Abstract
Motivation: While existing network visualization tools enable the exploration of cancer genomics data, most biologists prefer simplified, curated pathway diagrams, such as those featured in many manuscripts from The Cancer Genome Atlas (TCGA). These pathway diagrams typically summarize how a pathway is altered in individual cancer types, including alteration frequencies for each gene.
Results: To address this need, we developed the web-based tool PathwayMapper, which runs in most common web browsers. It can be used for viewing pre-curated cancer pathways, or as a graphical editor for creating new pathways, with the ability to overlay genomic alteration data from cBioPortal. In addition, a collaborative mode is available that allows scientists to co-operate interactively on constructing pathways, with support for concurrent modifications and built-in conflict resolution.
Availability and Implementation: The PathwayMapper tool is accessible at http://pathwaymapper.org and the code is available on Github (https://github.com/iVis-at-Bilkent/pathway-mapper).
Contact: ivis@cs.bilkent.edu.tr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Genomic alterations in cancer often affect a relatively small number of signaling pathways that are involved in cell proliferation, cell growth, apoptosis and DNA repair, among others (Garraway and Lander, 2013, Hanahan and Weinberg, 2011). The mechanism through which these pathways are altered, however, can be diverse, affecting many different genes through various mechanisms, such as mutations, gene fusions, epigenetic silencing and amplifications or deletions. During the last decade, as the pace of sequencing of tumor samples has accelerated dramatically, we have discovered many of these mechanisms and have developed a better understanding of the pathway alterations in many different tumor types. These alterations are commonly summarized visually in hand-drawn pathway diagrams, as exemplified by the publications of The Cancer Genome Atlas (TCGA), an effort that has comprehensively characterized the genomic alterations in more than 20 tumor types (The Cancer Genome Atlas Research Network, 2008, 2011, 2012a, 2012b, 2013, 2014a, 2014b, 2015, 2017).
Several pathway visualization and editing tools have been developed (Villaveces et al., 2015). However, none has the kind of simplified, curated pathway notation like those featured in TCGA manuscripts, specializes in cancer pathways, features visualization of alteration frequencies and allows real-time collaborative curation through a web-based editor.
To fill this void, we have developed PathwayMapper, a collaborative visual web editor for cancer pathways. PathwayMapper can be used for viewing pre-curated cancer pathways with the ability to overlay genomic alteration data, and as an interactive graphical editing tool for creating and modifying pathways. It also allows multiple users to co-operate on curation in real time, with support for concurrent modifications and built-in conflict resolution. Furthermore, users can import data from the cBioPortal for Cancer Genomics (Cerami et al., 2012; Gao et al., 2013) and export pathway images with alteration frequencies overlaid as scalable vector graphics for use in publications.
2 Materials and methods
PathwayMapper was designed with ease of use in mind using modern web technologies. It runs on most modern web browsers, without requiring any installations. PathwayMapper is based on the Cytoscape.js (Franz et al., 2016) graph library, uses backbone.js in the frontend, and node.js and the Google Realtime API in the backend.
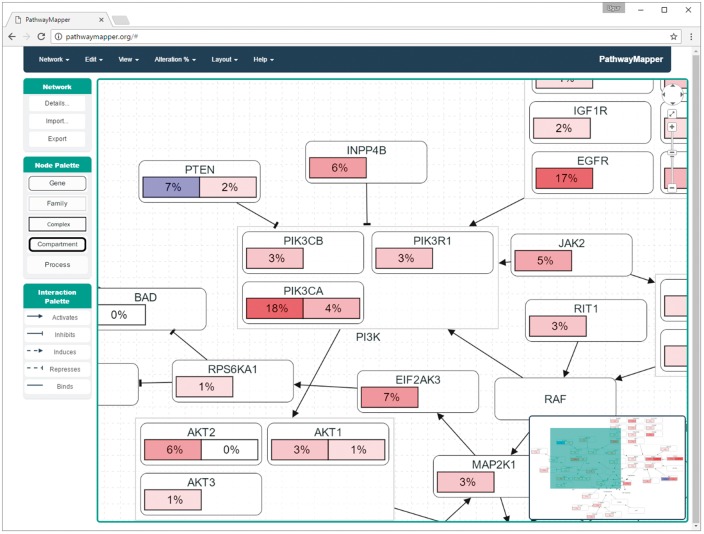
Through advanced customization of Cytoscape.js and newly developed extensions for this library, PathwayMapper is not only a state-of-the-art viewer supporting compound structures for complexes, families, processes, and cellular locations, but also an easy to use editor, due to tools such as automatic layout, node alignment, undo support, an overview window and real time collaborative editing support (Fig. 1).
Fig. 1.
A screenshot of PathwayMapper. The example shows part of the PI3-Kinase pathway, with alteration frequencies for two different tumor types overlaid. Frequencies in shades of red indicate gene activation; shades of blue indicate gene inactivation
3 Results
3.1 Interactive construction
PathwayMapper provides an easy to use drag-and-drop mechanism for quickly creating nodes and their interactions through Node and Interaction Palettes on the left. Nodes can be of the following types: gene, family, complex, compartment,and process. Interaction types are as follows: activates, inhibits, induces, represses and binds.
Standard zoom and scroll and drag facilities are also available for manual arrangement of the pathways. Undo support is available for all operations. In addition, pathway file name, title and description can be defined and persisted on disk as needed.
3.2 Import and export facilities
PathwayMapper supports persistence of constructed pathways as tab-delimited text. Manually or programmatically constructed text files using the pre-defined extension can be imported as drawings as well.
Pathways can be exported as static images (JPEG and PNG) or scalable vector graphics (SVG).
3.3 Rearranging pathways
PathwayMapper provides both static and incremental layout facilities, using the CoSE layout algorithm, which is specifically designed for graphs with compound structures (Dogrusoz et al., 2009). The former is especially useful for models inferred by computational methods or queried from a database, where no layout information is available. The latter, on the other hand, is useful during curation as incremental changes are made to a pathway and newly integrated genes and interactions need to be ‘tidied up’, conforming to overall graph drawing criteria such as no node overlaps and uniform edge length, while maintaining the user’s existing mental map.
PathwayMapper also supports alignment operations by showing guidelines as two nodes are aligned (e.g. center to center) as well as facilitating operations for horizontal and vertical alignment of two or more nodes based on the first selected node. Alignment of nodes with the help of a grid and ‘snap to grid’ support are also available.
3.4 Overlaying alteration frequencies
Users can upload and overlay alterations frequencies of genes in the current pathway, either from a local tab-delimited text file or from the cBioPortal database through its web service. Positive values signify percentage of samples with an activating alteration and are shown with a white-to-red color scale, whereas negative values signify frequencies of inactivation, shown with a white-to-blue color scale, similar to pathway representations in TCGA manuscripts (The Cancer Genome Atlas Research Network, 2008, 2011, 2012a, 2012b, 2013, 2014a, 2014b, 2015, 2017).
The alteration data file may contain an arbitrary number of data sets, and its view can be customized through the data view settings dialog. Similarly, users can retrieve any number of data sets from cBioPortal, and selectively overlay such data on the pathway map.
3.5 Pre-curated pathways
A collection of pre-curated pathways matching the results published in TCGA manuscripts are made available in PathwayMapper through the Network menu. Users can choose to modify the topology and/or layout of these drawings. Alteration frequencies from the cBioPortal database or other sources may be overlaid on these pathways as well.
3.6 Real-time collaborative editing
PathwayMapper supports remote users collaborating on pathway editing. Should the user choose the ‘Collaborative’ mode (as opposed to ‘Local’) on the welcome page, s/he will be first prompted for Google account authentication since the shared data model will be stored in a shared document in the Google Drive folder of the user. Then, the editing session will be given a unique ID and the user will have the option of sharing the URL containing this ID with desired others and construct or edit a pathway in real-time with support for concurrent modifications and built-in conflict resolution.
Thus, any changes made by a person working on the pathway with the same URL will be instantly synchronized with others currently editing the same pathway through Google’s Realtime API.
Funding
This work was supported by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, a National Cancer Institute Cancer Center Core Grant (P30-CA008748), the Robertson Foundation, and the Prostate Cancer Foundation. We are also grateful for the support of the Google Summer of Code program.
Conflict of Interest: none declared.
Supplementary Material
References
- Cerami E. et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogrusoz U. et al. (2009) A layout algorithm for undirected compound graphs. Inf. Sci., 179, 980–994. [Google Scholar]
- Franz M. et al. (2016) Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics, 32, 309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao. et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6(269), pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway L.A., Lander E.S. (2013) Lessons from the cancer genome. Cell, 153, 17–37. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature, 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2011) Integrated genomic analyses of ovarian carcinoma. Nature, 474, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2012a) Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2012b) Comprehensive genomic characterization of squamous cell lung cancers. Nature, 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2013) Integrated genomic characterization of endometrial carcinoma. Nature, 497, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2014a) Comprehensive molecular profiling of lung adenocarcinoma. Nature, 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2014b) Integrated genomic characterization of papillary thyroid carcinoma. Cell, 159, 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature, 517, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2017) Integrated genomic characterization of oesophageal carcinoma. Nature, 541, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaveces J.M. et al. (2015) Tools for visualization and analysis of molecular networks, pathways, and -omics data. Adv. Appl. Bioinform. Chem., 8, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.