Abstract
Quality control (QC) is a critical step in RNA sequencing (RNA-seq). Yet, it is often ignored or conducted on a limited basis. Here, we present a multi-perspective strategy for QC of RNA-seq experiments. The QC of RNA-seq can be divided into four related stages: (1) RNA quality, (2) raw read data (FASTQ), (3) alignment and (4) gene expression. We illustrate the importance of conducting QC at each stage of an RNA-seq experiment and demonstrate our recommended RNA-seq QC strategy. Furthermore, we discuss the major and often neglected quality issues associated with the three major types of RNA-seq: mRNA, total RNA and small RNA. This RNA-seq QC overview provides comprehensive guidance for researchers who conduct RNA-seq experiments.
Keywords: RNA-seq, small RNA-seq, total RNA-seq, quality control, raw data, alignment, gene expression
Introduction
The introduction of RNA sequencing (RNA-seq) technology has revolutionized gene expression research, and has replaced microarray technology as the tool of choice for high-throughput gene expression profiling studies [1–5]. RNA-seq technology provides several distinct advantages over microarray technology. A microarray is a hybridization-based technology, requiring the design of probes in complimentary sequence of previously identified genes. This limitation confines microarray technology to quantifying only the expression level of known genes. In contrast, an RNA-seq library is constructed using all or selected RNA present in a sample, thus enabling the discovery of novel transcripts and their associated genes. Furthermore, microarray gene expression profiling is conducted at the gene or exon level, which restricts the application of microarray technology to expression profiling only. Based on high-throughput sequencing technology, RNA-seq provides the sequence of the complementary DNA (cDNA) obtained from RNA through reverse transcription. Similar to high-throughput sequencing of DNA (exome sequencing, whole genome sequencing, etc.), RNA-seq data are at the single-nucleotide resolution, which offers various other applications, including the detection of single-nucleotide variation, gene fusion, alternative splicing and novel isoforms [6].
While RNA-seq data analysis provides substantially more information than microarrays, it simultaneously produces more bioinformatics challenges. One of the most overlooked aspects of RNA-seq data analysis is quality control (QC). QC is essential for successful RNA-seq experiments because the QC process improves the reproducibility of the biological results. Here, we advocate the idea of conducting QC for RNA-seq from the following four perspectives: (1) RNA quality, (2) raw read data (FASTQ), (3) alignment and (4) gene quantification. Although similar to the QC of DNA sequencing data, the QC of RNA-seq data has several distinct features that should guide the application of the process. Specifically, strong emphasis is given to focusing raw data QC on the initial stage of high-throughput sequencing technology, as evidenced by the numerous tools designed to tackle FASTQ QC exclusively [7–12]. It is now evident that this partial QC can lead to incorrect interpretations of the data [13]. For example, QC on the raw data do not guarantee a good alignment rate, and QC on the alignment data can detect library construction issues, but does not identify cross-sample contamination. To correctly evaluate the quality of the data and the results, a multi-perspective QC strategy needs to be applied, which extends throughout the full data processing course.
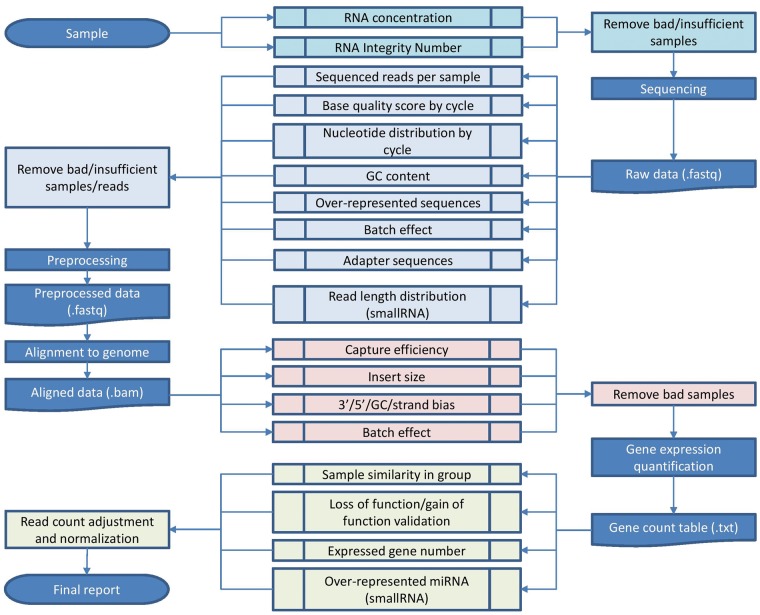
Illumina high-throughput sequencing allocates sample sources into two major categories: DNA and RNA. Within RNA-seq, there are two major distinctions: long RNA-seq and small RNA-seq (sRNA-seq). Long RNA-seq is usually denoted simply as RNA-seq without qualifier and has two main approaches for RNA library constructions: poly(A) and ribosomal RNA (rRNA)-depleted total RNA-seq. The most abundant species of RNA is rRNA (∼80%), which can hinder sequencing depth of less abundant species and therefore are eliminated during library construction. sRNA-seq is often referred to as microRNA-seq (miRNA-seq); however, the name miRNA-seq is rather misleading because sRNAs are selected by size, not by specific RNA type. Thus, sRNA libraries can contain small fragments and cleavage products from rRNA, transfer RNAs (tRNAs) and small nucleolar RNAs [6, 14–16]. We discuss the unique quality issues associated with each type of RNA-seq from multi-perspectives (Figure 1).
Figure 1.
The overall workflow of RNA-seq QC. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
RNA quality
In an RNA-seq experiment, the 1st QC is not performed on data, but rather on the RNA itself. The integrity of the RNA is the most important criterion for obtaining good quality data. Different RNA library construction kits have different input requirements for RNA quantity, and accurate quantitation is critical for selecting the appropriate platform. RNA concentration is typically measured by Qubit fluorometer (ThermoFisher Scientific). The Qubit HS assay is accurate for detecting RNA concentrations as low as 250 pg/µl. Most importantly, Qubit reagents are nucleic acid specific. Thus, assaying a sample using both the RNA and the DNA kits provides not only a highly accurate concentration, but also an indication of sample purity. An assessment of RNA integrity can be evaluated using the ratio of rRNAs 28s:18s. A higher ratio indicates better integrity. To test RNA quality, samples are tested on a 2100 Bioanalyzer (Agilent), which automatically measures the 28s:18s rRNA ratio and computes an RNA integrity number (RIN) [17]. Other equipment such as Agilent’s 4200 tape station and the Fragment Analyzer from Advanced Analytical can also be used to measure RNA quality. However, these are not widely adopted yet.
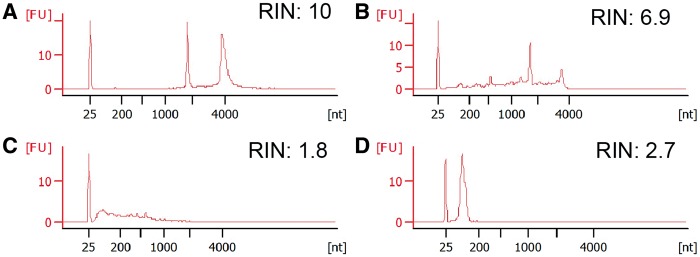
RINs provide a numerical score (range 1–10) for RNA quality (based on degradation), and a higher RIN indicates better RNA integrity. Indeed, RINs are highly correlated with 28s:18s ratios (Figure 2). In general, an RIN > 7 is considered good and appropriate for downstream analysis. For low-quality sources, e.g. formalin-fixed paraffin-embedded (FFPE) tissues, it is difficult to obtain good 28s:18s ratios because of RNA degradation. The RINs for FFPE samples usually range from 2 to 5. Unfortunately, there is currently no good assay for evaluating the success chance of FFPE tissue-based RNA-seq experiment. Instead, people determine the chance of success based on factors such as storage time, conditions, fixation time and specimen size [18]. Furthermore, traditional RIN numbers are not a good indicator of sRNA quality if RNA isolation are enriched for sRNA, as this approach will remove 28s or 18s RNA. Many RNA isolation methods will collect total RNA and not enrich for sRNA, and the RNA quality of these samples can be evaluated on the 2100 Bioanalyzer. An accurate assessment of RNA quality is a key factor for the success of downstream sequencing because this initial step determines which type of library preparation and sequencing parameters are required.
Figure 2.
(A) RNA with good quality, RIN = 10. (B) RNA with feasible quality, RIN = 6.9. (C) RNA with poor quality, RIN = 1.8. (D) Typical small RNA quality, RIN is usually <5 for small RNA. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Raw read data QC
Illumina sequencing raw read data are stored as a text file in the FASTQ format [19]. The FASTQ format stores each sequencing read in four lines of text: (1) identifiers, (2) nucleotide sequences, (3) currently not used and (4) base qualities [20] for each of the nucleotide in the read. The 1st identifier line contains useful information, e.g. machine name, run ID, lane ID and flow cell ID, which can be used for detecting batch effects. At the raw data level, the most common parameters to examine are the total number of reads sequenced, GC content and the overall base quality score, which are all commonly computed by standard raw data QC tools. A common inquiry from many people regarding RNA-seq is how many reads should be sequenced to achieve an expected coverage level. This inquiry is relevant for DNA sequencing where variant calling is the primary goal, but RNA-seq is designed to evaluate gene expression. Thus, the expected coverage of any individual gene is highly dependent on the level of that gene expression; therefore, researchers should inquire about how many reads are needed to successfully profile gene expression for their sample type. The general rule of thumb is that the recommended total number of reads for human messenger RNA-seq (mRNA-seq) is 30 million per sample and 40 million per sample for total RNA-seq. Nevertheless, successful gene expression profiling can be achieved with levels as small as 20 million reads [21]. Based on the ENCODE consortium’s recommendation, up to 100 million reads maybe needed to correctly detect gene expression of isoforms [21]. These read number targets are based on human data. For other species, the number of reads required should be scaled based on the genome size compared with the human genome size. Furthermore, human reference is the best annotated and the most complete reference. The alignment rate may be lower for species with less complete reference. Complications in assessing total read number can arise when counting pair-end reads. Usually, a pair can be considered as a single read or double read during counting. Investigators conducting pair-end sequencing studies need to be cautious of this discrepancy when counting reads.
The number of reads sequenced per sample is highly dependent on two factors: RNA quality and the number of samples being pooled on a single lane. Libraries constructed from low-quality RNA will generally produce fewer reads. Multiplexing is often used in Illumina high-throughput sequencing to reduce costs and avoid waste. As mentioned earlier, 20–40 million reads are typically needed to profile a sample’s transcriptome. On an Illumina HiSeq3000 lane, the number of reads produced is approximately 300 million. A cost-effective approach for sequencing multiple samples is to attach unique index (barcodes) sequences to each sample and then pool multiple samples together within a single lane. After the pool is sequenced, each index can be distinguished and assigned to the respective sample of origin. The process of assigning reads by barcode is called demultiplexing. For long RNA-seq, demultiplexing is part of Illumina’s pipeline (CASAVA: discontinued 31 December 2015, replaced by BaseSpace Isaac Enrichment). Thus, raw data are usually demultiplexed when received from a sequencing facility. In good practice, the 1st parameter to examine for raw data should be the total number of reads sequenced per sample, as it is the easiest, quickest and most intuitive parameter to detect failed sequencing.
Two useful plots often drawn for sequencing raw data QC are base quality score by cycle and nucleotide distribution by cycle. Cycle denotes the number of nucleotides sequenced per read. For example, if a read contains 100 nucleotides, then these nucleotides were generated from 100 cycles. The base quality score is in Phred scale, saved every 4th line within the FASTQ file. The base quality is computed using the formula where is the probability of the base being incorrect. Note that is determined by the Illumina sequencer, and the exact algorithm is not currently publically available. The base quality score is encoded with a single character from the ASCII table. The standard scale is Phred +33 (ASCII 0–62). The older Illumina pipelines (pre-CASAVA 1.3) used Phred +64 (ASCII 59–126). In Illumina sequencing data, the range of base quality is between 0 and 50. The expected base quality score is often between 30 and 40 [22]. Bases with quality scores <20 are usually considered low quality and filtered out by various analysis tools in the default setting.
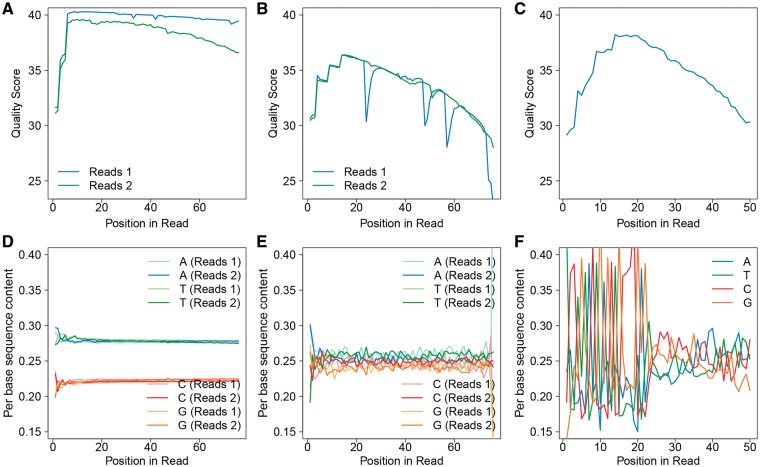
The base quality score by cycle plot is usually drawn as a plot of base quality score by each cycle. This helps to identify outlier samples (Figures 3A, B). For data generated from the discontinued Illumina sequencing platform GA II, the base quality score usually starts out high, then gradually degrades as the cycle increases, moving along the read. The drop in base quality score can be attributed to a variety of factors, including phasing/prephasing, a decreased signal to noise ratio and template damage over many cycles of laser imaging. However, because of changes in Illumina’s quality score algorithm for later sequencing platforms (HiSeq, MiSeq), the base quality score for the first 10–15 cycles is relatively lower as compared with the middle section of the read. To date, no alignment issues have been reported because of the lower-quality bases at the beginning of the reads. One approach of dealing with the low-quality bases at the end of the read is by trimming [23]. However, current aligners, such as Bowtie 2 [24], BWA [25] and STAR [26], are capable of soft clipping. Thus, these low-quality bases at the end of the reads usually do not affect the accuracy of alignment.
Figure 3.
This figure was produced from QC3. (A) Example of a long RNA-seq sample with expected base quality score. Read 2 tends to have a slightly lower median base score than read 1, but it is not usually a quality concern. (B) Example of a long RNA-seq sample with potential base quality problem, as denoted by the sudden drops of median base quality in read 2 of pair-end read sequencing. (C) Example of a sRNA-seq sample with expected quality score. Owing to trimming, the reads of sRNA-seq are of unequal length, causing the quality dropping more dramatically toward the end of the read cycles. (D) Example of a long RNA-seq sample with expected nucleotide distribution, as denoted by the stable nucleotide distribution across the samples. (E) Example of a long RNA-seq sample with a potential nucleotide distribution issue, as denoted by the unstable distribution across the cycles. (F) Example nucleotide distribution from a sRNA-seq sample (same sample as C). Large variation of nucleotide distribution can be observed which is typical for sRNA-seq data. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
The nucleotide distribution by cycle plot enables the identification of outlier samples with peculiar nucleotide distribution patterns. Ideally, the four nucleotides (A, T, C and G) should have stable distribution across all cycles. In practice, the large fluctuation in nucleotide distribution often indicates lower quality of data. For example, if one sRNA is overrepresented, then the distribution of bases at each position will reflect the sequence of that sRNA, and if there is equal distribution of bases at each position, then the sequencing is of high quality and not dominated by one RNA, i.e. adapters. There is usually a correlation between the base quality and nucleotide distribution. When we observe low quality in the base quality by cycle plot, we are likely to observe fluctuation in the nucleotide distribution at the same cycle (Figures 3D, E). An interesting phenomenon frequently associated with pair-end reads is that there is often a difference between the base quality score and percentage of nucleotides between the first read and second read in the pair. However, no analysis issues have been shown to be associated with this phenomenon. For the sRNA-seq, the base quality score by cycle and nucleotide distribution by cycle plots are usually more convoluted than long RNA-seq data (Figures 3C, 2F). Thus, they do not truly reflect the quality of the sRNA-seq data.
GC content is another useful parameter to examine the overall quality of the data. GC content is computed as the percentage of G + C in the data. The GC content of the reference sequence is an approximation of the expected GC content for the sequenced data. GC content varies by species and genomic regions, and a significant deviation (>10%) from the expected value may indicate contamination. In the latest human reference genome GRCh38, the GC content is 39.3% for the whole genome, 48.9% for coding RNA, 39.7% for long noncoding RNA (lncRNA), 50.2% for rRNA, 51.5% for miRNA, 55.7% for tRNA and 46.7% for other species of small RNAs. When performing total RNA-seq, the target is coding RNA plus other species of lncRNA; thus, the expected GC content falls somewhere between the expected GC content of lncRNA and coding RNA (39.7–48.9%). For sRNA-seq, all species of sRNAs are included in the library, and miRNA often has overrepresentation issues (discussed in depth later). Therefore, nucleotide distribution by cycle and the GC content are not good parameters to access sequencing quality for these RNA types.
Other critical parameters for RNA-seq raw read data QC are overrepresented sequences, adapter sequences and K-mer. A good sequencing library should contain a diverse set of RNA sequences. Overrepresentation of a particular sequence can be explained by two possibilities: (1) the sequence is highly biologically relevant (e.g. over expression of certain RNA because of a disease phenotype); and (2) the library is contaminated with sequences from adapters or other sources. Adapters are single-stranded RNA sequences ligated to sRNAs for cDNA synthesis. In Illumina high-throughput sequencing, an adapter serves three purposes: (1) to allow the cDNA to bind to the flow cell for sequencing; (2) to allow for polymerase chain reaction amplification of adapter-ligated DNA fragments; and (3) to allow for multiplexing (barcoding). Adapter contamination is the undesired sequencing of partial or complete adapter sequences. The best approach to solve adapter contamination is by size selection of libraries to remove adapter sequences. This can be achieved through standard gel electrophoresis or automated preparative electrophoresis with gel cassettes. In long RNA-seq data analysis, adapter trimming is usually not performed because RNA fragments are long, and the adapter is unlikely to be sequenced. Even with a partial sequencing of the adapter, the alignment will usually not be affected because of the soft clip functionality of current aligners. For sRNA-seq, the standard read length is 50 nucleotides (single-end 50 cycles), and as the majority of the sRNA-seq is <50 nucleotides, this increases the likelihood of sequencing of the attached adapter sequence. Thus, adapter trimming is required for sRNA-seq data analysis. The term K-mer refers to all possible nucleotide combinations of length K. A short K-mer (5–7-mers) computation evaluates the abundance of all possible K-mers, which can reveal additional short duplicated sequences that slip through the detection of overrepresented sequences that usually target long sequences.
One of the most overlooked QC aspects for sequencing data are batch effects. Batch effects are technical variations caused by processing data in separate batches. Batch effects, if not corrected, can cause incorrect interpretation of the data, especially for large data sets. Many studies have documented batch effects in high-throughput sequencing, including The Cancer Genome Atlas [27, 28]. Systematic sequencing failures often occur nonrandomly. A pattern of failure can be observed by lane, flow cell, run or machine. Some tools, e.g. QC3 [13], extract batch information from the ID lines of the FASTQ file and assess the significance of batch effect through statistical testing. However, sequencing libraries are often pooled on multiple lanes, and so, in the event that one lane fails, data for that library are still generated by other lanes. In such a scenario, batch effects of raw read data by lane cannot be assessed for the merged data (from different lanes). Instead, the batch effect analysis needs to be performed before the merging of raw read data by samples from multiple lanes.
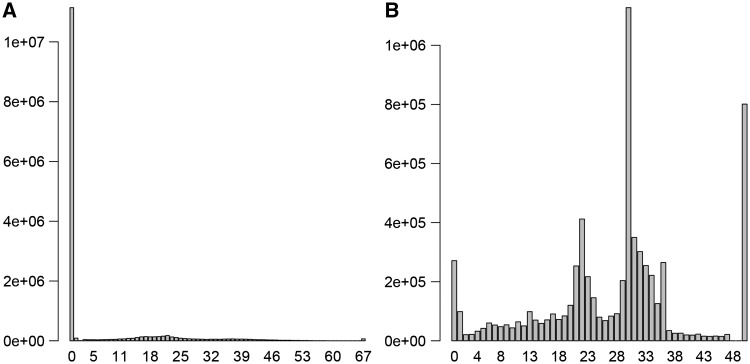
A unique QC parameter to check for sRNA-seq data is the read length distribution. Because the trimming of adapters and the sRNA-seq data contains multiple species of sRNAs with differing lengths, a read-length histogram can reveal potential sRNA distributions (Figure 4). Two peaks are generally observed. The 1st peak (approximately 22 nucleotides in length) indicates the likely abundance of miRNA, tRNA-derived fragments (tRF) and other sRNAs from longer noncoding RNAs. The 2nd peak (approximately 33 nucleotides in length) indicates the likely abundance of tRNA-derived halves (tRHs). Depending on the sample source, it is possible to have more tRHs than miRNAs and tRFs, and the 2nd peak may be greater than the 1st peak. tRHs are the result of parent tRNA cleavage by an RNase III enzyme, angiogenin [29, 30], close to the anticodon loop [31, 32]. tRFs arise from Dicer-dependent cleavage, or as an in vitro phenomenon by incubation with MgCl2 or nuclease S1 [32]. Both tRFs and tRHs are captured by sRNA-seq. A peak at the 0 nucleotide indicates overrepresentation of adapter sequences, and a peak at 50 nucleotides may indicate the sequencing of longer RNA.
Figure 4.
(A) Example of a small RNA sample with potential quality issue based on read length distribution after trimming. The high peak at zero indicates the majority of the reads are adapter sequences. (B) Example of a small RNA sample with expected read length after trimming. We observe a high peak at 22 for miRNA and another at 33 for tRNA.
Alignment QC
Sequencing data that pass raw read data QC should be further screened after alignment. In sequencing analysis, alignment is the process used to determine the best location (with least mismatches) for each read to the reference genome. It is a required step in RNA-seq analysis. QC on the alignment result file—Sequencing Alignment Map (SAM) or its equivalent compressed format of the Binary Alignment Map (BAM)—can yield additional insight into the quality of the sample and capture bad quality samples not detectable by raw data QC. For example, capture efficiency and contamination of RNA from an unwanted source (other than an adapter sequence) cannot be easily detected during raw data QC.
Capture efficiency is measured as the percentage of total sequenced reads mapped to the intended target region. For mRNA-seq, the target is the coding region; for total RNA-seq, the target is all species of long RNA except rRNA (which are removed); for sRNA-seq, the intended target is all species of sRNA. When counting the number of aligned reads, the user needs to be aware of the scenario of multiple best alignment. Multiple best alignment happens when a read can be aligned equally well (with the least mismatches) to more than one location on the reference genome. RNA aligners, e.g. TopHat [33] and STAR [26], will output multiple best alignments of a single read by default in the SAM/BAM file. Therefore, counting reads directly from the SAM/BAM file will inflate the aligned read number because of this multiple alignment. To solve this, some tools, such as QC3 [13], count a read by a fraction, so if a read has two best alignments, 0.5 will be added to both of the loci’s read counts. On the other hand, tools such as HTSeq [34], discard all reads with multiple best alignments during counting.
It is well known that the capture efficiency of the target of interest in high-throughput sequencing is not 100%. In exome sequencing, the capture efficiency ranges from 50 to 80% depending on the capture kit used [22, 35]. For RNA-seq, we should expect the capture efficiency to also range from 50 to 80%. The general rule is that a higher-quality sample should produce a higher capture efficiency. Capture efficiency is computed by QC tools, e.g. QC3 [13], Picard [36] and RNA-SeQC [37]. The command flagstat from SAMtools [38] can also produce a quick summary of mapped, unmapped, discordantly mapped and properly paired reads.
In the event that low capture efficiency is observed, there are three likely possibilities: (1) the sample quality is low; (2) rRNAs were not removed efficiently; and (3) the sequencing library is contaminated with RNA from another source. Usually, bad data because of low-quality samples are hard to salvage using bioinformatics methods. The best approach is to reconstruct the library and try for better quality. To detect rRNA abundance, rRNA quantification is needed. Contamination from other sources can be detected by aligning unaligned reads to a collection of reference genomes of other candidate species. The tool FastQ Screen [8] is designed to detect contamination using FASTQ data by aligning the reads against a collection of reference genomes using a small percent (2% by default) of reads for fast screening. The most likely causal candidates for contamination are bacteria. A good collection of bacteria reference genomes can be downloaded from the Human Microbiome Project [39]. For sRNA-seq, we expect a certain percentage of sRNAs from exogenous origin. For example, viruses have been detected readily in RNA-seq data [40–45]. It has been reported that plant miRNAs (rice, corn, etc.) can cross the GI tract and enter general circulation in humans [46, 47]. However, the small percentage of detected exogenous sRNA usually does not affect later data analysis.
A commonly used and confused QC parameter for alignment is mapping quality score (MAPQ)—the 5th column in a SAM/BAM file. The distribution of MAPQ can paint a picture of overall alignment quality. The official definition of MAPQ states that it is a Phred score, ranging from 0 to 255, with a value of 255 indicating that the MAPQ is not available. One of the earliest and most used aligner, BWA [25], uses MAPQ to represent the probability of the mapped read being correct. However, in frequently used RNA-seq aligners, e.g. TopHat [33] and STAR [26], MAPQ is used to denote the uniqueness of the alignment. A MAPQ of 255 in SAM/BAM files created by TopHat or STAR indicates the read is uniquely mapped, and a lower MAPQ indicates multiple best alignments. In DNA sequencing data analysis, MAPQ is often used as a filter for many tools [38, 48–50]. As aforementioned, RNA-seq data can be mined to obtain many types of variants typically found from DNA sequencing data. Many of the DNA sequencing analysis tools can be applied to RNA-seq data as well. One needs to be aware that the interpretation of MAPQ can be different for different aligners, and this might have an impact on the performance of the analysis tools. For RNA aligners, a more appropriate score for measuring MAPQ is alignment score, usually saved in the optional column of the SAM/BAM file.
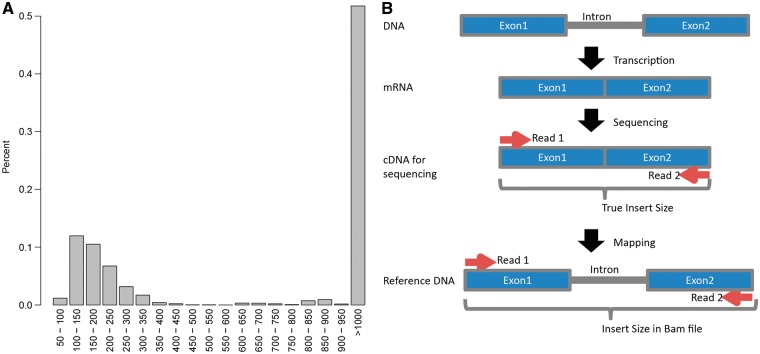
Insert size is a useful QC parameter that is often overlooked for pair-end sequencing data. Insert size is the length of the RNA fragments being sequenced. In the SAM/BAM format, the 9th column records the insert size. The distribution of insert size should follow a nonnormal distribution with a peak equal to the targeted size and a long right tail (Figure 5A). The long right tail is the result from longer RNA fragments that slipped through size selection, structural variants or alignment error. Small insert size can cause overrepresentation of the middle segment of the RNA fragments (sequenced twice by the two reads in one single pair). The insert size calculation in RNA-seq data is slightly inaccurate in comparison with DNA-seq data. During the transcription from DNA to RNA, introns are spliced out, and exons are ligated together to form mRNA. The cDNA library constructed from mRNA does not contain introns either. Thus, the intended insert size does not consider introns. After alignment, the insert size recorded in the SAM/BAM file includes introns, making it artificially longer than the intended insert size (Figure 5B). Summary statistics of the insert size also should be computed using the median rather than the mean, as the mean is easily swayed by any large outliers, causing mean insert size to be larger than the targeted insert size. The median insert size is usually more in line with the targeted insert size, in our experience.
Figure 5.
(A) Expected insert size distribution from RNA-seq data. A peak should be observed between 100 and 200 nucleotides, as it is the targeted fragment size in most RNA-seq kits. The high peak for insert size >1000 nucleotide is caused by improperly mapped pairs. These are usually errors caused by sequencing or alignment, or indicate structural variations. (B) A graphical explanation of the reason of inaccurate insert size estimation in the SAM/BAM file. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Nonuniform coverage of transcripts is a prevalent issue in poly(A)-selected RNA-seq [51, 52]. Because the poly(A) tail only occurs at the 3′ end of the mRNA, the 3′end of the RNA is usually overrepresented in poly(A)-based RNA-seq. Bias at the 5′ end of RNA can also happen because of various factors, such as the fragmentation method (the 5′ end of RNA is more stable), reverse transcription from RNA to cDNA and strand-oriented library construction protocol. GC content plays an important role in Illumina sequencing coverage [53, 54]. It has been observed that either high or low GC content will result in lower depth coverage [55]. The GC bias in RNA-seq has been thoroughly documented [56–58]. The 3′/5′ and GC bias can be computed and visualized in tools such as RNA-SeQC [37] and RSeQC [59]. Fortunately, various methods [51, 56, 57, 60, 61] have been developed to correct these biases, and a correction method should be applied if bias was observed.
Currently, the majority of the long RNA library construction is strand oriented, meaning that when performing sequencing, investigators should only obtain the reads from the strand from which the RNA was originally transcribed. However, the efficiency of the process to remove unwanted strands is not perfect, allowing the sequencing of a small percentage of anti-sense RNA. If a non-strand-specific protocol is used, then the reads representing sense and anti-sense RNA should be roughly 50–50. RNA-SeQC [37] has a built-in functionality to access the performance of strand-specific library construction. However, the efficiency of strand-specific library construction is not crucial for standard RNA-seq gene expression analysis, but can aid in identifying the originating strand for detected novel RNAs.
Batch effects can also be detected in alignment QC. Applying a similar method as the batch effect detection in raw data QC, we can detect if alignment rate and capture efficiency are associated with machine, run, lane, flow cell, etc. The tool QC3 [13] is equipped with the functionality for batch effect detection using SAM/BAM files. However, the limitation discussed earlier also applies when a sample/pool is sequenced on multiple lanes.
Expression QC
After quantifying gene expression using tools such as HTSeq [34] and Cufflinks [33], additional QC can be performed to further identify outlier samples. The overall goal for the majority of RNA-seq studies is to compare gene expression from two or more groups. The underlying assumption is that gene expression patterns should be relatively similar for samples within the same phenotype group. However, a sample from one group is often found to be more similar to the sample of another group based on gene expression. Clustering can be used to identify which samples are closely related based on gene expression. Tools such as Cluster 3.0 [62], Heatmap3 [63] and MultiRankSeq [64] can be used to perform cluster analysis. A heat map is a graphical representation of the data where expression values in a matrix are presented as colors. Heat mapping is often coupled with clustering analysis to present the gene expression patterns by cluster. Misleading clustering analysis often involves constructing the cluster based on genes that are differentially expressed. Such an approach is biased and will always produce clusters that can neatly separate the groups. Using clustering as a QC measure will require an unbiased and unsupervised method. Normalization of the count data is also an essential issue for cluster analysis. Reads per kilobase per million (RPKM) normalization may be affected by some extremely highly expressed genes. Variance stabilization offered in DESeq2 [65] is a good way to minimize the effect of unstable variance.
Observing samples clustered outside the intended phenotype group does not make the samples outliers. Instead, an attempt to improve the clustering should be made. A common technique that can be used to improve the clustering is to only use genes with a high variation instead of all of the genes because genes that lack variation among samples do not contribute to the clustering. In the scenario where outliers are detected even when using genes with the largest variation, there are four possible explanations: (1) the gene expression pattern of these outlier samples do not represent their phenotype; (2) the phenotype of these samples are not strong, or misclassified; (3) the samples are mislabeled; and (4) there is cross-contamination of samples. The decision of whether to remove or re-sequence the outlier samples is based on how strongly one believes the gene expression pattern represents the phenotype. If cross-contamination is suspected, one can validate it by performing a heterozygous genotype consistency analysis. To perform this analysis, we first need to perform genotype calls using the RNA-seq data and compute their pair-wise heterozygous genotype consistency. Consistency is similar to the measure of identity by descent in genetic studies, which is often used to evaluate the relatedness of subjects. For two independent subjects, the expected consistency is <30%; for two genetically related subjects, the expected consistency is >80%. High consistency rates between independent samples are a sign of cross-contamination. The tool QC3 [13] provides heterozygous genotype consistency analysis.
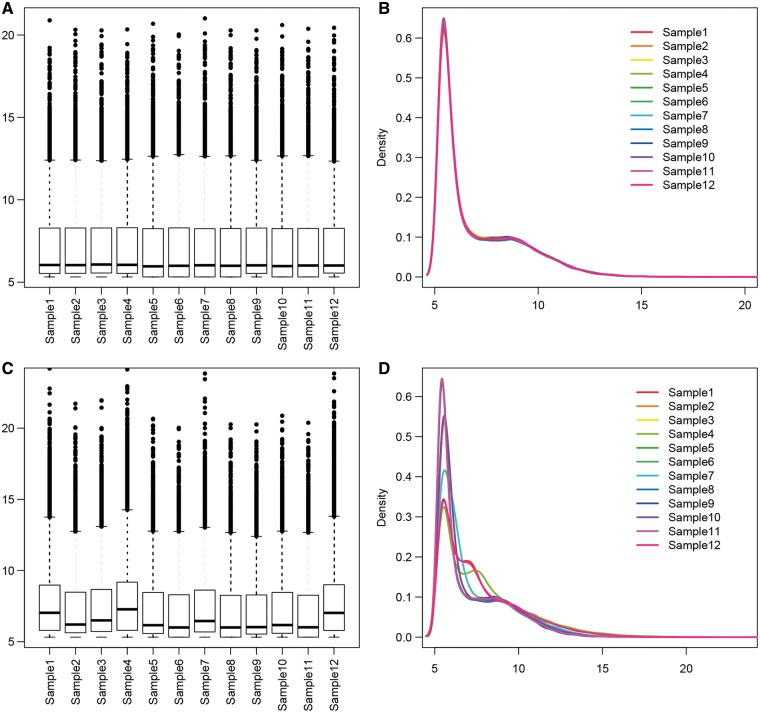
Many gene expression studies are based on loss of function (e.g. mouse knockouts) or gain of function (e.g. gene overexpression). It is crucial that one examines if the loss of function or gain of function was successful for the gene of interest. Of note, it is easier to achieve greater effect for overexpression than deletion. Thus, overexpression designs tend to produce larger fold changes than that of gene deletions. In humans, there are approximately 20 000 coding genes that have been identified, but only half are likely expressed at a given time within a sample [66, 67]. The number of expressed genes per sample is highly dependent on two criteria: (1) the total number of reads sequenced and mapped to genes; and (2) the threshold used for detection. It is assumed that the number of expressed genes should be roughly equal for all samples within the study within a phenotype category group (i.e. controls). The number of genes expressed and the magnitude of expression can be visualized in either box plots or density plots (Figure 6). Outlier samples may cause inaccurate differential expression analysis.
Figure 6.
(A and B) Examples of expected read count distribution after variance stabilization normalization, where all samples share similar distributions. (C and D) Examples of uneven read count distribution after normalization. This might indicate batch effect or outlier samples. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
To date, there has not been a consensus on what RPKM threshold to use for gene detection. Ad hoc filters have been applied in many studies. For example, the RPKM thresholds that have been used include 0.1 [68], 0.125 [69], 0.3 [70], 1.0 [71], etc. We recommend a minimum of a 1.0 RPKM detection threshold because smaller detection thresholds tend to generate large and misleading fold changes during differential expression analysis. Similarly, arbitrary thresholds have been used for read count data [72, 73]. Instead of using an ad hoc approach, a data-driven detection threshold [74] may be more appropriate. An alternative to discarding the low gene expression data is to compute the fold change based on an empirical Bayes shrinkage, which is implemented in DESeq2 [65].
In microarray data, one major source of batch effect is the fluorescence intensity level. In RNA-seq, fluorescence intensity is not relevant, but a unique characteristic of high-throughput sequencing can cause other kinds of bias: gene detection bias caused by an uneven read count among samples [75]. In microarrays, the hybridization of all probes happens simultaneously without bias toward any particular gene. In high-throughput sequencing, each RNA fragment in the sequencing library has equal probability of being sequenced. Thus, genes with high expression (more fragments presented in the library) are more likely to be sequenced, and genes with low expression (less fragments in the library) are less likely to be sequenced. Furthermore, when multiple samples are pooled together, an equal amount of material from each sample is added to the pool. Ideally, one hopes that each sample is represented equally by the total sequenced reads. However, in practice, the number of reads sequenced can differ by several folds between samples pooled together [76]. Many factors can contribute to the unevenness of read distribution, e.g. sample quality and human error [77]. The number of detectable genes is correlated with the number of reads sequenced before reaching saturation. Saturation is the point at which no more additional genes can be detected by sequencing more reads. Large differences in the number of reads sequenced between two groups can cause unreliable differential expression analysis on low-expressed genes. Samples with more sequenced reads will detect additional low-expressed genes (low RPKM or read count), and the group with less sequenced reads will not detect these low-expressed genes (0 RPKM or read count), causing the illusion of large or infinite fold changes. This is also one of the reasons to recommend a large threshold on RPKM for gene expression detection. Furthermore, packages such as PEER [78], ComBat [79] and svaseq [80] have been developed to correct for RNA-seq batch effect.
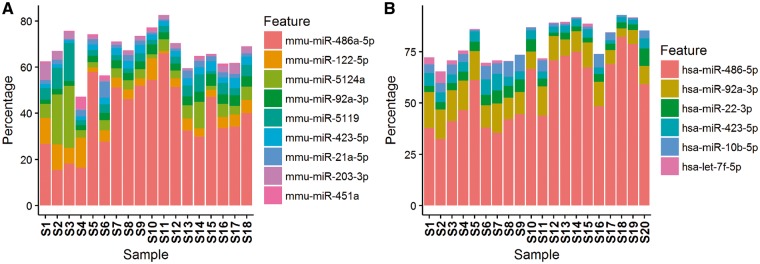
A unique quality issue associated with sRNA-seq is the overrepresentation of particular miRNAs—a phenomenon where certain miRNAs can account for up to 80% of the total miRNA reads. Currently, there are 35 828 annotated miRNAs in the miRBase [81]. The number of detectable miRNAs can range from 100 to 500 depending on sample source, sample quality and detection threshold [14, 15]. There are variations among miRNA expression; however, when a single miRNA occupies >20% of all miRNA reads, there is likely a selection bias caused by either the sRNA extraction kit or the library contraction kit. For example, miR-486-5p is often detected with high percentage in both human and mouse samples. A visual presentation that accounts for miRNA representation percentage can help to identify overexpressed miRNAs (Figure 7), and overrepresented miRNAs may be removed from further analysis.
Figure 7.
Top miRNAs detected in each sample. Left: human small RNA data set with high detected abundance of hsa-miR-486-5p. Right: mouse small RNA data set with high detected abundance of mmu-miR-486a-5p. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Discussion and conclusion
During our description of QC, many parameters can be computed either by the mean or the median. The median is in general a more robust measurement because of its immunity to outliers. However, for large data sets, the median will take a substantially longer time to compute because of the necessary sorting step. For example, we performed an experiment sorting an array of 50 million randomly generated numbers repeatedly for 1000 iterations. The median time for computing the mean was 0.5 s, and the median time for computing the median was 21.3 s. Thus, computing the median took 42.6 times longer than the mean. The longer time will be substantially amplified when accounting for many parameters from multiple samples. However, when computational time is not an issue, median values should be computed instead of mean values for more robust and accurate QC.
In this overview of RNA-seq QC, we advocate a multi-perspective QC strategy. The perspectives include RNA quality, raw data, alignment and gene expression. Each layer of proposed QC provides additional perspective into the true quality of the data and sample. Analysis for RNA-seq data is complex and involves multiple steps. Limited QC often fails to grasp the full picture of data quality, allowing problematic samples to convolute analysis results. Responsible analysts should always perform QC from multiple angles, ensuring the delivery of correct and reproducible results.
Key Points
QC is a required step in RNA-seq data analysis.
QC for RNA-seq data should be applied from multiple perspectives.
Different types of RNA-seq data have their unique challenges in QC.
Funding
This study was supported by NIH (P30 CA68485). This work was also supported by awards from the National Institutes of Health, National Heart, Lung and Blood Institute to K.C.V. (HL128996, HL113039 and HL116263). This work was also supported by an award from the American Heart Association to K.C.V. (CSA2066001). We would also like to thank Stephanie Page Hoskins for editorial support.
Biographies
Quanhu Sheng is a research assistant professor at the Department of Cancer Biology at Vanderbilt University. His research has been focused on small RNA sequencing and interpretation.
Kasey Vickers is an assistant professor at the Department of Cardiovascular Medicine, Vanderbilt University. He is an expert in small RNA sequencing data analysis.
Shilin Zhao is a research fellow at the Department of Cancer Biology, Vanderbilt University. His research has been focused on bioinformatics research.
Jing Wang is a research fellow at Department of Cancer Biology, Vanderbilt University. Her research is focused on enhancer RNA analysis.
David C. Samuels is an associate professor in the Department of Molecular Physics and Biology, Vanderbilt University. His research has been focused on mitochondria-related topics.
Olivia Koues is the manager of Next-Generation Sequencing, VANTAGE, a core faculty of Vanderbilt University. Her expertise lies in the technical aspect of high-throughput sequencing.
Yu Shyr is the Director of Center for Quantitative Sciences, and professor of Biostatistics, Vanderbilt University. His research focuses on biostatistics and bioinformatics analysis of big data.
Yan Guo is an assistant professor at the Department of Cancer Biology at Vanderbilt University. His research is focused on cancer bioinformatics. He has been serving as the Technical Director of Bioinformatics for Vanderbilt Technologies for Advanced Genomics Analysis and Research Design (VANGARD) since 2012.
References
- 1. Wang Z, Gerstein M, Snyder M.. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marioni JC, Mason CE, Mane SM, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 2008;18:1509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asmann YW, Klee EW, Thompson EA, et al. 3' tag digital gene expression profiling of human brain and universal reference RNA using Illumina genome analyzer. BMC Genomics 2009;10:531.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cloonan N, Forrest AR, Kolle G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 2008;5:613–9. [DOI] [PubMed] [Google Scholar]
- 5. Guo Y, Sheng Q, Li J, et al. Large scale comparison of gene expression levels by microarrays and RNAseq using TCGA data. PLoS One 2013;8:e71462.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han L, Vickers KC, Samuels DC, et al. Alternative applications for distinct RNA sequencing strategies. Brief Bioinform 2015;16:629–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. BabrahamBioinformatics. FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 8. BabrahamBioinformatics. FastQ Screen. http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/.
- 9. HannonLab. FASTX Toolkit. http://hannonlab.cshl.edu/fastx_toolkit/
- 10. Patel RK, Jain M.. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 2012;7:e30619.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmieder R, Edwards R.. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011;27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou Q, Su X, Wang A, et al. QC-chain: fast and holistic quality control method for next-generation sequencing data. PLoS One 2013;8:e60234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo Y, Zhao S, Sheng Q, et al. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics 2014;103:323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo Y, Bosompem A, Mohan S, et al. Transfer RNA detection by small RNA deep sequencing and disease association with myelodysplastic syndromes. BMC Genomics 2015;16:727.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo Y, Xiong Y, Sheng Q , et al. A micro-RNA expression signature for human NAFLD progression, J Gastroenterol 2016, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vickers KC, Roteta LA, Hucheson-Dilks H, et al. Mining diverse small RNA species in the deep transcriptome. Trends Biochem Sci 2015;40:4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 2006;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. von Ahlfen S, Missel A, Bendrat K, et al. Determinants of RNA quality from FFPE samples. PLoS One 2007;2:e1261.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cock PJ, Fields CJ, Goto N, et al. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 2010;38:1767–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ewing B, Hillier L, Wendl MC, et al. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998;8:175–85. [DOI] [PubMed] [Google Scholar]
- 21. Consortium TE, Standards, Guidelines and Best Practices for RNA-Seq. https://genome.ucsc.edu/ENCODE/protocols/dataStandards/ENCODE_RNAseq_Standards_V1.0.pdf.
- 22. Guo Y, Long J, He J, et al. Exome sequencing generates high quality data in non-target regions. BMC Genomics 2012;13:194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Q, Guo Y, Li J, et al. Steps to ensure accuracy in genotype and SNP calling from Illumina sequencing data. BMC Genomics 2012;13(Suppl 8):S8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010;11:733–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lauss M, Visne I, Kriegner A, et al. Monitoring of technical variation in quantitative high-throughput datasets. Cancer Inform 2013;12:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia-Silva MR, Cabrera-Cabrera F, Guida MC, et al. Hints of tRNA-derived small RNAs role in RNA silencing mechanisms. Genes (Basel) 2012;3:603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 2009;583:437–42. [DOI] [PubMed] [Google Scholar]
- 31. Cole C, Sobala A, Lu C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009;15:2147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harada F, Dahlberg JE.. Specific cleavage of tRNA by nuclease S1. Nucleic Acids Res 1975;2:865–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anders S, Pyl PT, Huber W.. HTSeq-a python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samuels DC, Han L, Li J, et al. Finding the lost treasures in exome sequencing data. Trends Genet 2013;29:593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. BroadInstitute. Picard. http://broadinstitute.github.io/picard/.
- 37. DeLuca DS, Levin JZ, Sivachenko A, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012;28:1530–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. NIH. Human Microbiome Project. http://hmpdacc.org/reference_genomes/reference_genomes.php.
- 40. Palacios G, Druce J, Du L, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 2008;358:991–8. [DOI] [PubMed] [Google Scholar]
- 41. Nakamura S, Yang CS, Sakon N, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One 2009;4:e4219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quan PL, Wagner TA, Briese T, et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 2010;16:918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Briese T, Paweska JT, McMullan LK, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog 2009;5:e1000455.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Isakov O, Modai S, Shomron N.. Pathogen detection using short-RNA deep sequencing subtraction and assembly. Bioinformatics 2011;27:2027–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khoury JD, Tannir NM, Williams MD, et al. Landscape of DNA virus associations across human malignant cancers: analysis of 3,775 cases using RNA-Seq. J Virol 2013;87:8916–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang K, Li H, Yuan Y, et al. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One 2012;7:e51009.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 2012;22:107–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo Y, Li J, Li CI, et al. MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis. Bioinformatics 2013;29:1210–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roberts A, Trapnell C, Donaghey J, et al. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 2011;12:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zheng W, Chung LM, Zhao HY.. Bias detection and correction in RNA-Sequencing data. BMC Bioinformatics 2011;12:290.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008;456:53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dohm JC, Lottaz C, Borodina T, et al. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res 2008;36:e105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo Y, Zhao S, Lehmann BD, et al. Detection of internal exon deletion with exon Del. Bmc Bioinformatics 2014;15:332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Risso D, Schwartz K, Sherlock G, et al. GC-content normalization for RNA-Seq data. BMC Bioinformatics 2011;12:480.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Benjamini Y, Speed TP.. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res 2012;40:e72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lahens NF, Kavakli IH, Zhang R, et al. IVT-seq reveals extreme bias in RNA sequencing. Genome Biol 2014;15:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang L, Wang S, Li W.. RSeQC: quality control of RNA-seq experiments. Bioinformatics 2012;28:2184–5. [DOI] [PubMed] [Google Scholar]
- 60. Li S, Labaj PP, Zumbo P, et al. Detecting and correcting systematic variation in large-scale RNA sequencing data. Nat Biotechnol 2014;32:888–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jones DC, Ruzzo WL, Peng XX, et al. A new approach to bias correction in RNA-Seq. Bioinformatics 2012;28:921–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao SL, Guo Y, Sheng QH, et al. Advanced heat map and clustering analysis using heatmap3. Biomed Res Int 2014;2014:986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Guo Y, Zhao S, Ye F, et al. MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. Biomed Res Int 2014;2014:248090.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ramskold D,, Wang ET,, Burge CB, et al. An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data, Plos Comput Biol 2009;5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Strausberg RL, Buetow KH, Greenhut SF, et al. The cancer genome anatomy project: online resources to reveal the molecular signatures of cancer. Cancer Invest 2002;20:1038–50. [DOI] [PubMed] [Google Scholar]
- 68. Lundberg E, Fagerberg L, Klevebring D, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol 2010;6:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hackett NR, Butler MW, Shaykhiev R, et al. RNA-Seq quantification of the human small airway epithelium transcriptome. BMC Genomics 2012;13:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Canovas A, Rincon G, Islas-Trejo A, et al. SNP discovery in the bovine milk transcriptome using RNA-Seq technology. Mamm Genome 2010;21:592–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mortazavi A, Williams BA, Mccue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5:621–8. [DOI] [PubMed] [Google Scholar]
- 72. Guo Y, Zhao SL, Sheng QH, et al. RNAseq by total RNA library identifies additional RNAs compared to Poly(A) RNA library. Biomed Res Int 2015;2015:862130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bottomly D, Walter NAR, Hunter JE, et al. Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS One 2011;6:e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sultan M, Schulz MH, Richard H, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 2008;321:956–60. [DOI] [PubMed] [Google Scholar]
- 75. Guo Y, Zhao S, Su PF, et al. Statistical strategies for microRNAseq batch effect reduction. Transl Cancer Res 2014;3:260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guo Y, Cai Q, Li C, et al. An evaluation of allele frequency estimation accuracy using pooled sequencing data. Int J Comput Biol Drug Des 2013;6:279–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guo Y, Samuels DC, Li J, et al. Evaluation of allele frequency estimation using pooled sequencing data simulation. Scientific World Journal 2013;2013:895496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stegle O, Parts L, Piipari M, et al. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 2012;7:500–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Johnson WE, Li C, Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 80. Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res 2014;42:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kozomara A, Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]