Abstract
Summary: Biologists produce large data sets and are in demand of rich and simple web portals in which they can upload and analyze their files. Providing such tools requires to mask the complexity induced by the needed High Performance Computing (HPC) environment. The connection between interface and computing infrastructure is usually specific to each portal. With Jflow, we introduce a Workflow Management System (WMS), composed of jQuery plug-ins which can easily be embedded in any web application and a Python library providing all requested features to setup, run and monitor workflows.
Availability and implementation: Jflow is available under the GNU General Public License (GPL) at http://bioinfo.genotoul.fr/jflow. The package is coming with full documentation, quick start and a running test portal.
Contact: Jerome.Mariette@toulouse.inra.fr
1 Introduction
Building rich web environments aimed at helping scientists analyze their data is a common trend in bioinformatics. Specialized web portals such as MG-RAST (Meyer et al., 2008), MetaVir (Roux et al., 2011) or NG6 (Mariette et al., 2012) provide multiple services and analysis tools in an integrated manner for specific experiments or data types. These applications require WMS features to manage and execute their computational pipelines.
Generic WMS, such as Galaxy (Goecks et al., 2010), Ergatis (Orvis et al., 2010) or Mobyle (Néron et al., 2009) provide a user friendly graphical interface easing workflow creation and execution. Unfortunately, such environments come with their own interface, complicating their integration within already existing web tools. Other WMS such as weaver (Bui et al., 2012), Snakemake (Koster et al., 2012), Ruffus (Goodstadt, 2010) or Cosmos (Gafni et al., 2014) provide a framework or a domain-specific language to developers wanting to build and run workflows. These software packages offer the flexibility and power of a high-level programming language, but they do not provide a user interface, enable component and workflow definition.
JFlow combines a user friendly interface with an intuitive python API. It is, to our knowledge, the only WMS designed to be embedded in any web application, thanks to its organization as jQuery (http://jquery.com/) plug-ins.
2 Methods
Jflow user interface gathers five jQuery plug-ins providing user oriented views.
availablewf lists all runnable workflows accessible to users,
activewf monitors all started, completed, failed, aborted and reseted workflows,
wfform presents workflow editable parameters in a form,
wfoutputs displays all outputs produced by the workflow organized per component,
wfstatus shows the workflow execution state as a list or an execution graph. The graph visualization uses the Cytoscape web JavaScript plug-in (Lopes et al., 2010).
The plug-ins give access to multiple communication methods and events. They interact with the server side through Jflow’s REST API, running under a cherrypy (http://www.cherrypy.org/) web server. The included server uses the JSONP communication technique enabling cross-domain requests.
To be available from the different jQuery plug-ins, the workflows have to be implemented using the Jflow API. A Jflow component is in charge of an execution step. Adding a component to the system requires to write a Python Component subclass. In Jflow, different solutions are available to ease component creation. To wrap a single command line, the developer can give a position or a flag for each parameter. Jflow also embeds an XML parser which allows to run genuine Mobyle (Néron et al., 2009) components. Finally, to allow developers to integrate components from other WMS, Jflow provides a skeleton class. This class only requires to implement the parsing step. A workflow chains components. It is represented by a directed acyclic graph (DAG) where nodes represent jobs and edges links between inputs and outputs. When paths are disjoint, jobs are run in parallel. A Jflow workflow is built as a Workflow subclass. Components are added to the workflow as variables and chained linking outputs and inputs.
To define the parameters presented to the final user, Jflow gives access to different class methods. Each parameter has at least a name, a user help text and a data type. For file or directory parameters, it is possible to set required file format, size limitation and location. Jflow handles server side files with regular expressions, but also URLs and client side files, in which case, it automatically uploads them. Before running the workflow, Jflow checks data type compliance for each parameter. Job submission, status checking and error handling, rely on Makeflow (Albrecht et al., 2012) and weaver (Bui et al., 2012). Therefore Jflow manages error recovery and supports most distributed resource management systems (Condor, SGE, Work Queue or a single multi core machine, …). Replacing Makeflow by an other job submitter requires to implement a new Engine subclass. This class creates and executes the workflow DAG.
3 Example
Jflow user interface has been designed to allow an easy integration in mash up web applications. Hereunder, we present its integration in NG6, which provides a user-friendly interface to process, store and download high-throughput sequencing data. The environment displays sequencing runs as a table. From this view, the user can add new data by running workflows in charge of loading the data and checking its quality. Different workflows are available considering data type and sequencing technology.
Workflows are listed by the availablewf plug-in built within a NG6 modal box. A select.availablewf event thrown by the availablewf plug-in is listened and caught to generate the parameter form using the wfform plug-in. Considering the parameter type, Jflow adapts its display. For example, a date is shown as a calendar and a boolean as a check box.
Biologists use NG6 to check sequencing reads quality, including experimental samples contamination measure. The first input of this analysis is the contaminant reference genome fasta file, displayed as a file selector. The second input is a parameter set describing the biological samples. It includes the read files and metadata such as sample name, tissue and development stage. To help biologists populate it, Jflow uses a structured data input rendered by the wfform plug-in as a spreadsheet. It allows to copy and paste multiple lines. Jflow iterates then on the table content to launch each sample processing in parallel.
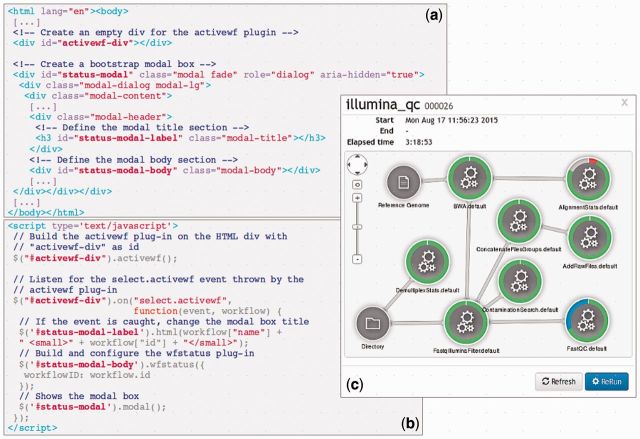
To monitor running workflows, NG6 provides a table in a specific page. The table is filled by the activewf plug-in. In the same way as described above, the wfstatus is built on a modal box when a select.activewf event is thrown by the activewf plug-in, as presented in Figure 1. This view shows the workflow’s execution graph where nodes represent components and edges links between inputs and outputs.
Fig. 1.
Jflow integration: (a) a piece of the NG6 HTML code source in which is positioned an empty div to build the activewf plug-in and a modal box for the wfstatus plug-in. (b) The jQuery code in charge to build Jflow plug-ins and manage user action. When the select.activewf event is thrown from activewf-div, a function is called with two parameters: event and workflow. The last parameter stores all the workflow’s information, such as its name and its id, used in this example to update the modal box title and to build the wfstatus plug-in. (c) The status of the illumina_qc workflow with the id 26 displayed as a graph in the NG6 application
NG6 was first implemented using the Ergatis (Orvis et al., 2010) WMS, which had a separate user interface. With Jflow, all actions are now available from the same application, which makes it user friendly.
4 Conclusion
Jflow is a simple and efficient solution to embed WMS features within a web application. It is, to our knowledge, the only WMS designed with this purpose. It is already embedded in RNAbrowse (Mariette et al., 2014) and NG6 (Mariette et al., 2012), where it has been used to process more than 2000 sequencing runs on a 5000 cores HPC environment.
Conflict of Interest: none declared.
References
- Albrecht M., et al. (2012) Makeflow: a portable abstraction for data intensive computing on clusters, clouds, and grids. SWEET at ACM SIGMOD, 20. doi: 10.1145/2443416.2443417. [Google Scholar]
- Bui P. (2012) Compiler Toolchain For Data Intensive Scientific Workflows. Ph.D. Thesis, University of Notre Dame. [Google Scholar]
- Gafni E., et al. (2014) COSMOS: python library for massively parallel workflows. Bioinformatics, 30, 2956–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J., et al. (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences, Genome Biol., 11, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstadt L. (2010) Ruffus: a lightweight Python library for computational pipelines. Bioinformatics, 26, 2778–2779. [DOI] [PubMed] [Google Scholar]
- Koster J., Rahmann S. (2012) Snakemake—a scalable bioinformatics workflow engine. Bioinformatics, 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Lopes C.T., et al. (2010) Cytoscape Web: an interactive web-based network browser, Bioinformatics, 26, 2347–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariette J., et al. (2012) NG6: Integrated next generation sequencing storage and processing environment. BMC Genomics, 13, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariette J., et al. (2014) RNAbrowse: RNA-seq de novo assembly results browser. PLoS ONE, 9, e96821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F., et al. (2008) The metagenomics RAST server a public resource for the automatic phylogenetic and functional analysis of metagenomes, BMC Bioinformatics, 9, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Néron B., et al. (2009) Mobyle: a new full web bioinformatics framework, 25, 3005–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvis J., et al. (2010) Ergatis: a web interface and scalable software system for bioinformatics workflows. Bioinformatics, 15, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S., et al. (2011) Metavir: a web server dedicated to virome analysis, Bioinformatics, 21, 3074–3075. [DOI] [PubMed] [Google Scholar]