Abstract
Motivation
Accurate identification of genotypes is an essential part of the analysis of genomic data, including in identification of sequence polymorphisms, linking mutations with disease and determining mutation rates. Biological and technical processes that adversely affect genotyping include copy-number-variation, paralogous sequences, library preparation, sequencing error and reference-mapping biases, among others.
Results
We modeled the read depth for all data as a mixture of Dirichlet-multinomial distributions, resulting in significant improvements over previously used models. In most cases the best model was comprised of two distributions. The major-component distribution is similar to a binomial distribution with low error and low reference bias. The minor-component distribution is overdispersed with higher error and reference bias. We also found that sites fitting the minor component are enriched for copy number variants and low complexity regions, which can produce erroneous genotype calls. By removing sites that do not fit the major component, we can improve the accuracy of genotype calls.
Availability and Implementation
Methods and data files are available at https://github.com/CartwrightLab/WuEtAl2017/ (doi:10.5281/zenodo.256858).
Supplementary information
Supplementary data is available at Bioinformatics online.
1 Introduction
Identifying genotypes from next-generation sequencing (NGS) data is an important component of modern genomic analysis. Accurate genotyping is key to identifying sequence polymorphisms, detecting de novo mutations, linking genetic variants with disease, and determining mutation rates (Awadalla et al., 2010; Goldstein et al., 2013; Koboldt et al., 2013; Peng et al., 2013; Sayed et al., 2009). Inaccurate genotyping affects the accuracy of identifying de novo mutations, as true mutations are rare compared to errors in sequencing and downstream analyses. Because putative SNPs are typically validated using another sequencing technology, high false positive rates increase the effort required for validation.
However, estimating genotypes from NGS data can be computationally and statistically challenging. A typical NGS experiment generates millions of short read fragments, 100–650 base-pairs in length that are aligned to a reference genome. If the only error was due to sampling, NGS methods would produce data that was perfectly representative of the underlying genotype; the base calls for homozygous sites would be identical, and the base calls for heterozygous sites would follow a 1:1 binomial distribution. For this reason, genotyping and variant calling software initially used binomial or multinomial distributions to model heterozygous base-counts (Cartwright et al., 2012; Goya et al., 2010; Hohenlohe et al., 2010; Li et al., 2008a, b; Lynch, 2009; Maruki and Lynch, 2015).
However, there are at least three experimental processes that are thought to affect the ratio of the alleles. (i) During library preparation, variation in amplification rates can cause some chromosomes to be replicated more than others (Heinrich et al., 2012). This variation is especially a concern if there is little starting material. (ii) NGS technologies can introduce sequencing errors into sequencing reads. Error rates are on the order of 0.1–1% per base-call. While this may seem small, 0.1% error is equivalent to the difference between an average human and the human reference genome (Fox et al., 2014; Wall et al., 2014). (iii) Bioinformatic methods that assemble reads with respect to a reference can misplace reads and penalize non-reference alleles (i.e. fewer non-reference alleles align to the reference genome) (Degner et al., 2009; Krawitz et al., 2010). Together these processes can shift the mean and increase the variance of sequencing read count distributions. These processes do not affect all parts of the genome equally. The genomic context of a site, including the presence of nearby indels, structural variants, or low complexity regions, influences the probability that reads generated from a given site will be subject to these processes (Li, 2014; Malhis and Jones, 2010). Thus, it is possible for both homozygotes and heterozygotes to have an intermediate ratio of two alleles, and it can be difficult to accurately identify genotypes using a binomial distribution (Malhis and Jones, 2010).
Current approaches to calling genotypes from NGS data deal with the issues described above to some degree. The increased variance and skewed allele ratios expected from mismapped reads can be partially controlled by including mapping quality data in a genotype-calling procedure. In the simplest approaches, reads with low quality scores are removed from an analysis. In Bayesian approaches to genotype calling, read quality data may be included when calculating genotype probabilities (e.g. Li et al., 2009b).
The increased variance expected from library preparation, sequencing and errors in mapping reads to a reference genome can be accommodated by modeling read-counts as coming from a beta-binomial distribution (Ramu et al., 2013). The beta-binomial distribution acts as an over-dispersed binomial, allowing the excess variance to be handled in a standard statistical framework. The Dirichlet-multinomial (DM) distribution is the general case of the beta-binomial, allowing for overdispersion and modeling of more than two outcomes. The DM has been used to model allele counts and the frequency of multi-allelic genotypes within tumor samples (Josephidou et al., 2015; Muralidharan et al., 2012; Tvedebrink, 2010). Such genotype calling procedures can be combined with machine learning algorithms that attempt to differentiate between true variants and those caused by sequencing artifacts (DePristo et al., 2011).
In this study, we evaluate the hypothesis that finite mixtures of DM distributions can produce a better fit to the underlying error processes than previous approaches. The mismatch between observed and expected read distributions created by the processes described above contributes to observed false-positive single nucleotide polymorphism (SNP) discovery rates of 3–15% (Farrer et al., 2013; Harismendy et al., 2009). We posit that improving the expected read distribution (i.e. the model) will reduce false positives. As a first step toward this goal in this study we model the distribution of base counts produced from NGS using a mixture of DM distributions (MDMs). Furthermore, we model sites that are the most likely to be true heterozygotes (THs) (rather than false positives). By modeling these sites exclusively, we determine the true effect of variation in amplification rates, error and mismapping.
Fitting MDMs to sequencing data improves existing genotyping methods in two important ways. First, we can account for the context-dependent nature of genotyping errors by allowing multiple DM distributions, each with different parameter values, to be estimated for a given dataset. Additionally, by using more than two categories, we explicitly model the presence of bases that are neither reference nor the likely alternative allele at a given site. Thus, we are able to directly estimate the probability of sequencing errors in a given DM model.
We first demonstrate the value of our approach by fitting MDMs to sequencing data derived from a haploid human cell line (CHM1). The MDM produces a superior fit to this dataset compared to other models, showing that even relatively simple genetic datasets can be the result of heterogeneous processes, and thus benefit from a mixed-model approach. We then fit MDMs to diploid data generated by the 1000 Genomes Project (1000 Genomes Project Consortium et al., 2010, 2015). For these datasets, the MDM also improves the fit compared to other models. Most interestingly, when we limit our data to sites of known polymorphism, most of the data fit a binomial distribution, rather than the expectation of over-dispersed, reference-biased data. Using a model that fits the data should lead to significant improvements in genotyping, which in turn should lead to fewer candidate mutations that are found to be inaccurate when validated in a lab. When we assign sites to different model components and retain those that fit the major component, we find that our approach is often better than other methods at correctly calling putative heterozygotes while minimizing false-positive calls.
2 Materials and methods
2.1 Data
We extracted datasets from two types of data. The first dataset is the haploid human sequence from a hydatidiform mole cell line (CHM1hTERT SRR1283824 from samples: SRS605680, experiment: SRX540665 and study: SRP017546). We refer to this dataset as the CHM1 dataset in this paper. Second, we obtained Illumina sequences from the 1000 Genomes Project for three individuals, a woman (NA12878) and both of her parents (NA12891 and NA12892). Sequencing was repeated for these individuals in different years using different technologies (2010, 2011, 2012 and 2013). The 2010 dataset was generated during pilot 2 studies with short and variable read length between 36bp and 76bp. The 2011 and 2012 dataset were not part of a release; they contain the same sequencing data, 101 bp reads, with slightly different bioinformatics. The 2013 dataset was part of the phase 3 release and was sequenced without PCR, with the longest read length of 250bp (1000 Genomes Project Consortium et al., 2010, 2015). We refer to this dataset as the CEU dataset. If the release year is appended, for example CEU13, then we refer to the specific release in that year (e.g. 2013).
For each of these five datasets (CHM1 and each of the four releases of CEU), we analyzed two genomic regions, the whole chromosome 21 and a subregion of chromosome 10, from positions 85534747 to 135534747, which is approximately the same size as chromosome 21 (48 million base pairs). For CHM1, we called genotypes by first obtaining allele counts for each base at each site using the mpileup function in Samtools v1.2 (Li, 2011; Li et al., 2009a) and the human reference genome (Genome Reference Consortium human genome build 37). We then used BCFTools v1.2 (Li, 2011; Li et al., 2009a) to identify sites called as homozygous reference. For each of these sites, we calculated the frequency of the reference allele and the frequency of all non-reference alleles (error). We filtered this dataset based on the read depth for each site: we only kept sites with total read counts of greater than 10 and less than 150, and we refer this dataset as CHM1’s full dataset (FD). The upper limit is calculated using a method based on Warr et al. (2015), which takes the median plus twice the standard deviation of coverage per site; sites with zero coverage are ignored. Sites with high numbers of reads are likely from copy-number-variable loci that have aligned to a single region of the genome. In sites with extreme coverage, apparent heterozygotes are more likely to be due to paralogs rather than variation within a site. The low read filter limits the data to calls with enough coverage to provide a reasonably accurate call and proportion of reads for each base.
We also focused on modeling sites that were very likely to be homozygous reference sites in order to understand what the distribution of reads for those sites looks like. To generate our reference dataset (RD), we removed sites for which less than 80% of the reads contained the reference allele from the FD dataset. The cutoff point was picked after examining the empirical distributions; there were only a few sites for which 50–80% of the reads contained the reference allele.
For the CEU data, we obtained allele counts as above for all three individuals. We then called genotypes as above on individual NA12878 (the daughter of the trio) and by using the BCFtools trio caller with the data from all three individuals. We limited the dataset we used for subsequent analyses to sites that were found by both the trio caller and the individual caller. Sites that were found only by the trio caller, but not by the individual caller were likely identified by the pedigree with limited data for the daughter; thus, these low coverage sites were not included in subsequent analysis. We removed sites with read counts of less than 10 or greater than 150, as for CHM1. We call this CEU’s potential heterozygote (PH) dataset. For each of these sites we calculated the frequency of the reference allele, alternate allele, and any other alleles (error). We compared the frequencies of each allele category (reference, alternate, error) for each possible genotype combination. Because we found no differences in frequencies for different genotypes, all subsequent analyses were only performed on the general reference–alternate–error dataset.
We created an additional dataset by removing sites from the PH dataset not found to be heterozygous by the 1000 Genomes Project (1000 Genomes Project Consortium et al., 2010, 2015). We then discarded sites for which the alternate allele differed from the one previously identified by the 1000 Genomes Project. We call this the TH dataset.
Because the CHM1 RD dataset was larger than the CEU TH dataset, we randomly subsampled the CHM1 dataset to have an approximately equal number of sites (40 000 sites) as the CEU TH dataset.
2.2 Model fitting and parameter estimation
We fit seven models to each CHM1 dataset, and eight to each CEU dataset. The models included a multinomial, a multinomial with reference bias (CEU only), DM and MDM distributions, ranging from 2 to 6 components. We estimated the parameters and calculated the genotype likelihood for each model. Additional details of these methods are available in the GitHub repository for this paper (see Availability section).
The genotype likelihood measures the likelihood of a sample’s genotype, G, given a set of base-calls, R, and is proportional to the probability of observing R if the genotype was G, i.e. . We derived genotype likelihoods using MDM distributions. The DM is a compound distribution generated when a Dirichlet distribution is used as a prior for the probabilities of success of a multinomial distribution: and , where is a vector of concentration parameters, is a vector of proportions, is a vector of counts and N is the sample size. After integrating out , the resulting probability mass function can be trivially expressed as a product of ratios of gamma functions:
| (1) |
where i is one of the nucleotides, and . Furthermore,
| (2) |
where and . It is helpful to reparameterize the distribution by letting , where represents the pairwise correlation between samples. As a result, and is a parameter controlling the amount of excess variation in the DM. When φ approaches 0, the DM converges to a multinomial. Thus, the DM can be interpreted as an over-dispersed multinomial distribution. As φ increases the distribution becomes more overdispersed, and as φ approaches 1, every observation contains only one type of nucleotide.
For a single-component DM, we estimated the maximum-likelihood model starting with a method-of-moments estimation and optimizing using the Newton–Raphson method. For MDM, the maximum-likelihood estimate was computed using an EM algorithm and random starting points. This procedure was repeated 1000 times to search for the global maximum-likelihood estimate.
For the DM distribution, we estimated φ as a measure of the overdispersion of the data. In addition, we estimated ρ, the mixing proportion of sites belonging to each DM component. For the homozygous CHM1 dataset, we estimated the proportion of the reference allele and error terms for each model or model component. For each CEU dataset, we estimated the proportion of the reference allele, alternate allele, and the error terms. The reference and alternate allele were determined by Samtools.
To determine the optimal number of components in the MDM model, we calculated the Bayesian information criterion (BIC) for each dataset. The model with the lowest BIC is considered the model that best optimizes both goodness-of-fit and simplicity. Similarly, we also calculated the Akaike information criterion (AIC). The BIC and the AIC for each model are calculated by the following formulas:
where L is the maximum-likelihood estimate from the model, k is the number of free parameters and n is the number of sites in the dataset.
2.3 Visualizing model fit
We visualized the fit of the data to each model and compared the fit between models using quantile–quantile (QQ) plots. QQ plots display the quantile of the observed read count frequencies from the dataset against the quantile of the expected read count frequencies. The expected read count frequencies are simulated from parameters estimated from the EM by Monte Carlo simulation with 100 replicates. Two separate QQ plots were used to illustrate the fit of models for the CHM1 dataset, one for the reference allele and one for the error term. Three QQ plots were used for the CEU datasets, one each for reference, alternate and error terms.
2.4 Frequency of sites in copy number variable regions and low complexity regions
In order to explore the composition of each component, the likelihood of every site was calculated under each component of the model in each of the CEU datasets. The likelihood for each component was evaluated using the parameters estimated from the EM. By comparing the likelihood between all components, each site was assigned to the component with the highest likelihood.
We extracted all the known copy number variable regions (CNVs) sites for NA12878 in the CEU dataset (Mills et al., 2011). We calculated the number and proportion of sites belonging to the known CNVs for the major and minor components (combined, because some components only contain a small proportion of sites) for the best fit model for each of the eight datasets. We used a Fisher’s exact test to determine whether there is a significant difference between the proportion of CNVs in each component.
We extracted all known repetitive/low complexity regions (LCRs) in the human reference from the UCSC Genome Bioinformatics with the Table Browser data retrieval tool (Karolchik et al., 2004), and repeated the same analyses performed for the CNV regions. We calculated the number and proportion of LCRs in the major and minor components for the best fit model and used a Fisher’s exact test to determine whether there is a significant difference between these two components.
2.5 Assignment of sites to model components
Sites can be assigned to components based on the posterior probability that they were generated by each component. We developed a classification algorithm that called sites as high-quality heterozygotes (positive) or low-quality (negative) based on the probability that they belonged to the major component of estimated two-component models. Sites in our potential heterozygote datasets were classified into these categories and validated using the corresponding TH dataset, e.g. true positives were any potentially heterozygous sites identified as high quality by our classifier and were known to be polymorphic in humans.
We varied the cut-off probability used in the classifier and constructed the receiver operating characteristic (ROC) curve, where sensitivity is plotted against specificity, to examine the performance of our model as a classifier across a range of classification thresholds. The area under the ROC curve (AUC) summarizes the performance of this classification method across a range of cutoff points. An AUC of 1 represents a perfect classifier, while an AUC of 0.5 suggests the prediction is close to random.
Three different optimal cutoff points were used to evaluate the performance of our classifier (López-Ratón et al., 2014). (i) Cost-benefit (CB) method, where the default costs ratio of 1 was used. (ii) Youden’s Index, which is equivalent to maximizing the sum of sensitivity and specificity. (iii) ROC01, which finds the point on the ROC curve closest to the point (0,1).
For comparison, the default GATK variant discovery workflow was applied to the full dataset (DePristo et al., 2011; McKenna et al., 2010; Van der Auwera et al., 2013). In short, we used HaplotypeCaller from GATK and set the minimum phred-scaled confidence threshold to 0 in order to obtain all possible calls. The GATK ROC curve is generated by altering this threshold and calculated the sensitivity and specificity for each threshold. We reported the sensitivity and specificity from each cutoff and GATK.
3 Results
3.1 Haploid dataset—homozygous reference
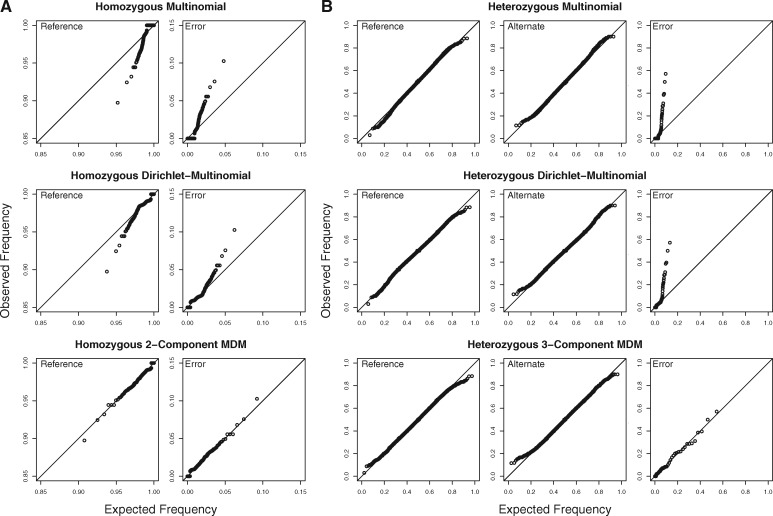
Using the expectation maximization (EM) algorithm, we fit seven models to each genomic region in each CHM1 dataset: a multinomial, a DM and MDM models with two to six components. The best model for each dataset was the two-component MDM. The fit of the two-component MDM model to the Chr21 RD dataset is shown in Figure 1a (see Supplementary figures for additional results.)
Fig. 1.
Mixtures of Dirichlet-multinomials provide the best fits to genomic datasets. QQ plots evaluate the fit of three different models to (a) the CHM1 chromosome 21 RD dataset. (b) The CEU 2013 chromosome 21 TH dataset. The quantiles of the observed read count frequencies are calculated from the datasets, and the quantiles of the expected read count frequencies are estimated from the fitted model. A model that fits the data well produces points that fall along the diagonal
In all cases, one component contained a substantial majority of sites (approximately 75% of sites for Chr21 RD and 95% of the sites for other datasets). We will refer to the component to which the highest proportion of sites was assigned as the ‘major component’ and all other components as ‘minor components’.
For FD, the major component had relatively little overdispersion ( and 0.004 for chromosome 21 and chromosome 10, respectively), while the minor component displayed strong overdispersion ( and 0.948). When we fitted MDMs to the reference datasets (in which sites with a high proportion of non-reference alleles were removed) the major component had less overdisperson compared with the full dataset ( and 0.003). The minor components of models fitted to this dataset were slightly overdispersed ( and 0.048) (Table 1 and Supplementary Tables).
Table 1.
The number of components (c) in the best MDM model according to BIC values for CHM1 and CEU datasets, and parameters estimated for these models
| Dataset | c | πref | πalt | πerr | φ | ρ |
|---|---|---|---|---|---|---|
| CHM1 RD Chr21 | 2 | 1.00 | NA | 2.20e−4 | 0.000 | 0.751 |
| 1.00 | NA | 3.61e−4 | 1.53e−2 | 0.249 | ||
| CHM1 RD Chr10 | 2 | 1.00 | NA | 2.60e−4 | 2.69e−3 | 0.942 |
| 0.999 | NA | 8.33e−4 | 4.75e−2 | 5.85e−2 | ||
| CHM1 FD Chr21 | 2 | 1.00 | NA | 2.49e−4 | 2.52e−3 | 0.972 |
| 0.982 | NA | 1.76e−2 | 0.892 | 2.78e−2 | ||
| CHM1 FD Chr10 | 2 | 1.00 | NA | 2.82e−4 | 4.15e−3 | 0.984 |
| 0.975 | NA | 2.54e−2 | 0.948 | 1.64e−2 | ||
| CEU13 TH Chr21 | 3 | 0.504 | 0.496 | 3.53e−04 | 2.46e−04 | 0.939 |
| 0.508 | 0.491 | 5.26e−04 | 6.89e−02 | 6.04e−02 | ||
| 0.239 | 0.483 | 0.278 | 6.56e−02 | 5.87e−04 | ||
| CEU12 TH Chr21 | 2 | 0.509 | 0.491 | 3.15e−04 | 1.29e−04 | 0.961 |
| 0.541 | 0.457 | 2.04e−03 | 7.73e−02 | 3.90e−02 | ||
| CEU11 TH Chr21 | 2 | 0.509 | 0.491 | 3.15e−04 | 1.31e−04 | 0.961 |
| 0.541 | 0.457 | 1.98e−03 | 7.82e−02 | 3.91e−02 | ||
| CEU10 TH Chr21 | 2 | 0.533 | 0.465 | 2.25e−03 | 1.52e−03 | 0.922 |
| 0.670 | 0.327 | 2.70e−03 | 0.000 | 7.83e−02 | ||
| CEU13 TH Chr10 | 2 | 0.502 | 0.498 | 3.36e−04 | 4.28e−04 | 0.922 |
| 0.504 | 0.490 | 6.21e−03 | 1.24e−01 | 7.54e−03 | ||
| CEU12 TH Chr10 | 2 | 0.508 | 0.491 | 3.19e−04 | 5.53e−04 | 0.986 |
| 0.540 | 0.457 | 3.05e−03 | 7.60e−02 | 1.38e−02 | ||
| CEU11 TH Chr10 | 2 | 0.508 | 0.491 | 3.19e−04 | 5.56e−04 | 0.986 |
| 0.542 | 0.455 | 3.01e−03 | 7.37e−02 | 1.39e−02 | ||
| CEU10 TH Chr10 | 2 | 0.534 | 0.463 | 3.05e−03 | 0.000 | 0.684 |
| 0.550 | 0.449 | 6.45e−04 | 1.29e−02 | 0.316 |
Note: Each row represents a different component in the model. πref, πalt and πerr are the proportion of the reference, alternative and error terms respectively. φ is the overdispersion parameter. When φ approaches 0, the distribution approaches a multinomial, and when φ approaches 1, the distribution is nearly completely overdispersed. ρ is the proportion of sites in each component.
3.2 Diploid dataset—heterozygous sites
We fit eight models to each of the 16 CEU datasets (two genomic regions, four release years, two levels of filters): a multinomial, multinomial with reference bias, a DM and MDMs with two to six components. As expected, the addition of model components increased the likelihood of the MDM model for all cases (Supplementary Tables).
For the TH datasets, the best-fitting models had either two or three components. The fit of the three-component MDM model to the CEU13 Chr21 TH dataset is shown in Figure 1b (see Supplementary figures for additional results.) The majority of the sites (68–99%) were assigned to one component in the model (Table 1 and Supplementary Tables). This major component of the model had little overdispersion (–0.0014). For the 2011, 2012 and 2013 datasets, the major component had an approximately equal proportion of reference and alternate alleles (50.2–50.9% reference versus 49.1–49.8% alternate, and a relatively small error term (). Thus, for these datasets, the majority of sites fall into a component that is well approximated by a binomial distribution with equal probabilities of reference and alternative alleles. However, the best-fitting model for the 2010 dataset has a slightly larger error term (0.2% and 0.3%), and the reference and alternate terms are 53% and 46%, respectively.
The proportion of the reference allele in the second component of the model for each dataset ranged from 50.4% to 67.0%. This component also had greater overdispersion in most cases (–0.124). For the dataset with a three component model (CEU13 chromosome 21), the third component has an elevated proportion of the error term.
Comparing the PH dataset to the TH dataset, the best fitting model in the PH dataset had three to six components (Supplementary Tables and Fig. S1). However, when models have more than four components, the additional components contained a very small proportion of the data (). Thus, a model with more than four components was likely overfitting the data from a biological perspective. The major components all had little overdispersion, which is similar to the TH dataset. However, there is huge variation in the value of φ within each of the minor components. PH datasets tend to have minor components with the proportion of reference allele deviating from 50%. Overall, the major component in the PH dataset is similar to the major components in the TH dataset, but minor components showed very different parameter distributions.
3.3 Copy number variants and low complexity regions
We assigned each site from the eight CEU PH datasets to a component in the best-fitting model based on the site likelihood. Repetitive regions of the genome (e.g. SINEs, ALUs, LINEs, LTRs and retroposons), low-complexity regions (LCRs), and copy-number variable genes (CNVs) are known to have higher genotyping error rate (Li, 2014; Malhis and Jones, 2010). The read counts of these sites are likely to deviate from the expected distributions and differ from other genomic regions. Therefore, we predict that the minor components of our models will be enriched for these elements.
The minor component of the best-fitting model for each dataset was enriched for CNVs, repetitive regions and LCRs. In chromosome 21, the proportion of CNVs is 3.7–6.7% in the minor component and 0.5–1.5% in the major component. In chromosome 10, the proportion of CNVs is 0.3–0.5% in the minor component and 0.2–0.3% in the major component. Fisher’s exact tests show a significantly higher proportion of CNVs sites in the minor component than major component for all datasets () except for CEU10 chromosome 10 (P = 0.198). In the minor component, 53.5–60.7% of sites were in repetitive regions/LCRs, compared with 45.9–51.1% in the major component. Fisher’s exact tests show a significantly higher proportion of repetitive regions/LCRs in the minor component (; Table 2).
Table 2.
The minor components have a higher percentage of copy number variable regions (CNVs) and repetitive/low complexity regions (LCRs)
| Dataset | Non-CNV/CNV | % | P value | Non-LCR/LCR | % | P value | |
|---|---|---|---|---|---|---|---|
| CEU13 Chr21 | Major | 26 924/415 | 1.5 | 4.11e−143 | 13 362/13 977 | 51.1 | 2.91e−48 |
| Minor | 10 851/773 | 6.7 | 4746/6878 | 59.2 | |||
| CEU13 Chr10 | Major | 32 012/94 | 0.3 | 4.19e−3 | 16 043/16 063 | 50.0 | 5.39e−06 |
| Minor | 5864/32 | 0.5 | 2756/3140 | 53.3 | |||
| CEU12 Chr21 | Major | 26 891/193 | 0.7 | 4.54e−87 | 13 362/13 722 | 50.7 | 2.67e−57 |
| Minor | 7760/331 | 4.1 | 3178/4913 | 60.7 | |||
| CEU12 Chr10 | Major | 32 560/90 | 0.3 | 1.58e−3 | 16 204/16 446 | 50.4 | 2.08e−16 |
| Minor | 7035/37 | 0.5 | 3129/3943 | 55.8 | |||
| CEU11 Chr21 | Major | 26 858/191 | 0.7 | 1.11e−88 | 13 344/13 705 | 50.7 | 3.11e−54 |
| Minor | 7743/333 | 4.1 | 3194/4882 | 60.5 | |||
| CEU11 Chr10 | Major | 32 539/89 | 0.3 | 1.01e−3 | 16 195/16 433 | 50.4 | 3.19e−16 |
| Minor | 7060/38 | 0.5 | 3144/3954 | 55.7 | |||
| CEU10 Chr21 | Major | 21 968/109 | 0.5 | 3e−90 | 11 936/10 141 | 45.9 | 3.62e−71 |
| Minor | 8518/326 | 3.7 | 3790/5054 | 57.1 | |||
| CEU10 Chr10 | Major | 26 380/49 | 0.2 | 0.198 | 14 305/12 124 | 45.9 | 1.68e−53 |
| Minor | 10 197/26 | 0.3 | 4617/5606 | 54.8 |
Note: The number and percent (%) of CNV regions and of LCRs are shown for the major and minor components (combined) for the best fit model for each CEU PH dataset. P value was calculated for the difference between the proportion of CNVs or LCRs in each component.
3.4 Identification of high-quality heterozygous genotypes
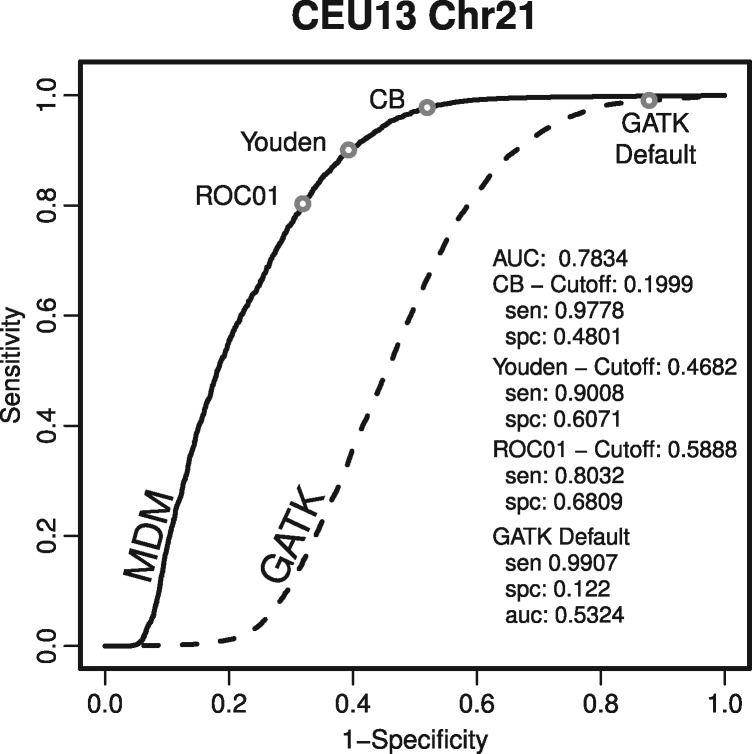
Figure 2 and Supplementary figures examine the performance of adapting our MDM model into a classifier of heterozygotes. AUC was between 0.63 and 0.81 (Supplementary Table). Based on the ROC curve, we examined three optimal threshold values (the probability cutoff for assigning a site to the major component). These thresholds demonstrate different ways to utilize this classifier to balance sensitivity and specificity. For example, using cost–benefit criteria for the CEU13 chromosome 21 dataset we could use our model to filter out half of the false-positive heterozygous sites without losing THs (Fig. 2).
Fig. 2.
The receiver operating characteristic (ROC) curve for the CEU13 chromosome 21 dataset demonstrates better classification of heterozygous sites using our approach. This dataset is shown as an example of how the model can be used to classify sites. Sensitivity and specificity are calculated for three possible optimizing criteria from the MDM heterozygotes site classifier: cost benefit (CB), closest point to (0,1) (ROC01), and Youden’s Index (Youden). For comparison, the output from the GATK recommended workflow is also shown
We compared our results to genotype calls produced using the Genome Analysis Tool Kit (GATK) variant discovery pipeline. For chromosome 21, our approach had greater sensitivity than GATK for any given specificity value, and thus produced higher AUC (Fig. 2). For chromosome 10, AUC values were similar between GATK and our method. Our approach is more sensitive than the GATK pipeline for high-specificity values, but less sensitive than GATK for lower values of specificity (Supplementary figures and Tables). Thus, while GATK output includes almost exclusively true positive heterozygotes, it also excludes many additional TH sites that we are able to include in our output.
4 Discussion
In this paper, we have shown that standard models inaccurately represent the distribution of reads for homozygotes and heterozygotes. For every dataset we considered, our MDM approach provided a better fit to NGS data than any single-component model. Indeed, the best-fitting models generated for a haploid human cell line contained two components, demonstrating that NGS datasets generated from relatively simple biological samples (i.e. no THs and a high-quality reference genome) can benefit from the approach we describe here. Similarly, our MDM model provides a better fit to more complex data, including potentially heterozygous sites in data arising from the 1000 Genomes Project. In order to take into account the excess variation in the PH dataset, the MDM models fit more complex models to the PH datasets than to the TH datasets.
4.1 Overdispersion and reference bias are not universal in NGS data
Previous work on the statistical properties of NGS data have emphasized the presence of reference bias due to errors in read mapping (Degner et al., 2009; Heinrich et al., 2012; Krawitz et al., 2010), and overdispersion due to correlated errors during library preparation and sequencing (Ramu et al., 2013). Both these processes would move the distribution of reads in an NGS experiment away from a standard binomial distribution with probabilities reflecting the underlying genotype. Our results demonstrate that not all sites in an NGS experiment are subject to these processes. The results are likely to vary between different datasets and different regions within the same dataset. The best-fitting model for every dataset that we considered contained a major component with relatively little overdispersion or allelic bias. Sites that fall in these components are thus well approximated by a binomial. On the other hand, a substantial minority of sites in all cases fall into components that do display allelic bias, a high rate of apparent sequencing error, and/or overdispersion. Our mixture model approach allows us to accurately model read distributions for different types of sites and to avoid applying inappropriate model parameters to the majority of sites.
4.2 Copy number variants and low complexity regions
The minor components of our MDM models are enriched for CNVs and low-complexity regions (LCRs). CNVs can cause misalignment of reads in repetitive regions, which is thought to be a cause of genotyping error (Muralidharan et al., 2012). LCRs are particularly prone to misalignment as a result of compositional biases and the alignment of paralogous sequences (Frith, 2011; Li, 2014; Wootton and Federhen, 1996). We believe that these genomic regions resulted in misalignment of some reads and altered the profile of the read counts for some sites. Sites with this erroneous read profile logically need to be modeled differently; thus, our approach of using an MDM avoids applying inappropriate model-parameters to the majority of sites.
4.3 Distinguishing true and false-positive heterozygotes
One of the applications of this model is to improve the accuracy of genotype calling and reduce the number of false-positive variant calls produced from NGS data. We investigated the performance of MDM models as classifiers for putatively heterozygous sites. The sensitivity and specificity of this classifier can be tailored to a particular application by altering the probability value at which a site is assigned to a minor component. Used in this way, the MDM classifier shows promise for reducing the number of falsely called heterozygotes. Unless high sensitivity without regard to specificity is a priority (i.e. false positives are unacceptable, but false negatives are unimportant), our method outperforms the standard GATK variant calling workflow for this purpose. It should be noted the GATK pipeline includes a complex genotype-calling algorithm and local realignment of reads, both of which reduce the number of false positive SNP calls. The approach we describe here could be included in such a pipeline, and thus further increase the accuracy of genotype calling.
The size of the dataset can influence parameter estimation. The two cases with relatively low-quality classification, CEU10 chromosome 10 and CEU13 chromosome 10, are likely due to an extremely low proportion of false positives in the dataset: with only 5% false positive sites, it is a challenging task for the classification algorithm to identify these sites. In these cases, it can be helpful to examine the 95% confidence intervals of the estimated parameters to determine whether there is enough power for accurate estimation.
4.4 Differences among datasets
Although all the diploid datasets contained reads produced from Illumina sequencing, a variety of different library preparations and sequencing platforms were used to produce the data. The 2010 data was generated using short paired reads (35–67 bp) and relatively error-prone sequencing. The 2011 and 2012 datasets contain reads of intermediate length (101 bp) mapped to a reference genome designed to lower the rate of misalignment. The 2013 dataset contains the longest reads (250 bp) and was produced using a PCR-free library preparation.
Our approach was able to fit each of these datasets well, regardless of the library preparations and sequencing technologies. The parameter values estimated by EM in each dataset reflect the technology used. Not surprisingly, the 2010 dataset appears to have a different error profile from other years: the major component of this model has higher overdispersion and stronger reference bias than that of any other dataset. The major components of datasets from 2011 and 2012 fit more closely to the expectation of a binomial distribution with equal frequencies of reference and non-reference alleles. Thus, we conclude that advances in sequencing technologies have led to the majority of sites in NGS datasets better fitting basic expectations of how alleles are amplified, sequenced, and mapped, while the remaining sites contain a high rate of error in genotype calling.
We expected the 2013 datasets, which was produced with a PCR-free protocol, to be ‘cleaner’ than other datasets. This was not the case. Our estimates of the overdispersion parameter do not appear to be lower for the 2013 datasets than others. In fact, for chromosome 21 the 2013 data produces a higher estimate of φ than either the 2012 or 2011 data (Table 1 and Supplementary Fig. S2).
It is possible that steps taken to avoid PCR-error in producing the CEU13 dataset introduced other errors or biases that are reflected in these estimates. We suggest reconsidering the benefit of the PCR-free approach. Alternatively, as CEU13 is the only dataset we examined that used 250 bp reads it is possible that the sequencing chemistry used to produce these longer reads has a unique error profile.
We conclude that read distributions for a majority of sites fit a basic binomial distribution. This observation contrasts with recent approaches to improve genotyping that fit a more complex beta-binomial. A minority of sites are subject to processes that drive read count distributions away from a binomial distribution. The PH dataset shows different parameter distributions comparing to TH dataset. Most of the minor components in the PH dataset also suggested these sites are deviated from binomial distributions. Our approach using a mixture of distributions allows us to correctly model these sites without applying inappropriate models to the majority of the genome. This approach also results in a better fit of the model to the data. In future, it may be possible to identity genomic features associated with unusual sequencing data and ultimately develop improved models for these sites in particular. The approach we describe here will be included in future versions of the mutation-detecting software DeNovoGear and accuMUlate (Long et al., 2016; Ramu et al., 2013).
Funding
This work was supported by National Institutes of Health Grants [R01-GM101352 to R.A. Zufall, R.B.R. Azevedo and R.A. Cartwright and R01-HG007178 to D.F. Conrad and R.A. Cartwright]; and National Science Foundation Grant [DBI-1356548 to R.A. Cartwright].
Conflict of Interest: none declared.
Supplementary Material
References
- 1000 Genomes Project Consortium. et al. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. et al. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla P. et al. (2010) Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet, 87, 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright R.A. et al. (2012) A family-based probabilistic method for capturing de novo mutations from high-throughput short-read sequencing data. Stat Appl Genet Mol Biol, 11, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J.F. et al. (2009) Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics, 25, 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet, 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer R.A. et al. (2013) Using false discovery rates to benchmark SNP-callers in next-generation sequencing projects. Sci Rep, 3, 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox E.J. et al. (2014) Accuracy of next generation sequencing platforms. Next Gener Seq Appl, 1, 1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M.C. (2011) Gentle masking of low-complexity sequences improves homology search. PloS One, 6, e28819.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D.B. et al. (2013) Sequencing studies in human genetics: design and interpretation. Nat Rev Genet, 14, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goya R. et al. (2010) SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics, 26, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O. et al. (2009) Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol, 10, R32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich V. et al. (2012) The allele distribution in next-generation sequencing data sets is accurately described as the result of a stochastic branching process. Nucleic Acids Res, 40, 2426–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe P.A. et al. (2010) Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLOS Genetics, 6, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephidou M. et al. (2015) multiSNV: a probabilistic approach for improving detection of somatic point mutations from multiple related tumour samples. Nucleic Acids Res, 43, e61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D. et al. (2004) The UCSC Table Browser data retrieval tool. Nucleic Acids Res, 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D.C. et al. (2013) The next-generation sequencing revolution and its impact on genomics. Cell, 155, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz P. et al. (2010) Microindel detection in short-read sequence data. Bioinformatics, 26, 722–729. [DOI] [PubMed] [Google Scholar]
- Li H. (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2014) Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics, 30, 2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. (2008a) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res, 18, 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. (2009a) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. et al. (2008b) SOAP: short oligonucleotide alignment program. Bioinformatics, 24, 713–714. [DOI] [PubMed] [Google Scholar]
- Li R. et al. (2009b) SNP detection for massively parallel whole-genome resequencing. Genome Res, 19, 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H. et al. (2016) Low base-substitution mutation rate in the ciliate Tetrahymena thermophila. Genome Biol Evol, 8, 3629–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ratón M. et al. (2014) OptimalCutpoints: an R package for selecting optimal cutpoints in diagnostic tests. Journal of Statistical Software, 61, 1–36. [Google Scholar]
- Lynch M. (2009) Estimation of allele frequencies from high-coverage genome-sequencing projects. Genetics, 182, 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhis N., Jones S.J.M. (2010) High quality SNP calling using Illumina data at shallow coverage. Bioinformatics, 26, 1029–1035. [DOI] [PubMed] [Google Scholar]
- Maruki T., Lynch M. (2015) Genotype-frequency estimation from high-throughput sequencing data. Genetics, 201, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res, 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.E. et al. (2011) Mapping copy number variation by population-scale genome sequencing. Nature, 470, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan O. et al. (2012) A cross-sample statistical model for SNP detection in short-read sequencing data. Nucleic Acids Res, 40, e5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G. et al. (2013) Rare variant detection using family-based sequencing analysis. Proc Natl Acad Sci U S A, 110, 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu A. et al. (2013) DeNovoGear: de novo indel and point mutation discovery and phasing. Nat Methods, 10, 985–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed S. et al. (2009) Extremes of clinical and enzymatic phenotypes in children with hyperinsulinism caused by glucokinase activating mutations. Diabetes, 58, 1419–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvedebrink T. (2010) Overdispersion in allelic counts and θ-correction in forensic genetics. Theor Popul Biol, 78, 200–210. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G.A. et al. (2013) From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Current Protocols in Bioinformatics 43:11.10.1-11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J.D. et al. (2014) Estimating genotype error rates from high-coverage next-generation sequence data. Genome Res, 24, 1734–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr A. et al. (2015) Identification of low-confidence regions in the pig reference genome (Sscrofa10.2). Front Genet, 6, 338.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J.C., Federhen S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol, 266, 554–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.