Abstract
Histone methylation is an epigenetic modification of chromatin undergoing dynamic changes and balancing tissue-specific demands of proliferation and differentiation. In cancer, aberrant histone methylation can facilitate oncogenic and tumor suppression programs by modulating gene expression. Histone remodelers such as lysine methyltransferases and lysine demethylases are seemingly opposite or contrary forces but may be part of an interconnected network complementing each other. We identify several layers of molecular communication where epigenetic master regulators engage in crosstalk between tumor metabolism and histone remodeling. Epigenetic master regulators have the ability to cooperate with members of the transcriptional machinery, DNA methyltransferases, as well as other histone modifiers. High-throughput sequencing and omics data in combination with cancer systems biology analysis have the power to prioritize regulatory events epigenome-wide.
Keywords: cancer systems biology, melanoma, cancer metabolism, epigenomics, epigenetics; epigenetic drug; precision medicine, histone demethylase, histone methyltransferase, driver, master regulator; oncometabolite; oncogene; tumor suppressors, KDM, KMT, EZH2, JMJD, jumonji
Introduction
One of the great mysteries of biology is how the human genome is programmed in each cell to activate only the relevant genes. Eukaryotic chromatin is structurally organized in nucleosome particles, which are composed of a histone octamer core around which 147 base pairs of DNA are wrapped in 1.65 turns of a tight superhelix [1, 2]. Chromatin is not an inert structure, but rather an instructive DNA scaffold that can respond to external cues to regulate the many uses of DNA. The methylation of histone proteins at specific residues plays a major role in the maintenance of active and silent states of gene expression in developmental processes and disease (Figure 1). Pioneering experiments by Allfrey and Mirsky >50 years ago identified methylation and acetylation of histones by isotope incorporation and showed that histone modification can influence whether RNA synthesis of genes is switched on or off [3].
Figure 1.
Structural properties and functional impact of epigenetic histone lysine modification on the nucleosome octamer. (A) Methylation of lysine residues on solvent-accessible histone tails increases hydrophobicity and compaction of nucleosome assembly. (B) Epigenetic transitions of histone lysine methylation pattern mediated by histone KMT and histone KDM families. Methylation of sites K9, K27 and K36 (red) is associated with transcriptional repression, while demethylation of these sites leads to transcriptional activation (blue). Methylation of other histone lysine residues (gray) is associated with promoter activation, DNA recombination, replication, repair, and enhancer functions.
Histone methylation mainly occurs on the side chains of solvent-accessible lysines and arginines of termini of histone H3 and H4. Unlike acetylation and phosphorylation, however, histone methylation does not alter the charge of the histone protein but its hydrophobicity. Hydrophobic CH3- groups may be sequentially added and result in mono-, di- or trimethylated ε-lysine (Figure 1A) or mono- or symmetrically or asymmetrically dimethylated arginine [4]. Such distinct patterns of covalent histone marks introduced the idea of a histone code, a language edited and read by proteins and communicated in addition to four-letter base code of DNA [5, 6]. Epigenetic control via histone modification is largely operated by lysine methyltransferases (KMTs) [7] and lysine demethylases (KDMs) [8–10], which were discovered to take key roles in gene expression (Figure 1B, Table 1) [11]. An important aspect of epigenetic regulation is that the process leading to transcriptional changes in a tissue does not rely on an isolated event governed by a solitary enzyme modifying a monomolecular substrate. A network of different histone-modifying enzymes writes, erases and reads epigenetic marks (Figure 1B). Epigenetics focuses on factors and processes that regulate how and when certain genes are turned on and off, whereas epigenomics refers to analysis of global epigenetic changes across many genes, made possible by high-throughput sequencing methods. In particular, chromatin immunoprecipitation with next-generation sequencing (ChIP-Seq) provided breakthroughs to globally detect and characterize protein–DNA interactions and epigenetic modifications [12–14]. Aberrant methylation of histones, in particular hypermethylation, is thought to influence the pathobiology of cancer by disrupting the same pathways as are affected by deleterious mutations and promoter cytosine-phosphate-guanine (CpG) site DNA hypermethylation [15, 16].
Table 1.
Deregulation of the epigenetic network of histone methyltransferases and demethylases and deregulation in cancer
| HK | KMT | KMT | SYMBOL | ALIASES | me1 | me2 | me3 | MUT | mRNA | AMP | DEL | KDM | KDM | SYMBOL | ALIASES | me1 | me2 | me3 | MUT | mRNA | AMP | DEL | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H3K4 | KMT2 | KMT2A | KMT2A | MLL1 | x | x | MUT # | DEL * | 2 | KDM1 | KDM1A | KDM1A | LSD1 | MUT | UP | AMP | DEL * | |||||||
| KMT2B | KMT2B | MLL4 | x | x | MUT *,# | UP | AMP ^ | KDM1B | KDM1B | LSD2 | MUT | AMP ^ | ||||||||||||
| KMT2C | KMT2C | MLL3 | x | MUT *,# | DOWN | AMP ^ | KDM2 | KDM2A | KDM2A | JHDM1A | x | x | MUT | UP | AMP ^ | |||||||||
| KMT2D | KMT2D | MLL2 | x | MUT *,# | UP | AMP ^ | KDM2B | KDM2B | JHDM1B | x | x | MUT | UP | AMP | ||||||||||
| KMT2E | KMT2E | MLL5 | MUT | DOWN | AMP * | KDM5 | KDM5A | KDM5A | JARID1A | x | x | x | MUT | AMP ^ | ||||||||||
| KMT2F | SETD1A | SET1A | x | x | MUT | UP | AMP * | KDM5B | KDM5B | JARID1B, FBXL | x | x | x | MUT * | UP | AMP * | ||||||||
| KMT2G | SETD1B | SET1B | x | x | MUT # | UP | AMP | KDM5C | KDM5C | JARID1C | x | x | x | MUT # | AMP | DEL | ||||||||
| KMT2H | ASH1L | ASH1 | x | x | MUT * | AMP * | KDM5D | KDM5D | JARID1D | x | x | x | MUT | |||||||||||
| KMT3 | KMT3D | SMYD1 | ZMYND18 | MUT * | DOWN | AMP ^ | RIOX1 | JMJD9, NO66 | x | x | AMP | DEL | ||||||||||||
| KMT3C | SMYD2 | ZMYND14 | MUT | AMP * | ||||||||||||||||||||
| KMT3E | SMYD3 | ZMYND1 | MUT | UP | AMP * | |||||||||||||||||||
| 1 | KMT3F | WHSC1L1 | NSD3 | x | x | x | MUT | UP | AMP | DEL | ||||||||||||||
| 1 | KMT3G | WHSC1 | NSD2, MMSET | x | x | x | MUT | UP | AMP | DEL | ||||||||||||||
| KMT7 | KMT7 | SETD7 | SET7, SET9 | MUT | DOWN | AMP | DEL * | |||||||||||||||||
| KMT8 | KMT8B | PRDM9 | MEISETZ | MUT * | UP | AMP * | ||||||||||||||||||
| H3K9 | KMT1 | KMT1A | SUV39H1 | x | MUT | AMP | DEL | KDM3 | KDM3A | KDM3A | JMJD1A, JHDM2A | x | x | MUT | UP | AMP ^ | ||||||||
| KMT1B | SUV39H2 | x | MUT | AMP | DEL | KDM3B | KDM3B | JMJD1B | x | x | MUT | AMP | ||||||||||||
| KMT1C | EHMT2 | G9A | x | x | MUT | UP | AMP ^ | KDM3C | JMJD1C | TRIP8 | x | x | MUT | DOWN | AMP | DEL | ||||||||
| KMT1D | EHMT1 | GLP | x | x | MUT $ | UP | AMP | 3 | KDM4 | KDM4A | KDM4A | JMJD2A, JHDM3A | x | MUT | AMP | |||||||||
| KMT1E | SETDB1 | ESET | x | MUT | UP | AMP * | KDM4B | KDM4B | JMJD2B | MUT $ | UP | AMP | DEL | |||||||||||
| KMT1F | SETDB2 | CLLD8 | x | MUT # | DOWN | DEL * | KDM4C | KDM4C | JMJD2C, GASC1 | x | MUT | AMP | DEL | |||||||||||
| KMT8 | KMT8 | PRDM2 | RIZ1 | MUT | AMP | DEL * | KDM4D | KDM4D | JMJD2D | x | x | MUT | UP | AMP | DEL * | |||||||||
| KDM4E | KDM4E | JMJD2E | MUT | AMP | DEL * | |||||||||||||||||||
| 4 | KDM7 | KDM7A | KDM7A | JHDM1D | x | x | MUT | AMP * | ||||||||||||||||
| KDM7B | PHF8 | JHDM1F | x | x | MUT | AMP | DEL | |||||||||||||||||
| KDM7C | PHF2 | JHDM1E | x | x | MUT | DEL | ||||||||||||||||||
| RIOX2 | JMJD10, MINA | x | x | MUT | AMP | |||||||||||||||||||
| H3K27 | 2 | KMT6 | KMT6A | EZH2 | ENX1 | x | x | x | MUT *,$ | UP | AMP * | KDM6 | KDM6A | KDM6A | UTX | x | x | MUT *,# | DEL | |||||
| KMT6B | EZH1 | x | x | x | MUT *,$ | AMP * | DEL | KDM6B | KDM6B | JMJD3 | x | x | MUT | DEL * | ||||||||||
| KDM6C | UTY | |||||||||||||||||||||||
| H3K36 | KMT3 | KMT3A | SETD2 | SET2 | x | x | x | MUT *,# | DEL * | KDM2 | KDM2A | KDM2A | JHDM1A, FBXL11 | x | x | MUT | UP | AMP ^ | ||||||
| KMT3B | NSD1 | SOTOS | MUT *,#,$ | AMP | DEL | KDM2B | KDM2B | JHDM1B, FBXL10 | x | x | MUT | UP | AMP | |||||||||||
| KMT3C | SMYD2 | ZMYND14 | MUT | AMP * | KDM8 | KDM8 | KDM8 | JMJD5 | x | MUT | DOWN | AMP ^ | ||||||||||||
| KMT3H? | SETMAR | METNASE | MUT | DEL * | KDM4 | KDM4A-D | x | x | x | |||||||||||||||
| H3K79 | KMT4 | KMT4 | DOT1L | DOT1 | x | x | x | MUT | UP | AMP | DEL * | |||||||||||||
| H4K20 | KMT5 | KMT5A | SET8 | SETD8 | x | UP | AMP | KDM7 | KDM7A-B | x | x | |||||||||||||
| KMT5B | SUV420H1 | CGI85 | x | x | MUT *,$ | AMP ^ | ||||||||||||||||||
| KMT5C | SUV420H2 | x | AMP ^ |
Systematic nomenclature of histone lysine (HK) methylation and gene symbols of their modifiers. Predominant structural aberrations of histone methyl transferases and demethylases and deregulation in cancer are classified by somatic mutations (MUT), transcriptional regulation of messenger RNA (mRNA), somatic copy number amplification (AMP) or deletion (DEL) in The Cancer Genome Altas [23, 31]. Transcriptional upregulation (UP) and downregulation (DOWN) display consistent trends of gene expression in tumor progression from normal to primary tumor to metastatic cancer.
1Also H3K27 and H3K36 activity reported.
2Also H3K9 activity reported.
3Also H3K36 activity reported.
4Also H3K27 activity reported.
Significant enrichment with deviation from mean based on 95% confidence interval.
#Enrichment of nonsense mutations indicative of tumor suppressor function.
$Enrichment of hotspot mutations.
^Enrichment in some cancer tissues.
Epigenetic master regulators use reversible chemical modifications of chromatin, histone or nucleotide marks, and affect gene activity without altering the core DNA sequence. In cancer, such epigenetic master regulators are found at the top of regulatory hierarchies, particularly in pathways related to cellular proliferation, survival, fate and differentiation. For the manifestation of a genomic or non-genomic aberration of an epigenetic master regulator, it is a necessity that its own activity is affected by somatic mutation, copy number alteration, expression levels, protein cofactors or methylation status. Epigenetic master regulators often accomplish target specificity of their phenotypic program by cooperation with members of the transcriptional machinery and therefore may depend on tissue-specific expression of such auxiliary factors. In cancer, an epigenetic master regulator populates an extreme state and is either permanently switched on or off. An epigenetic master regulator will become a cancer driver, if it is not functionally neutral but contributes to tumorigenesis or disease progression in its hyperactive or deactivated state. A defined challenge in the field of epigenetic master regulators is to identify cancer-specific vulnerabilities in gene targets and biological pathways that are frequently and consistently perturbed under the control of an epigenetic driver.
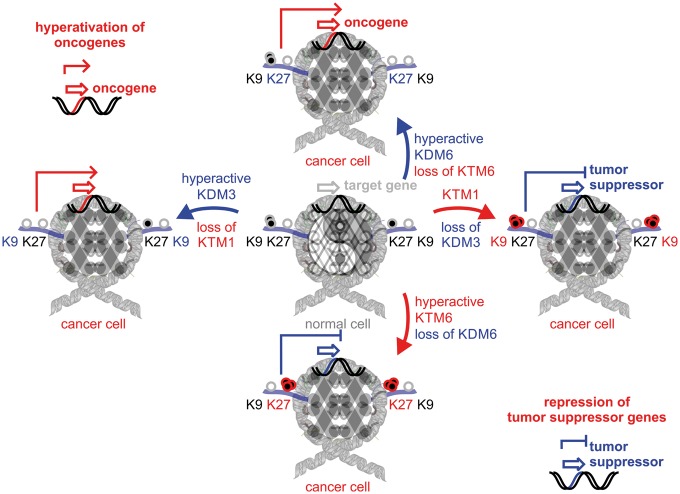
Looking at the distribution of somatic alterations of epigenetic modifiers in cancer, trends of protein up- or down-regulation are not clearly established. The idea that aberrant histone methylation can throw tissue homeostasis off balance is a simple resolution to this conundrum (Figure 2). Comparable with Yin and Yang, where complementary forces keep each other in check, activity of histone modifiers has to be maintained in dynamic steadiness and otherwise result in fatal transcriptional outcomes [10, 17]. On the one hand, hyperactive KMT or inactive KDM can cause repression of tumor suppressor genes owing to accumulation of histone lysinemethylation marks [18]. On the other hand, inactive KMTs or hyperactive KDMs can contribute to transcriptional activation of oncogenes. The idea of balanced epigenetic homeostasis explains why extreme, epigenome-wide histone hypo- or hypermethylation have been observed in cancer (Table 1) [19].
Figure 2.
Yin and Yang of histone demethylases and methyltransferases in cancer—epigenetic control of oncogenes and tumor suppressors. Consequences of hyper-activation or loss of histone modifiers is shown at the example of H3K9 regulators, KDM3/KTM1 and H3K27 modifiers, KDM6/KTM6=EZH2. Both regulatory events, transcriptional activation of oncogenes or transcriptional repression of tumor suppressor genes can contribute to tumor initiation and progression.
Recent multiomics data have shown that the H3K27-KMT6 family member, EZH2, is hyperactivated in many cancers including prostate cancer, lymphomas and melanomas with poor prognosis owing to immune evasion and repression of tumor suppressors [20, 21]. In synergy with EZH2, the counteracting demethylases KDM6A and KDM6B show frequent loss-of-function mutations as well as gene deletions resulting in accumulation of repressive H3K27 marks [17, 22–24]. KDM3 and KMT1 family members focused on remodeling of H3K9 marks display a great range of dysregulation in cancer. In prostate cancer, sarcoma, lung cancer and melanoma, copy number and transcriptional upregulation of KDM3A take a predominant role as amplifier by transcriptionally activating oncogenic target genes [9, 25]. Other KDM3 family members, KDM4C and KDM4D, are equally up- and downregulated in cancer specimen and may have diverse targets in oncogenic or tumor suppressor functions [10, 26–28]. The H3K9-KMT1 family member SETDB1 has been identified in malignant melanoma to be amplified and to increase the aggressiveness of the disease [29]. SETDB1 forms a multimeric complex with SUV39H1 and other H3K9 methyltransferases to maintain gene silencing of tumor suppressor genes [30]. Taken together, precision medicine in combination with cancer systems biology has the ability to elucidate genome- and epigenome-wide alterations and identifies molecular pathways suitable for rational drug targeting [31].
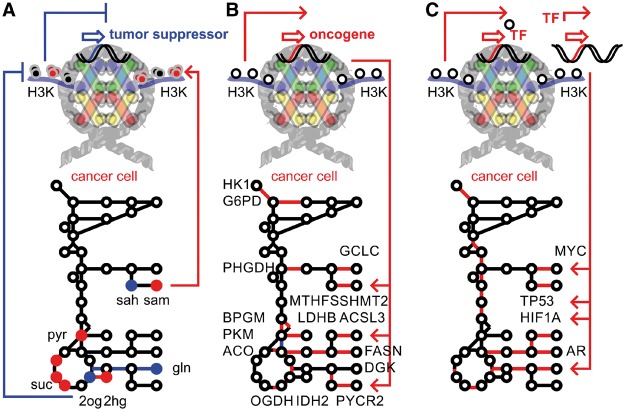
In addition to being carbon sources for covalent histone modifications, metabolites play key roles as signaling molecules in the regulation of epigenetics (Figure 3). KDMs are tightly linked to tricarboxylic acid cycle metabolites by their cofactor requirements and ability to bind organic ketoacids. Demethylation by jumonji-domain containing dioxygenase enzymes requires oxygen and 2-oxoglutarate (2OG) as substrates; complexed iron(II) and ascorbate as enzyme-cofactors; and generates formaldehyde and succinate as products. Following the oxidative decarboxylation of 2OG to succinate, a highly reactive ferryloxo species is generated that hydroxylates the methyllysine protein substrate. Because the intermediate, carbinolamine, is unstable, subsequent release of formaldehyde from the reaction intermediate produces demethylated lysine. The metabolic requirements for the 2OG-dependent dioxygenase cycle provide clues as to the physiological conditions that might affect KDM activity. Another complication arises in cancer cells where somatic active site mutations cause isocitrate dehydrogenase to increase an enzymatic sleeper-activity and to produce predominantly the oncometabolite 2-hydroxyglutarate (2HG) [32, 33]. Numerous organic acids, in particular 2-oxoketoacids including 2HG, pyruvate, oxaloacetate, malate, fumarate or succinate, have the ability to populate the enzymatic pocket of KDMs and act as competitive inhibitors (Figure 3A). Several metabolic conditions such as mitochondrial dysfunction, nutrient limitation, oxidative stress or hypoxia typical for cancer cells can modulate the activity of KDMs [34, 35]. Metabolic regulation of histone methylation is induced by low 2OG as a result of glutamine depletion, ascorbate or iron deprivation, high succinate, high fumarate or high 2HG, and potentially leads to globally altered histone lysine methylation. Importantly, such direct metabolic regulation impacts a wide range of effectors and is not restricted to a specific transcriptional program or a distinct histone lysine demethylation activity. Enzymes involved in cytosine demethylation or synthesis of signaling transmitters, carnitine and collagen have protein domains similar to jumonji dioxygenases and will be responsive to the same metabolic cues. Active KDMs have numerous metabolic enzymes as part of their target gene repertoire shown by their binding and demethylase activity in matched ChIP-Seq assays [25]. Activation of metabolic pathways supporting glycolysis, amino acid, organic acid and lipid synthesis ultimately leads to proliferative advantages of cancer cells (Figure 3B). Further, the ability of KMTs and KDMs to cooperate with the transcriptional machinery can amplify or mute specific transcriptional responses (Figure 3C). In the case of nuclear hormone receptors, targets of transcriptional cooperation include steroid biosynthesis as well as lipogenesis and enforces therapy resistance and steroid-dependent signaling in adenocarcinomas [25, 36]. However, an unambiguous mode of interaction is not identified and ranges from coexpression, protein contacts, to synergy in local chromatin environment, or even posttranslational modification. Indeed, non-histone protein modification by histone demethylases adds complexity to the regulatory ability of demethylases. Protein demethylation of TP53 suppressed proapoptotic functions by inhibiting its transcriptional activity [37–39]. Presence of regulatory POLYCOMB, COMPASS or COREST protein complexes can control, stabilize or shift the epigenetic balance [40]. Interestingly, KDMs cooperate with a network of transcription factors rather than an isolated partner, while maintaining and accomplishing gene target and DNA sequence specificity. Prominent players in cooperation with the epigenomic landscape of histone lysine methylation are MYC oncogene, androgen receptor, estrogen receptor, TP53 tumor suppressor protein, and hypoxia inducible factors, HIFs. HIF-hydroxylases are closely related to KDM dioxygenases and responsive to changes in physiological oxygen concentration. Even though these hydroxylases are cellular oxygen sensors, it is unknown whether 2OG-dependent dioxygenases like KDMs are similarly sensitive. KDMs are operated downstream of HIFs and direct gene targets of the transcription factors [41]. Such a synergistic relationship is counterintuitive, as hypoxia would expect to abolish KDM function. However, whether the KDM dioxygenase cycle is blocked in hypoxia is yet to be shown.
Figure 3.
Crosstalk between tumor metabolism and epigenetic master regulators. Epigenetic histone lysine modifiers regulate and interact with metabolism on at least three distinct levels: (A) Nutrient limitations of oxygen, glutamine and alpha ketoglutarate concentrations or oncometabolites such as 2-HG reduce KDM activity and shift the histone code toward a hypermethylated state. (B) Hyperactivity of histone modifiers by mutation, somatic copy number amplification or transcriptional regulation can target specific oncogenes and contribute to rewiring of central carbon metabolism. (C) Cooperation with transcription factors amplifies the transcriptional program underlying the specific response and network. It takes a cancer systems biology approach to decipher regulatory loops and signaling mechanisms comprising all involved levels.
Although cancer and stem cells can be challenged by enzyme inhibition [42–46], mechanistic details of factors and 2-oxoacid metabolites that are relevant under physiological conditions in the development of disease are yet to be defined. With both fields maturing, it is evident that further synergy between epigenetic and metabolomics will deliver new therapeutic agents as well as fundamental insights into how cellular chemistry regulates gene expression essential for tissue homeostasis.
Key Points
Histone methylation is part of the central histone code. Histone remodelers such as lysine methyltransferases (KMTs) and lysine demethylases (KDMs) are complementary but interconnected forces in the network of different histone-modifying enzymes that write, erase and read epigenetic marks. Histone methylation of distinct epigenetic marks leads to condensed, transcriptionally silent heterochromatin, whereas distinct loss of epigenetic marks by histone demethylation causes local formation of transcriptionally active euchromatin.
Epigenetic master regulators are at the top of cellular hierarchies and control a distinct phenotypic program via chromatin modification without altering the core DNA sequence. Epigenetic oncogenes or tumor suppressors can arise when an epigenetic master regulator is somatically activated or lost and contributes to cancer initiation and progression.
Deregulation of KMTs and KDMs turns epigenetic master regulators into cancer drivers (Table 1): histone hypomethylation can cause transcriptional activation of oncogenes; histone hypermethylation can cause transcriptional repression of tumor suppressor genes.
Histone methylation and DNA methylation are tightly linked and rely mechanistically on each other. Lysine methylation initiates, targets or maintains DNA methylation, and vice versa. In addition, there is a strong cooperation of epigenetic factors with the transcriptional complex. Cooperation with transcription factors or other members of the epigenetic machinery can target, amplify or mute specific transcriptional responses.
High-throughput technology in combination with multiomics systems biology is necessary to decipher the dynamic interplay between epigenomics and functional output in biological and biomedical settings. In particular, a solid bridge between next-generation sequencing platforms such and metabolomics has not yet been established and remains a future challenge.
Acknowledgements
Parents rarely let go of their children, so children let go of them. They move on; they move away. The moments that used to define them—a mother's approval, a father's nod—are covered by moments of their own accomplishments. It is not until much later, as the skin wrinkles and the bones weaken, that children understand; their stories, and all their accomplishments, sit atop the stories of their parents, stones upon stones, beneath the waters of their lives. Dedicated to Leland Volker, Franziska Violet, Lennart Curt, Frederic Carl Wolf, and Annabelle Clara Nia, beloved grandchildren of Ingrid Liselotte and Volker Eduard Filipp.
Funding
F.V.F. is grateful for the support of grant CA154887 from the National Institutes of Health, National Cancer Institute. This work is supported by University of California, Cancer Research Coordinating Committee CRN-17-427258, NSF, UC Senate Graduate Research Council, and HSRI program grants.
Biography
Fabian V. Filipp is a professor for Systems Biology and Cancer Metabolism focused on the connection between epigenomic master regulators and their control over central metabolism as an opportunity for diagnosis and treatment of therapy-resistant cancers.
References
- 1. Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science 1974;184:868–71. [DOI] [PubMed] [Google Scholar]
- 2. Luger K, Mader AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997;389:251–60. [DOI] [PubMed] [Google Scholar]
- 3. Allfrey VG, Faulkner R, Mirsky AE.. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA 1964;51:786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murray K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry 1964;3:10–5. [DOI] [PubMed] [Google Scholar]
- 5. Turner BM. Decoding the nucleosome. Cell 1993;75:5–8. [PubMed] [Google Scholar]
- 6. Strahl BD, Allis CD.. The language of covalent histone modifications. Nature 2000;403:41–5. [DOI] [PubMed] [Google Scholar]
- 7. Rea S, Eisenhaber F, O'Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000;406:593–9. [DOI] [PubMed] [Google Scholar]
- 8. Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119:941–53. [DOI] [PubMed] [Google Scholar]
- 9. Yamane K, Toumazou C, Tsukada Y, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006;125:483–95. [DOI] [PubMed] [Google Scholar]
- 10. Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006;125:467–81. [DOI] [PubMed] [Google Scholar]
- 11. Sun ZW, Allis CD.. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002;418:104–8. [DOI] [PubMed] [Google Scholar]
- 12. Solomon MJ, Larsen PL, Varshavsky A.. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 1988;53:937–47. [DOI] [PubMed] [Google Scholar]
- 13. Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005;120:169–81. [DOI] [PubMed] [Google Scholar]
- 14. Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007;129:823–37. [DOI] [PubMed] [Google Scholar]
- 15. Baylin SB, Makos M, Wu JJ, et al. Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer Cells 1991;3:383–90. [PubMed] [Google Scholar]
- 16. Laird PW, Jackson-Grusby L, Fazeli A, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 1995;81:197–205. [DOI] [PubMed] [Google Scholar]
- 17. Shpargel KB, Starmer J, Yee D, et al. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet 2014;10:e1004507.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kazanets A, Shorstova T, Hilmi K, et al. Epigenetic silencing of tumor suppressor genes: paradigms, puzzles, and potential. Biochim Biophys Acta 2016;1865:275–88. [DOI] [PubMed] [Google Scholar]
- 19. Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 2008;40:741–50. [DOI] [PubMed] [Google Scholar]
- 20. Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002;419:624–9. [DOI] [PubMed] [Google Scholar]
- 21. Visser HP, Gunster MJ, Kluin-Nelemans HC, et al. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol 2001;112:950–8. [DOI] [PubMed] [Google Scholar]
- 22. Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 23. Guan J, Gupta R, Filipp FV.. Cancer systems biology of TCGA SKCM: efficient detection of genomic drivers in melanoma. Sci Rep 2015;5:7857.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tiffen J, Wilson S, Gallagher SJ, et al. Somatic copy number amplification and hyperactivating somatic mutations of EZH2 correlate With DNA methylation and drive epigenetic silencing of genes involved in tumor suppression and immune responses in melanoma. Neoplasia 2016;18:121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilson S, Fan L, Sahgal N, et al. The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells. Oncotarget 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klose RJ, Yamane K, Bae Y, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 2006;442:312–6. [DOI] [PubMed] [Google Scholar]
- 27. Cloos PA, Christensen J, Agger K, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 2006;442:307–11. [DOI] [PubMed] [Google Scholar]
- 28. Kim TD, Oh S, Shin S, et al. Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS One 2012;7:e34618.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011;471:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tchasovnikarova IA, Timms RT, Matheson NJ, et al. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 2015;348:1481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Filipp FV. Precision medicine driven by cancer systems biology, Cancer Metastasis Rev 2017;36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Struys EA, Salomons GS, Achouri Y, et al. Mutations in the D-2-hydroxyglutarate dehydrogenase gene cause D-2-hydroxyglutaric aciduria. Am J Hum Genet 2005;76:358–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wellmann S, Bettkober M, Zelmer A, et al. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun 2008;372:892–7. [DOI] [PubMed] [Google Scholar]
- 35. Xia X, Lemieux ME, Li W, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA 2009;106:4260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilson S, Qi J, Filipp FV.. Refinement of the androgen response element based on ChIP-Seq in androgen-insensitive and androgen-responsive prostate cancer cell lines. Sci Rep 2016;6:32611.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chuikov S, Kurash JK, Wilson JR, et al. Regulation of p53 activity through lysine methylation. Nature 2004;432:353–60. [DOI] [PubMed] [Google Scholar]
- 38. Huang J, Perez-Burgos L, Placek BJ, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006;444:629–32. [DOI] [PubMed] [Google Scholar]
- 39. Ramadoss S, Guo G, Wang CY.. Lysine demethylase KDM3A regulates breast cancer cell invasion and apoptosis by targeting histone and the non-histone protein p53. Oncogene 2017;36:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee MG, Wynder C, Cooch N, et al. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005;437:432–5. [DOI] [PubMed] [Google Scholar]
- 41. Krieg AJ, Rankin EB, Chan D, et al. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol 2010;30:344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 2012;483:484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012;483:474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Losman JA, Looper RE, Koivunen P, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013;339:1621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013;340:626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang F, Travins J, DeLaBarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 2013;340:622–6. [DOI] [PubMed] [Google Scholar]