Abstract
Summary
The Illumina Infinium HumanMethylationEPIC BeadChip is the new platform for high-throughput DNA methylation analysis, effectively doubling the coverage compared to the older 450 K array. Here we present a significantly updated and improved version of the Bioconductor package ChAMP, which can be used to analyze EPIC and 450k data. Many enhanced functionalities have been added, including correction for cell-type heterogeneity, network analysis and a series of interactive graphical user interfaces.
Availability and implementation
ChAMP is a BioC package available from https://bioconductor.org/packages/release/bioc/html/ChAMP.html.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
DNA methylation is the most studied epigenetic modification. Illumina’s new EPIC BeadChip can measure methylation at over 850 000 sites with single-nucleotide resolution. The EPIC BeadChip includes over 90% of probes present on the 450 K array, shows high reproducibility, and will become a common tool for epigenome-wide association studies (Moran et al., 2016).
ChAMP is an integrated analysis pipeline published in 2014 (Morris et al., 2014), which includes functions for filtering low-quality probes, adjustment for Infinium I and Infinium II probe design, batch effect correction, detecting differentially methylated positions (DMPs), finding differentially methylated regions (DMRs) and detection of copy number aberrations (CNA).
The new version of ChAMP, extends and improves this analysis pipeline, adding novel and enhanced functionalities, including detection of differentially methylated genomic blocks (DMB), gene set enrichment analysis (GSEA), a method for correcting cell-type heterogeneity and detection of differentially methylated gene modules. Notably, the new package provides a series of web-based graphical user interfaces (GUIs), which facilitate analyses and enhance user-experience.
2 Description
ChAMP is an R package and currently requires R(≥3.4). ChAMP loads data from IDAT files using it’s novel loading function, or though minfi loading function (Aryee et al., 2014). Probes can be filtered based on detection P-values, chromosomal location, presence of single nucleotide polymorphisms in the probe sequence (Zhou et al., 2016) and cross-hybridization. Multi-dimensional scaling, density and clustering plots allow exploratory analysis. For normalization, functional normalization (Fortin et al., 2014) has been added as an option alongside beta-mixture quantile normalization (Teschendorff et al., 2013). Singular value decomposition is used to correlate principal components to biological and technical factors, helping the user decide if there are batch effects or confounding factors that need to be adjusted for.
For supervised analysis, besides limma-based DMP and ProbeLasso-based DMR analysis functions (Butcher and Beck, 2015), there is now added functionality for DMR detection using Bumphunter (Jaffe et al., 2012) and DMRcate (Peters et al., 2015). Large-scale differentially methylated blocks (DMB) can also be identified. These DMBs are large-scale genomic regions (10 kb–Mb) containing hundreds of inter-genic CpG sites (Fig. 1B), and which often exhibit hypomethylation in aging and cancer (Yuan et al., 2015). We also added functionality to allow users to detect differentially methylated hotspots in user-defined gene networks (Jiao et al., 2014). In addition, ChAMP incorporates GSEA capability on DMP and DMR results (Young et al., 2010).
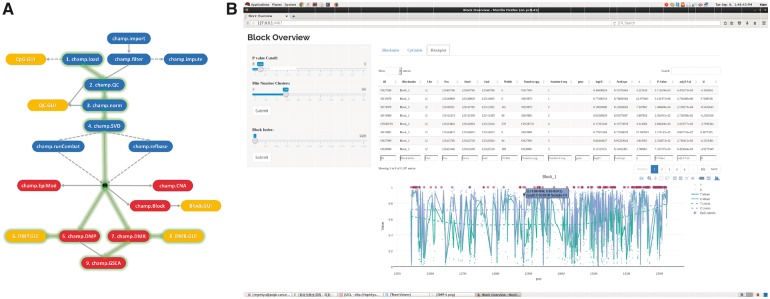
Fig. 1.
The ChAMP pipeline. (A) All functions included in ChAMP. Blue functions used for data preparation. Red functions used to generate analysis results. Yellow functions are GUI functions for visualization. Functions and edges with light green gleam stands for main pipeline (markers are steps for using ChAMP). Dash lines mean functions may not necessarily required. (B) GUI function for visualization of a DMB. The left panel displays parameters for controlling the plot and the table
In ChAMP, correction for cell-type heterogeneity in blood can be performed with the reference-based RefbaseEWAS (Houseman et al., 2012). Another unique feature of ChAMP is a function for detecting CNA (Feber et al., 2014). As a result of all these functionalities, ChAMP is now a much more powerful and comprehensive tool for DNA methylation analysis (Fig. 1A).
Besides making all above functions applicable to EPIC BeadChips, there are two other technical improvements which will benefit users. First, ChAMP accepts multiple data input formats, including IDATS, beta-valued matrices and phenotype data files. Second, a series of javascript-based GUIs are provided. This allows easy checking of results, and generating figures for DMR or DMBs. Shiny, a web application framework for R, suitable for creating simple interactive webpages, and Plotly, an open source JavaScript graphing library, are integrated with ChAMP results, allowing users to view, select, and zoom in and out from results obtained with ChAMP. All GUIs use the results of ChAMP functions as parameters (Fig. 1B).
Full details and an example workflow of ChAMP are provided (Supplementary Material).
3 Conclusion
In summary, ChAMP provides a much improved, powerful and comprehensive pipeline for Illumina HumanMethylation BeadChip analysis.
Funding
Royal Society and Chinese Academy of Sciences (Newton Advanced Fellowship 164914) [to A.E.T.]; Chinese Scholarship Council (CSC) [to Y.T.]; MRC [MR/M025411/1 to A.F.] and the UCLH/UCL Comprehensive Biomedical Research Centre [to A.F.]; and National Institute for Health Research (NIHR) Blood & Transplant Research Unit (BTRU) [NIHR-BTRU-2014-10074 to A.P.W. and S.B.].
Conflict of Interest: none declared.
Supplementary Material
References
- Aryee M.J. et al. (2014) Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher L.M., Beck S. (2015) Probe Lasso: a novel method to rope in differentially methylated regions with 450K DNA methylation data. Methods, 72, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feber A. et al. (2014) Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol, 15, R30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.P. et al. (2014) Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol, 15, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E.A. et al. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.E. et al. (2012) Significance analysis and statistical dissection of variably methylated regions. Biostatistics, 13, 166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y. et al. (2014) A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics, 30, 2360–2366. [DOI] [PubMed] [Google Scholar]
- Moran S. et al. (2016) Validation of a DNA methylation microarray for 850, 000 cpg sites of the human genome enriched in enhancer sequences. Epigenomics, 8, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris T.J. et al. (2014) Champ: 450k chip analysis methylation pipeline. Bioinformatics, 30, 428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T.J. et al. (2015) De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin, 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A.E. et al. (2013) A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics, 29, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D. et al. (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol, 11, R14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T. et al. (2015) An integrative multi-scale analysis of the dynamic DNA methylation landscape in aging. PLoS Genet, 11, e1004996.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. et al. (2016) Comprehensive characterization, annotation and innovative use of infinium DNA methylation Beadchip probes. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.