Abstract
Motivation
One of the main challenges with bioinformatics software is that the size and complexity of datasets necessitate trading speed for accuracy, or completeness. To combat this problem of computational complexity, a plethora of heuristic algorithms have arisen that report a ‘good enough’ solution to biological questions. However, in instances such as Simple Sequence Repeats (SSRs), a ‘good enough’ solution may not accurately portray results in population genetics, phylogenetics and forensics, which require accurate SSRs to calculate intra- and inter-species interactions.
Results
We present Kmer-SSR, which finds all SSRs faster than most heuristic SSR identification algorithms in a parallelized, easy-to-use manner. The exhaustive Kmer-SSR option has 100% precision and 100% recall and accurately identifies every SSR of any specified length. To identify more biologically pertinent SSRs, we also developed several filters that allow users to easily view a subset of SSRs based on user input. Kmer-SSR, coupled with the filter options, accurately and intuitively identifies SSRs quickly and in a more user-friendly manner than any other SSR identification algorithm.
Availability and implementation
The source code is freely available on GitHub at https://github.com/ridgelab/Kmer-SSR.
1 Introduction
Simple sequence repeats (SSRs) are short repetitive regions of DNA where at least one base is tandemly repeated many times due to slipped-strand mispairing and errors occurring in DNA replication, repair, or recombination (Levinson and Gutman, 1987). For decades, SSRs have been studied to determine phenotypic differences caused by increased copy numbers of short repetitive sequences (Kashi and King, 2006). Moreover, SSRs account for quantitative genetic variation and phenotypic differences without lowering species fitness (Kashi et al., 1997). SSR concentration varies not only between different species, but also between different chromosomes within the same species, and cannot be explained by assessing the nucleotide composition of sequences (Katti et al., 2001). Because SSRs reveal characteristic functions of DNA replication, recombination and repair, they are important in studying biological systems interactions, as well as studying repeat expansion-based diseases with next-generation sequencing data (Kashi and King, 2006).
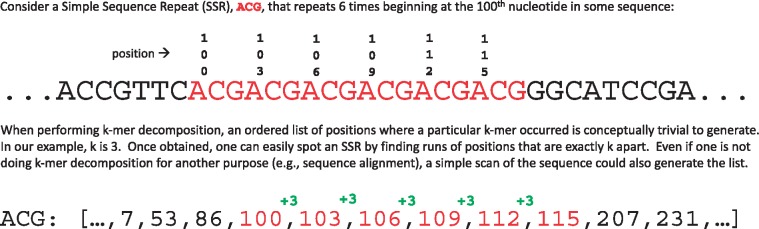
Many different approaches have been used to identify SSRs. Here, we propose the use of k-mers. The term k-mer refers to a subsequence of length ‘k’ derived from a given sequence, while k-mer decomposition refers to all possible substrings of length ‘k’ that can be made from a sequence. Uses for k-mer decomposition have previously been outlined in instances such as genome assembly and machine learning (Chikhi and Medvedev, 2014; Ghandi et al., 2014). Although k-mers have been used to identify similar subsequences as in (Han et al., 2007), to our knowledge SSR identification has never been attempted through k-mer decomposition.
2 Materials and methods
2.1 Overview
Kmer-SSR utilizes k-mer decomposition to provide an exhaustive or filtered approach to finding all SSRs in a given sequence (Figs 1 and 2). Our version of k-mer decomposition works by identifying all subsequences of length ‘k’ while tracking the start position of each k-mer. K-mer lengths are defined by the user as the SSR period length. Kmer-SSR minimizes the usage of random access memory (RAM) by performing k-mer decomposition and only storing k-mers that are the same as the preceding k-mer (SSR period length). If a k-mer is not identical to a k-mer found k bases previously, the previously identified k-mers will be discarded and k-mer decomposition will occur for the rest of the sequence.
Fig. 1.
Conceptual representation of Kmer-SSR. Although we implement some filters and tricks to speed up Kmer-SSR runtime, each SSR is identified through kmer decomposition, which allows the identification of instances when the same SSR period occurs k bases from the previously identified SSR period
2.2 Memory requirements
We used the following techniques to limit memory requirements:
Identify SSRs from left to right: Kmer-SSR checks each position starting at the leftmost position of the sequence for each SSR period size (i.e. k-mer length) given by the user. This method allowed us to store only a single potential SSR and immediately either discard it if it was not repeated or write it to a file if it was a valid SSR.
Identify SSRs with the largest period size first: Since Kmer-SSR does not store previously identified SSRs in memory, it is necessary to search for SSRs in a specific order, or else risk reporting SSRs fully enclosed within larger SSRs. To avoid this issue, we take the period sizes given by the user and search for SSRs from the longest period size to the smallest (e.g. if the user wants to search for 2-mers and 7-mers, we search for all 7-mer SSRs in the sequences before we search for 2-mer SSRs). When an SSR is discovered, an atomicity check is conducted to determine if the k-mer can be broken down to a smaller subsequence. An SSR is considered atomic if no smaller SSRs exist inside the first period. For example, ATATATAT would be identified as a 4-mer (ATAT) repeated twice, but ATAT is not atomic because AT (repeated twice) occurs within the first period. Thus, it is ignored because it is an invalid 4-mer and, if the user requested searching for 2-mers, it would be discovered again as a 2-mer (AT) repeated four times. If the atomicity check fails, the SSR is not reported. When an atomic (i.e. valid) SSR is discovered, the iterator moves just past the SSR, minus the current period size being searched, to ensure that overlapping SSRs are identified. For example, ACAACAACACACACAC has ACA repeated three times starting at position 0. Additionally, AC repeats five times starting at position 6. After finding the ACA repeat, we would miss the full AC repeat if we skipped to the end of the ACA repeat and resumed searching from there. Only by backtracking as described above (9–3 = 6), do we find the full AC repeat. Note that each of the nucleotides between positions 0 and 5 need not be searched for SSRs because Kmer-SSR has already found SSRs with larger period sizes than the current period size. In other words, since Kmer-SSR has already found SSRs with larger period sizes, the maximum possible overlap with the current SSR (ACA) and an adjacent following SSR is k (which is three in this example), removing the need to search for SSRs from the start of a valid SSR to k bases from the end of that SSR.
Create a Boolean filter array: To ensure that SSRs are unique and do not end in the same positions, we created a Boolean filter array of the same length as the sequence being analyzed, which is initiated to false. In C ++, the implementation of this array only requires one bit per position, so the memory requirement is nominal. When an SSR is discovered, we first ensure that at least one position in the first or last SSR period size on either end of the SSR is false in the Boolean array. If one position is false, we assign all values within the array that correspond to all positions in the SSR to true. The filter allows us to ignore completely overlapping SSRs because overlapping SSRs will be set to ‘true’ at the positions at the ends of the SSR.
By utilizing the above-mentioned methods, we were able to limit the amount of RAM needed to O(n), where n is the sequence length, and the constant value is slightly more than one byte (one byte to store each sequence base and one bit allocated in the Boolean filter for each base).
2.3 SSR filters
Next, we implemented a comprehensive filter that allows users to control the output of Kmer-SSR based on atomicity, cyclic duplicates, enclosed SSRs, minimum SSR length and specific SSR period sizes. Pseudocode for Kmer-SSR is in Figure 2. The following are different filters that are optionally applied to the output of Kmer-SSR:
Atomicity check: The atomicity check ensures that the smallest period size for each SSR is reported. For instance, if an ATAT repeats four times, it would be reported as an AT repeated eight times because AT is the smallest period size within ATAT.
Cyclic duplicates: Many SSRs create equally viable SSRs with slightly different positions reported. For instance, in the sequence ATATATATATATATATA, it is arguably equally valid to report the AT repeated eight times starting at position zero as it would be to report TA repeating eight times starting at position one. To avoid duplicate reporting of cyclic duplicates and ensure the longest SSR is always reported, we choose and report only the leftmost SSR. So, in this instance, only the AT repeated eight times would be reported.
Enclosed SSRs: Occasionally, SSRs might be completely enclosed within other SSRs. For example, in the sequence TAAAATTAAAATTAAAAT, the SSR TAAAAT is repeated three times, but within each TAAAAT there is an A that repeats four times. In this case, we only report the longest SSR, TAAAAT, repeated three times.
SSR length: We allow the user to input minimum and maximum SSR lengths via command line options. By default, SSRs are only reported if they are at least 16 nucleotides long.
Set specific period sizes: We allow the user to input specific period sizes to be checked (e.g. 1, 3, 5 would look for SSRs with period sizes of one, three and five), or ranges of period sizes (e.g. 1–7 would look for SSRs with period sizes one through seven). By default, Kmer-SSR reports SSRs of period sizes one through seven. SSRs outside of the user specified range are not reported.
Number of repeats: We allow the user to input minimum and maximum numbers of repeats via command line options. By default, SSRs must repeat at least twice to be reported.
Enumerated SSRs: If the user is interested in a very limited set of SSRs, they may specify those via a command line option and no other SSRs will be reported.
Sequence length: The user may specify minimum and maximum bounds on the length of an input sequence, outside of which the program will not search or report SSRs. By default, if a sequence is less than 100 bases or more than 500 megabases, it will be ignored.
Fig. 2.
Pseudocode for the Kmer-SSR algorithm. The function passesBooleanFilter ensures SSRs are not duplicates of previously reported SSRs. The function passesUserFilters (function not shown) completes other user-specified options, which may include: minimum SSR length, minimum and maximum number of periods, finding specific SSRs and sequence length bounds
3 Results
We conducted pairwise comparisons of Kmer-SSR against the following SSR identification algorithms: GMATo (Wang et al., 2013), MREPS (Kolpakov et al., 2003), PRoGeRF (Lopes et al., 2015), QDD (Meglécz et al., 2014), SA-SSR (Pickett et al., 2016), SSR-Pipeline (Miller et al., 2013), SSRIT (Temnykh et al., 2001) and TRF (Benson, 1999). These comparisons were performed on DNA sequences from six different species (whole genome assembly unless otherwise noted): Anolis carolinensis chromosome 6 (CM000942.1), Chlamydomonas reinhardtii (assembly v5.5) (Merchant et al., 2007), Danio rerio chromosome 25 (CM002909.1), Dictyostelium doscoideum (GCA_0000044695.1), Physcomitrella patens chromosome 1 (assembly v3.3) and Saccharomyces cerevisiae (GCA_001634645.1). Table 1 displays the computational time of each algorithm and the number of SSRs correctly identified for each dataset (CPU Time and Real Time columns).
Table 1.
Comparisons of all nine SSR-identification algorithms across six genomes with period sizes of 1–7 and a minimum SSR length of 16 bases
| Comparison with Kmer-SSR |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPU Time (mm:ss) | Real Time (mm:ss) | SSRs Reported | aSSRs After Adjustments | bSSRs In Range | cNumber Correct | dNumber Correct & Fixed | ePercent Correct & Fixed | SSRs Unique to Software | SSRs Unique to Kmer-SSR | SSRs Shared | ||
| Anolis carolinensis (chr 6) | GMATo | 2:38 | 2:38 | 20 623 008 | 16 369 297 | 16 871 | 16 871 | 16 870 | 100 | 0 | 8194 | 10 090 |
| Kmer-SSR | 2:24 | 0:24 | 18 284 | 18 284 | 18 284 | 18 284 | 18 284 | 100 | NA | NA | NA | |
| MREPS | 0:09 | 0:09 | 25 639 | 25 639 | 18 284 | 18 284 | 18 284 | 100 | 0 | 0 | 18 284 | |
| PRoGeRF | 18:07 | 18:07 | 16 841 656 | 16 840 821 | 17 763 | 17 762 | 17 763 | 100 | 0 | 610 | 17 674 | |
| QDD | 19:11 | 19:11 | 60 994 | 60 994 | 18 009 | 18 009 | 18 009 | 100 | 0 | 732 | 17 552 | |
| SA-SSR | 338:47 | 33:55 | 18 166 | 18 166 | 18 166 | 18 166 | 18 166 | 100 | 0 | 442 | 17 842 | |
| SSR-Pipeline | 611:55 | 611:55 | 19 173 282 | 17 301 120 | 18 044 | 18 044 | 18 044 | 100 | 0 | 913 | 17 371 | |
| SSRIT | 1:29 | 1:29 | 87 073 | 74 121 | 18 284 | 18 284 | 18 284 | 100 | 0 | 0 | 18 284 | |
| TRF | 2:09 | 2:09 | 422 851 | 411 644 | 42 157 | 13 872 | 17 307 | 41.05 | 0 | 1560 | 16 724 | |
| Chlamydomonas reihardtii | GMATo | 3:30 | 3:30 | 26 512 280 | 21 624 294 | 50 401 | 50 401 | 50 139 | 99 | 0 | 23 086 | 34 416 |
| Kmer-SSR | 3:26 | 0:19 | 57 502 | 57 502 | 57 502 | 57 502 | 57 502 | 100 | NA | NA | NA | |
| MREPS | 0:14 | 0:14 | 94 875 | 94 875 | 57 502 | 57 502 | 57 502 | 100 | 0 | 0 | 57 502 | |
| PRoGeRF | 37:55 | 37:55 | 8 071 102 | 8 020 213 | 32 043 | 31 989 | 32 004 | 100 | 0 | 25 588 | 31 914 | |
| QDD | 8:51 | 8:51 | 216 943 | 216 943 | 55 470 | 55 470 | 55 470 | 100 | 0 | 3002 | 54 500 | |
| SA-SSR | 1324:33 | 167:48 | 56 833 | 56 833 | 56 833 | 56 833 | 56 833 | 100 | 0 | 1214 | 56 288 | |
| SSR-Pipeline | 632:10 | 632:10 | 26 973 434 | 23 032 838 | 56 729 | 56 729 | 56 729 | 100 | 0 | 1793 | 55 709 | |
| SSRIT | 2:00 | 2:00 | 310 109 | 252 223 | 57 502 | 57 502 | 57 502 | 100 | 0 | 0 | 57 502 | |
| TRF | 8:52 | 8:52 | 1 022 145 | 990 316 | 181 973 | 25 451 | 45 773 | 25.15 | 0 | 14 546 | 42 956 | |
| Danio rerio (chr 25) | GMATo | 1:12 | 1:12 | 9 501 860 | 7 535 749 | 22 546 | 22 546 | 22 362 | 99 | 0 | 8463 | 13 636 |
| Kmer-SSR | 1:10 | 0:13 | 22 099 | 22 099 | 22 099 | 22 099 | 22 099 | 100 | NA | NA | NA | |
| MREPS | 0:05 | 0:05 | 26 862 | 26 862 | 22 099 | 22 099 | 22 099 | 100 | 0 | 0 | 22 099 | |
| PRoGeRF | 8:14 | 8:14 | 7 696 269 | 7 695 012 | 21 729 | 21 668 | 21 684 | 100 | 0 | 494 | 21 605 | |
| QDD | 7:43 | 7:43 | 49 016 | 49 016 | 21 805 | 21 805 | 21 805 | 100 | 0 | 908 | 21 191 | |
| SA-SSR | 2075:03 | 648:00 | 21 862 | 21 862 | 21 862 | 21 862 | 21 862 | 100 | 0 | 690 | 21 409 | |
| SSR-Pipeline | 1958:54 | 1958:54 | 8 948 450 | 7 954 899 | 21 857 | 21 857 | 21 857 | 100 | 0 | 987 | 21 112 | |
| SSRIT | 0:43 | 0:43 | 69 645 | 58 065 | 22 099 | 22 099 | 22 099 | 100 | 0 | 0 | 22 099 | |
| TRF | 5:03 | 5:03 | 293 378 | 283 764 | 40 343 | 11 255 | 16 911 | 41.92 | 0 | 6144 | 15 955 | |
| Dictyostelium doscoideum | GMATo | 1:02 | 1:02 | 8 810 607 | 7 126 425 | 82 643 | 82 643 | 82 526 | 100 | 0 | 28 714 | 62 967 |
| Kmer-SSR | 1:12 | 0:08 | 91 681 | 91 681 | 91 681 | 91 681 | 91 681 | 100 | NA | NA | NA | |
| MREPS | 0:05 | 0:05 | 121 835 | 121 835 | 91 681 | 91 681 | 91 681 | 100 | 0 | 0 | 91 681 | |
| PRoGeRF | 11:42 | 11:42 | 4 629 786 | 4 604 499 | 60 176 | 60 174 | 60 174 | 100 | 0 | 31 707 | 59 974 | |
| QDD | 3:44 | 3:44 | 171 686 | 171 686 | 88 017 | 88 017 | 88 017 | 100 | 0 | 5295 | 86 386 | |
| SA-SSR | 723:31 | 236:01 | 90 700 | 90 700 | 90 700 | 90 700 | 90 700 | 100 | 0 | 1635 | 90 046 | |
| SSR-Pipeline | 246:35 | 246:35 | 9 292 900 | 7 397 561 | 90 810 | 90 810 | 90 810 | 100 | 0 | 1759 | 89 922 | |
| SSRIT | 0:42 | 0:42 | 265 894 | 202 531 | 91 681 | 91 681 | 91 681 | 100 | 0 | 0 | 91 681 | |
| TRF | 17:30 | 17:30 | 642 904 | 602 301 | 178 902 | 40 772 | 75 742 | 42.34 | 0 | 18 962 | 72 719 | |
| Physcomitrella patens (chr 1) | GMATo | 0:59 | 0:59 | 7 981 869 | 6 500 395 | 7739 | 7739 | 7736 | 100 | 0 | 3259 | 5528 |
| Kmer-SSR | 0:58 | 0:10 | 8 787 | 8 787 | 8787 | 8787 | 8787 | 100 | NA | NA | NA | |
| MREPS | 0:04 | 0:04 | 12 885 | 12 885 | 8787 | 8787 | 8787 | 100 | 0 | 0 | 8787 | |
| PRoGeRF | 7:32 | 7:32 | 6 639 989 | 6 639 933 | 8669 | 8668 | 8668 | 100 | 0 | 131 | 8656 | |
| QDD | 4:29 | 4:29 | 27 774 | 27 774 | 8319 | 8319 | 8319 | 100 | 0 | 621 | 8166 | |
| SA-SSR | 642:36 | 91:59 | 8719 | 8719 | 8719 | 8719 | 8719 | 100 | 0 | 152 | 8635 | |
| SSR-Pipeline | 1498:06 | 1498:06 | 7 763 141 | 6 874 175 | 8720 | 8720 | 8720 | 100 | 0 | 253 | 8534 | |
| SSRIT | 0:35 | 0:35 | 39 472 | 35 941 | 8787 | 8787 | 8787 | 100 | 0 | 0 | 8787 | |
| TRF | 1:53 | 1:53 | 223 938 | 215 818 | 22 730 | 6132 | 8192 | 36.04 | 0 | 891 | 7896 | |
| Saccharyomyces cerevisiae | GMATo | 0:23 | 0:23 | 3 281 592 | 2 674 303 | 1101 | 1101 | 1101 | 100 | 0 | 588 | 887 |
| Kmer-SSR | 0:23 | 0:04 | 1475 | 1475 | 1475 | 1475 | 1475 | 100 | NA | NA | NA | |
| MREPS | 0:02 | 0:02 | 2293 | 2293 | 1475 | 1475 | 1475 | 100 | 0 | 0 | 1475 | |
| PRoGeRF | 3:43 | 3:43 | 1 065 515 | 1 065 510 | 492 | 492 | 492 | 100 | 0 | 988 | 487 | |
| QDD | 0:47 | 0:47 | 8672 | 8672 | 1368 | 1368 | 1368 | 100 | 0 | 139 | 1336 | |
| SA-SSR | 338:50 | 60:55 | 1430 | 1430 | 1430 | 1430 | 1430 | 100 | 0 | 57 | 1418 | |
| SSR-Pipeline | 9:32 | 9:32 | 3 124 288 | 2 820 560 | 1427 | 1427 | 1427 | 100 | 0 | 73 | 1402 | |
| SSRIT | 0:14 | 0:14 | 12 276 | 10 386 | 1475 | 1475 | 1475 | 100 | 0 | 0 | 1475 | |
| TRF | 0:26 | 0:26 | 62 616 | 61 038 | 4634 | 755 | 1242 | 26.80 | 0 | 290 | 1185 | |
| Combined | GMATo | 9:44 | 9:44 | 76 711 216 | 61 830 463 | 181 301 | 181 301 | 180 734 | 100 | 0 | 72 304 | 127 524 |
| Kmer-SSR | 9:33 | 1:18 | 199 828 | 199 828 | 199 828 | 199 828 | 199 828 | 100 | NA | NA | NA | |
| MREPS | 0:39 | 0:39 | 284 389 | 284 389 | 199 828 | 199 828 | 199 828 | 100 | 0 | 0 | 199 828 | |
| PRoGeRF | 87:13 | 87:13 | 44 944 317 | 44 865 988 | 140 872 | 140 753 | 140 785 | 100 | 0 | 59 518 | 140 310 | |
| QDD | 44:45 | 44:45 | 535 085 | 535 085 | 192 988 | 192 988 | 192 988 | 100 | 0 | 10 697 | 189 131 | |
| SA-SSR | 5443:20 | 1238:38 | 197 710 | 197 710 | 197 710 | 197 710 | 197 710 | 100 | 0 | 4190 | 195 638 | |
| SSR-Pipeline | 4957:12 | 4957:12 | 75 275 495 | 65 381 153 | 197 587 | 197 587 | 197 587 | 100 | 0 | 5778 | 194 050 | |
| SSRIT | 5:43 | 5:43 | 784 469 | 633 267 | 199 828 | 199 828 | 199 828 | 100 | 0 | 0 | 199 828 | |
| TRF | 35:53 | 35:53 | 2 667 832 | 2 564 881 | 470 739 | 98 237 | 165 167 | 35.09 | 0 | 42 393 | 157 435 | |
Note: This table shows that Kmer-SSR reports all possible SSRs in reasonable runtime with more refined user control and filtering options relative to the other softwares. We ran all comparisons on a 2.3 Ghz Intel Haswell processor. Although each algorithm was given the same amount of memory and CPUs, due to hardware variability of the CPU, runtimes could vary by up to 20%. Also, MREPS required pre-processing of the fasta files, which typically added anywhere from a few seconds to several minutes to the runtime (not depicted in the table), depending on the pre-processing approach used. Similarly, we did not include the time required to edit SSRIT and QDD’s source code in order for their programs to function over the period sizes in these tests. SSR-Pipeline could not finish searching for 1-mers in chromosome 6 of the Anolis carolinensis in 21 days of runtime. Accordingly, the chromosome was split into 24 approximately equal sized chunks (i.e. approximately 3.3 Mb each) and each chunk was searched for 1-mers separately by SSR-Pipeline. The required time for each chunk was summed (approximately 5 hours) and used in place of 504 hours (21 days).
The SSRs After Adjustments column reflects the number of SSRs that we did not remove or alter for purposes of making the comparison simpler. SSRs that were exact duplicates, duplicates with only the repeat number varying, duplicates that varied only by cycle (e.g. ACG versus CGA with the same number of repeats right next to each other), entirely surrounded by another SSR, or not atomic (e.g. ATAT repeated 2 times instead of AT repeated 8 times) were removed. SSRs that shared the same base and overlapped were combined into one SSR (e.g. AT repeated 8 times at position 1 and AT repeated 6 times at position 11 would be combined to AT repeated 11 times at position 1).
The SSRs In Range column is the number of SSRs from the previous column that were 16 nt or longer and had a period size of 1–7 (inclusive).
The Number Correct column is the number of SSRs In Range that were actually present in the sequence.
The Number Correct and Fixed is the Number Correct plus a few incorrect SSRs that we are able to fix (e.g. a program might report an AT repeated 30 times, but it only repeated 20 times in the sequence).
The Percent Correct and Fixed is the percent of SSRs in Range that were correct or fixed.
Because Kmer-SSR is multithreaded and robust to fasta files with unknown nucleotides, the real time for SSR identification using Kmer-SSR is faster than any other algorithm. Although MREPS reports a faster real time identification of SSRs, the program does not usually run with sequences containing unknown characters. With the addition of the time necessary to make the input fasta files usable for MREPS, it underperformed Kmer-SSR in all six datasets (Table 1, RealTime column). We found that with the exception of TRF, all algorithms tested were 100% accurate in identifying SSRs; however, only Kmer-SSR, MREPS and SSRIT reported all possible filtered SSRs within the range specified for each dataset (Table 1, SSRs In Range column). Although SSRIT has a faster CPU time than Kmer-SSR, it does not have the multithreading capabilities of Kmer-SSR, nor does it allow for querying of SSRs other than period sizes 2–4 without directly editing the algorithm’s source code.
4 Discussion
SSR identification is important in many biological comparisons. It is important to have 100% accuracy in SSR identification because primers often depend on the exact SSR sequence with conserved flanking sequences (Robinson et al., 2004), and phenotypic variations associated with SSRs require an accurate portrayal of a genome. Furthermore, determining the exact SSR copy number is important in species identification and aids in the identification of discrete families and individuals. Kmer-SSR fills a usability gap in SSR identification. While many SSR identification algorithms exist, it is often difficult to install, use and read the output from the algorithms available. Two of the main strengths of Kmer-SSR are its usability and the SSR filters that are easily accessible to help answer biological questions. Installing Kmer-SSR is at least as easy to install as other algorithms. Kmer-SSR was implemented in C ++. It does not require any editing of the source code to find SSRs of different lengths or filter overlapping SSRs, and provides a robust documentation for its command line options. Step-by-step instructions for installation and implementation of Kmer-SSR are available with the algorithm’s source code at http://github.com/ridgelab/Kmer-SSR.
The filters available in Kmer-SSR help answer primary biological questions. Instead of inundating a researcher with duplicate SSRs, Kmer-SSR eliminates overlapping SSRs by only reporting the left-most SSR in each sequence when multiple SSRs are equally valid. Furthermore, longer SSRs are typically more biologically interesting, so completely enclosed SSRs are not included in the output. Importantly, these filters still allow for overlapping SSRs where at least one period size is completely outside of the previously reported SSR. These filters set Kmer-SSR apart from all other SSR identification algorithms because of its ease of use as well as its utility.
As we compared other algorithms, a few difficulties arose that made it challenging to directly compare the output from each program. We learned that QDD does not allow the sequence header line to contain the vertical bar [|] (and possibly other characters that have special meaning in a regular expression). Also, analysis of 1-mers in longer sequences, such as the lizard genome, exceeded 21 days in SSR-pipeline. MREPS also required pre-splitting of the input sequence files because the algorithm does not accept any characters besides A, T, C and G in the sequence lines (it will accept a very limited number of well-distributed Ns). SSRIT requires directly editing the source code to query period sizes other than lengths two through four. Similarly, QDD requires directly editing its source code to retrieve different period lengths and different SSR lengths. QDD defaults to 1-mers that must be 1 million bases long and 2-mers through 6-mers that must repeat at least 5 times. Furthermore, unlike some other algorithms, the output format for Kmer-SSR is easily parsable, and can be exported directly to an Excel spreadsheet or another tab delimited parser. GMATO, ProGeRF, SSRIT and SA-SSR have similar output formats (although, ProGeRF and SSRIT do not provide column headers). MREPS and TRF are text-based reports with embedded tables. QDD provides a semicolon-separated value report with a few fixed columns followed by a variable number of columns thereafter depending on the number of SSRs found in a given sequence. SSR-Pipeline provides FASTA formatted output where the SSRs are encoded in the header (see Table 2). MREPS, PRoGeRF and TRF attempt to identify SSRs through heuristics. Heuristics is a common approach to achieve an adequate solution to a problem that is either too computationally intensive to check all possible solutions, or does not have a good approach to calculate the exact solution (Clancey, 1985). Table 2 displays features of each software package per each software package’s documentation (Benson, 1999; Kolpakov et al., 2003; Lopes et al., 2015; Meglécz et al., 2014; Miller et al., 2013; Pickett et al., 2016; Temnykh et al., 2001; Wang et al., 2013).
Table 2.
We documented each SSR algorithm’s basic usages and options based on the documentation from each algorithm
| GUI | Output | Language | Algorithm | Type | Period | Repeats | Multi-threaded | Search for Specific SSRs | |
|---|---|---|---|---|---|---|---|---|---|
| Kmer-SSR | TSV | C ++ | K-mer Decomposition | Exact | 1+ | 2+ | X | X | |
| SA-SSR | TSV | C ++ | Combinatorial | Exact | 1+ | 2+ | X | X | |
| GMATo | X | TSV | Perl & Java | Regular Expressions | Exact | 1–10 | 2+ | ||
| MREPS | Text | C | Combinatorial | Inexact | 1+ | 2+ | |||
| PRoGeRF | Web | TSV | Perl | ? | Inexact | 1–12 | 2+ | ||
| QDD | SCSV | Perl | ? | Exact | 1–6 | 5+ | |||
| SSR-Pipeline | FASTA | Python | ? | Exact | 1–25 | 2+ | |||
| SSRIT | TSV | Perl | Regular Expressions | Exact | 2–4 | 2+ | |||
| TRF | X | Text | ? | Heuristic | Inexact | 1+ | 2+ |
Note: All algorithms can run in a Linux environment, accept command line options and take a fasta file as input.
Columns in the table are as follows: GUI= Graphical user input available. The algorithms create either a text file, tab separated values (TSV), semicolon separated values (SCSV), or fasta file. The language in which the program is written is followed by the method that the algorithm uses and the type of SSRs it can find (exact or inexact). Minimum SSR period sizes and SSR repeat numbers are also listed. Finally, we list if the algorithm is multithreaded or configurable to search for specific SSRs. Only Kmer-SSR and SA-SSR are multithreaded and configurable to search for specific SSRs.
While Kmer-SSR provides a substantially better user experience with more filters and options than all other algorithms, Kmer-SSR has several weaknesses. First, since Kmer-SSR is an exact algorithm, it is not as fast as the heuristic approach of MREPS when there are only canonical nucleotides in a sequence. Second, due to the kmer decomposition approach used in Kmer-SSR, it is unable to identify fuzzy repeat regions where only one or two nucleotides differ from an exact repeat. Although not necessary for many applications, fuzzy repeats would provide Kmer- SSR with increased functionality that is not currently possible with the algorithm’s implementation. Third, Kmer-SSR has no web interface.
Unlike all other algorithms, Kmer-SSR offers the convenience of a completely exhaustive search in linear time (though with a larger constant factor than normal). This truly exhaustive search is entirely filter- free. As an example, that means it would report an ACG repeated seven times at position 1, six times at position 4, five times at position 7, etc. This is likely not necessary for most applications. However, with the exhaustive option, we set an upper limit for all SSR identifications. Furthermore, since genome complexity is important in primer design and predicting recombination events (Murray et al., 1999), the exhaustive option could be used as an easy approach to determine the proportion of a sequence that repeats.
Acknowledgements
We appreciate Brigham Young University and the Fulton Supercomputing Laboratory (https://marylou.byu.edu) for their continued support of our research. We thank the US Department of Energy Joint Genome Institute for granting access to Chromosome 1 of Physcomitrella patens.
Funding
This work has been supported by funds provided by Brigham Young University and the Department of Biology.
Conflict of Interest: none declared.
References
- Benson G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res., 27, 573.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi R., Medvedev P. (2014) Informed and automated k-mer size selection for genome assembly. Bioinformatics, 30, 31–37. [DOI] [PubMed] [Google Scholar]
- Clancey W.J. (1985) Heuristic classification. Artif. Intell., 27, 289–350. [Google Scholar]
- Ghandi M. et al. (2014) Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol., 10, e1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W.-S. et al. (2007) Ranked subsequence matching in time-series databases. In: Proceedings of the 33rd international conference on Very large data bases. VLDB Endowment. p. 423-434.
- Kashi Y. et al. (1997) Simple sequence repeats as a source of quantitative genetic variation. Trends Genet., 13, 74–78. [DOI] [PubMed] [Google Scholar]
- Kashi Y., King D.G. (2006) Simple sequence repeats as advantageous mutators in evolution. Trends Genet., 22, 253–259. [DOI] [PubMed] [Google Scholar]
- Katti M.V. et al. (2001) Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol., 18, 1161–1167. [DOI] [PubMed] [Google Scholar]
- Kolpakov R. et al. (2003) mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res., 31, 3672–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G.A. (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol., 4, 203–221. [DOI] [PubMed] [Google Scholar]
- Lopes RdS. et al. (2015) ProGeRF: Proteome and Genome Repeat Finder Utilizing a Fast Parallel Hash Function. BioMed. Res. Int., 2015, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meglécz E. et al. (2014) QDD version 3.1: a user‐friendly computer program for microsatellite selection and primer design revisited: experimental validation of variables determining genotyping success rate. Mol. Ecol. Resources, 14, 1302–1313. [DOI] [PubMed] [Google Scholar]
- Merchant S.S. et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science, 318, 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.P. et al. (2013) SSR_pipeline: A bioinformatic infrastructure for identifying microsatellites from paired-end Illumina high-throughput DNA sequencing data. J. Hered., est056. [DOI] [PubMed] [Google Scholar]
- Murray J. et al. (1999) Comparative sequence analysis of human minisatellites showing meiotic repeat instability. Genome Res., 9, 130–136. [PMC free article] [PubMed] [Google Scholar]
- Pickett B.D. et al. (2016) SA-SSR: a suffix array-based algorithm for exhaustive and efficient SSR discovery in large genetic sequences. Bioinformatics, 32, 2707–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A.J. et al. (2004) Simple sequence repeat marker loci discovery using SSR primer. Bioinformatics, 20, 1475–1476. [DOI] [PubMed] [Google Scholar]
- Temnykh S. et al. (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res., 11, 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. (2013) GMATo: A novel tool for the identification and analysis of microsatellites in large genomes. Bioinformation, 9, 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]