Abstract
Summary
Foldit Standalone is an interactive graphical interface to the Rosetta molecular modeling package. In contrast to most command-line or batch interactions with Rosetta, Foldit Standalone is designed to allow easy, real-time, direct manipulation of protein structures, while also giving access to the extensive power of Rosetta computations. Derived from the user interface of the scientific discovery game Foldit (itself based on Rosetta), Foldit Standalone has added more advanced features and removed the competitive game elements. Foldit Standalone was built from the ground up with a custom rendering and event engine, configurable visualizations and interactions driven by Rosetta. Foldit Standalone contains, among other features: electron density and contact map visualizations, multiple sequence alignment tools for template-based modeling, rigid body transformation controls, RosettaScripts support and an embedded Lua interpreter.
Availability and Implementation
Foldit Standalone is available for download at https://fold.it/standalone, under the Rosetta license, which is free for academic and non-profit users. It is implemented in cross-platform C ++ and binary executables are available for Windows, macOS and Linux.
1 Introduction
Graphical, interactive tools for molecular modeling are becoming accessible to the general public through citizen science projects. The interface of the game Foldit, one such project, was designed to make manipulating protein structures accessible to non-experts, while still maintaining scientific accuracy (Cooper et al., 2010). In order to make Foldit’s interface more useful as a biochemistry application, we have developed Foldit Standalone by removing game-specific features while adapting and adding features catering to experienced biochemists, including allowing users to load and manipulate their own molecular structures—mainly proteins. Figure 1 shows a Foldit Standalone screenshot.
Fig. 1.
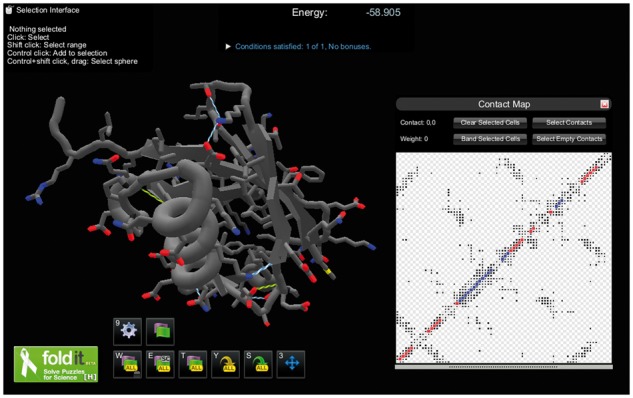
A screenshot of the Foldit Standalone interface, with the contact map shown for the currently loaded structure. There are a variety of options to customize the visualization
Foldit Standalone’s manipulations are driven by the powerful Rosetta molecular modeling package (Leaver-Fay et al., 2011; Rohl et al., 2004), which has been used in a variety of computational modeling applications. This includes the prediction and design of protein structures (Kuhlman et al., 2003), protein–protein interfaces (Gray et al., 2003) and protein-small molecule interfaces (Meiler and Baker, 2006). As Rosetta is implemented in C ++, a number of other front-ends have been developed to support easier authoring and use of scientific protocols. These include PyRosetta (Chaudhury et al., 2010), which provides Python bindings for much of the Rosetta C ++ API, and RosettaScripts (Fleishman et al., 2011), which allows various aspects of protocols to be specified in XML format. Graphical interfaces to Rosetta include InteractiveROSETTA (Schenkelberg and Bystroff, 2015) and the PyRosetta toolkit (Adolf-Bryfogle and Dunbrack, 2013), both of which provide extensive means for setting up, running and viewing the results of Rosetta protocols. In contrast, Foldit Standalone focuses on providing simplicity of use and does not require previous knowledge of Rosetta to get started.
2 Features
Foldit Standalone is implemented in C ++ within the Rosetta codebase. It consists of additional libraries and an executable in the architectural style of the rest of Rosetta. The code is cross-platform and builds and runs on Windows, macOS and Linux. Foldit Standalone contains a custom rendering engine built with OpenGL that handles display of 2D interface elements and 3D geometry, along with a custom input event system. Most features are shared with the Foldit game; Foldit Standalone replaces the game’s terminology with more standard terminology. Although the interface, visualizations and tools are focused on proteins, there is basic support for DNA, RNA and small molecules.
Foldit Standalone supports a variety of visualization options, including different geometries (including line, sphere and cartoon); different colorings (including energy-based, CPK and rainbow); and different levels of abstraction (hiding or showing hydrogens and protein sidechains). Additional custom visualizations include clashes where atoms’ van der Waals volumes overlap; voids showing empty, desolvated space; marking solvent-exposed hydrophobics; and hydrogen bonds and disulfides. Electron density maps can be loaded and visualized with their own set of visualization options.
Foldit Standalone’s custom interactive manipulations have been implemented specifically for real-time molecule manipulation. These manipulations include clicking and dragging to pull on the backbone or sidechains; adding rubber bands or freezing portions of a molecule, which use soft and hard constraints that impact other manipulations; fragment insertion; amino acid insertion, deletion and mutation; and rigid body controls for docking. Users can also launch interactive versions of minimization and rotamer optimization, which update the display as they run and can be canceled. Long-lived or computationally intensive manipulations run in a background thread so that the interface remains responsive. Using Foldit Standalone’s selection interface, the user can select regions of the molecule, and then perform different manipulations on their selection. Additional tools allow more advanced manipulations, including a Ramachandran map panel, a multiple sequence alignment panel, and a contact map panel. Foldit Standalone supports protocol automation through both RosettaScripts and an embedded Lua interpreter.
Foldit Standalone can import several standard and Rosetta-specific file formats. A session can be started by importing at least a PDB or FASTA file; other files imported at the same time—such as constraints, electron density, Rosetta-based symmetry definitions, or template structures—will customize the session setup. Session states can be saved and loaded using a custom file format, and PDBs can be exported. While it is possible to load large structures into Foldit Standalone, we have found that smaller structures (fewer than 500 residues) are preferable for smooth interactions.
3 Conclusion
Throughout the years, a number of expert-oriented tools for visualizing and interacting with molecular structures have been developed, including Sculpt (Surles et al., 1994), ProteinShop (Crivelli et al., 2004) and Coot (Emsley and Cowtan, 2004). PyMOL (DeLano, 2002), a widely popular tool for molecule visualization, also supports some manipulation of input molecules. Foldit Standalone goes well beyond this body of software, providing an accessible graphical structure manipulation interface coupled to the powerful Rosetta energy function and sampling methods. As the tools improve and broader participation is elicited, there will almost certainly be new and significant scientific discoveries enabled through this innovative interface for modeling and design.
Acknowledgements
The authors would like to thank the Rosetta developers and RosettaCommons for their contributions to the Rosetta software used by Foldit Standalone. The authors also thank the Foldit development team for their work, and the Foldit players for providing feedback on the Foldit game interface.
Funding
This work was supported by the National Science Foundation [1629879]; the National Institutes of Health [1UH2CA203780]; and RosettaCommons.
Conflict of Interest: none declared.
References
- Adolf-Bryfogle J., Dunbrack R.L. Jr. (2013) The PyRosetta toolkit: a graphical user interface for the Rosetta software suite. PLoS One, 8, e66856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S. et al. (2010) PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics, 26, 689–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S. et al. (2010) Predicting protein structures with a multiplayer online game. Nature, 466, 756–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivelli S. et al. (2004) ProteinShop: a tool for interactive protein manipulation and steering. J. Comput. Aided Mol. Des., 18, 271–285. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA, USA. [Google Scholar]
- Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Struct. Biol., 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fleishman S.J. et al. (2011) RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One, 6, e20161.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.J. et al. (2003) Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol., 331, 281–299. [DOI] [PubMed] [Google Scholar]
- Kuhlman B. et al. (2003) Design of a novel globular protein fold with atomic-level accuracy. Science, 302, 1364–1368. [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A. et al. (2011) ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol., 487, 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler J., Baker D. (2006) ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins: Struct., Funct., Bioinf., 65, 538–548. [DOI] [PubMed] [Google Scholar]
- Rohl C.A. et al. (2004) Protein structure prediction using Rosetta. Methods Enzymol., 383, 66–93. [DOI] [PubMed] [Google Scholar]
- Schenkelberg C.D., Bystroff C. (2015) InteractiveROSETTA: a graphical user interface for the PyRosetta protein modeling suite. Bioinformatics, 31, 4023–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surles M.C. et al. (1994) Sculpting proteins interactively: continual energy minimization embedded in a graphical modeling system. Protein Sci., 3, 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]


