [ANR-10-IDEX-0001-02 PSL]), ANR-12-BSV6-0009, and the Fondation ARC, ANR-11-PDOC-0011 to J.G.; the Fond d'Intervention of the University Grenoble Alpes to J.G.; and the European Union FP7 Marie Curie Action “Career Integration Grant” (304003) to J.G. Proteomic experiments were partly supported by the Proteomics French Infrastructure (ANR-10-INBS-08-01 grant) and GRAL (Grenoble Alliance for Integrated Structural Cell Biology; ANR-10-LABX-49-01).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.308510.117.
References
- Agarwal S, Roeder GS. 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Zickler D. 2004. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Gendrot G, Vezon D, Doutriaux MP, Mercier R, Grelon M. 2007. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet 3: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jomaa A, Ortega J, Alani EE. 2014. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc Natl Acad Sci 111: E44–E53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Suhandynata RT, Sandhu R, Rockmill B, Mohibullah N, Niu H, Liang J, Lo H-C, Miller DE, Zhou H, et al. 2015. Phosphorylation of the synaptonemal complex protein Zip1 regulates the crossover/noncrossover decision during yeast meiosis. PLoS Biol 13: e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. 2006. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev 20: 2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. 1998. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani EH, Joenje H, McDonald N, de Winter JP, et al. 2007. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell 25: 331–343. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. 2008. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem 77: 259–287. [DOI] [PubMed] [Google Scholar]
- Coulthard R, Deans AJ, Swuec P, Bowles M, Costa A, West SC, McDonald NQ. 2013. Architecture and DNA recognition elements of the Fanconi anemia FANCM–FAAP24 complex. Structure 21: 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea L, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem Sci 28: 655–662. [DOI] [PubMed] [Google Scholar]
- Dehé P-M, Gaillard P-HL. 2017. Control of structure-specific endonucleases to maintain genome stability. Nat Rev Mol Cell Biol 18: 315–330. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. 1998. DNA structural elements required for ERCC1-XPF endonuclease activity. J Biol Chem 273: 7835–7842. [DOI] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y, Kumar R, Ranjha L, Adam C, Guérois R, Md Muntaz K, Marsolier-Kergoat M-C, Dingli F, Laureau R, Loew D, et al. 2017. Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion. Elife 6: e21900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzlin JH, Schärer OD. 2002. The active site of the DNA repair endonuclease XPF–ERCC1 forms a highly conserved nuclease motif. EMBO J 21: 2045–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwon GH, Kim Y, Liu Y, Watson AT, Jo A, Etheridge TJ, Yuan F, Zhang Y, Kim Y, Carr AM, et al. 2014. Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5′ flap DNA: basis of interstrand cross-link repair by FAN1. Genes Dev 28: 2276–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. 2001. The single-end invasion. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Jessop L, Rockmill B, Roeder GS, Lichten M. 2006. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet 2: 1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, DeMuyt A, Lichten M. 2015. Top3–Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol Cell 57: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. 2008. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol Cell 29: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levikova M, Klaue D, Seidel R, Cejka P. 2013. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc Natl Acad Sci 110: E1992–E2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Börner GV. 2007. ZMM proteins during meiosis: crossover artists at work. Chromosom Res 15: 591–605. [DOI] [PubMed] [Google Scholar]
- Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R. 2008. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18: 1432–1437. [DOI] [PubMed] [Google Scholar]
- Macaisne N, Vignard J, Mercier R. 2011. SHOC1 and PTD form an XPF–ERCC1-like complex that is required for formation of class I crossovers. J Cell Sci 124: 2687–2691. [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K-I. 2002. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J Cell Physiol 190: 287–296. [DOI] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC. 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51: implications for crossover control in meiotic recombination. Cell 117: 47–56. [DOI] [PubMed] [Google Scholar]
- Medhi D, Goldman AS, Lichten M. 2016. Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. Elife 5: e19669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej J, Frigo A, Le Crom S, Camadro J-M, Devaux F, Lelandais G. 2014. bPeaks: a bioinformatics tool to detect transcription factor binding sites from ChIPseq data in yeasts and other organisms with small genomes. Yeast 31: 375–391. [DOI] [PubMed] [Google Scholar]
- Nishino T, Komori K, Ishino Y, Morikawa K. 2003. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease. Structure 11: 445–457. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PYH, Taylor AF, Smith GR, Hunter N. 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke A, Anderson CM, Yam P, Fung JC. 2014. Controlling meiotic recombinational repair—specifying the roles of ZMMs, Sgs1 and Mus81/Mms4 in crossover formation. PLoS Genet 10: e1004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. [DOI] [PubMed] [Google Scholar]
- Perry J, Kleckner N, Börner GV. 2005. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc Natl Acad Sci 102: 17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820. [DOI] [PubMed] [Google Scholar]
- Qiao H, Prasada Rao HBD, Yang Y, Fong JH, Cloutier JM, Deacon DC, Nagel KE, Swartz RK, Strong E, Holloway JK, et al. 2014. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet 46: 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjha L, Anand R, Cejka P. 2014. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J Biol Chem 289: 5674–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato G, Cannavo E, Cejka P. 2017. Physiological protein blocks direct the Mre11–Rad50–Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev 31: 2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, Söding J. 2011. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9: 173–175. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, et al. 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet 45: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Hirota K, Fukuda T, Kakusho N, Kugou K, Kawasaki Y, Shibata T, Masai H, Ohta K. 2008. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev 22: 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrentino ME, Chaplais E, Sommermeyer V, Borde V. 2013. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet 9: e1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, Shinohara A. 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40: 299–309. [DOI] [PubMed] [Google Scholar]
- Smith AV, Roeder GS. 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol 136: 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. 2004. hMSH4–hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Söding J. 2005. Protein homology detection by HMM–HMM comparison. Bioinformatics 21: 951–960. [DOI] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang R-R, Kim D, Miles C, Brunette T, Thompson J, Baker D. 2013. High-resolution comparative modeling with RosettaCM. Structure 21: 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. 2010. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141: 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Huang L, Markowitz TE, Blitzblau HG, Chen D, Klein F, Hochwagen A. 2015. Transcription dynamically patterns the meiotic chromosome-axis interface. Elife 4: e07424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Wu MKY, Zhang R, Hunter N. 2015. Pervasive and essential roles of the top3-rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol Cell 57: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS. 2006. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev Cell 10: 809–819. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T, MacQueen AJ, Roeder GS. 2008. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev 22: 3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Niu H, Futcher B, Zhang C, Shokat KM, Boulton SJ, Hollingsworth NM. 2008. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev 5: 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lambert MW. 2010. The Fanconi anemia protein, FANCG, binds to the ERCC1–XPF endonuclease via its tetratricopeptide repeats and the central domain of ERCC1. Biochemistry 49: 5560–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne AJ, Chen C, Zhang W, Timofejeva L, Ma H. 2005. The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol Biol Cell 17: 1331–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Sung P, Zhao X. 2015. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev 29: 1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Höög C, McLaughlin KJ, et al. 2008. Meiotic failure in male mice lacking an X-linked factor. Genes Dev 22: 682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R. 2012. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20: 397–405. [DOI] [PubMed] [Google Scholar]
- Zhu X, Keeney S. 2015. High-resolution global analysis of the influences of Bas1 and Ino4 transcription factors on meiotic DNA break distributions in Saccharomyces cerevisiae. Genetics 201: 25–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



