Abstract
Motivation
Whole-genome sequencing (WGS) data are being generated at an unprecedented rate. Analysis of WGS data requires a flexible data format to store the different types of DNA variation. Variant call format (VCF) is a general text-based format developed to store variant genotypes and their annotations. However, VCF files are large and data retrieval is relatively slow. Here we introduce a new WGS variant data format implemented in the R/Bioconductor package ‘SeqArray’ for storing variant calls in an array-oriented manner which provides the same capabilities as VCF, but with multiple high compression options and data access using high-performance parallel computing.
Results
Benchmarks using 1000 Genomes Phase 3 data show file sizes are 14.0 Gb (VCF), 12.3 Gb (BCF, binary VCF), 3.5 Gb (BGT) and 2.6 Gb (SeqArray) respectively. Reading genotypes in the SeqArray package are two to three times faster compared with the htslib C library using BCF files. For the allele frequency calculation, the implementation in the SeqArray package is over 5 times faster than PLINK v1.9 with VCF and BCF files, and over 16 times faster than vcftools. When used in conjunction with R/Bioconductor packages, the SeqArray package provides users a flexible, feature-rich, high-performance programming environment for analysis of WGS variant data.
Availability and Implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
As the cost of DNA sequencing rapidly decreases, whole-genome sequencing (WGS) is generating data at an unprecedented rate (Goodwin et al., 2016; Metzker, 2010). Scientists are being challenged to manage data sets that are terabyte-sized, contain diverse types of data and complex data relationships. Data analyses of WGS require a general file format for storing genetic variants including single nucleotide variations (SNVs), insertions and deletions and structural variants. The variant call format (VCF) is a generic and flexible format for storing DNA polymorphisms developed for the 1000 Genomes Project (Danecek et al., 2011; 1000 Genomes Project Consortium, 2010) that is the standard WGS format in use today. VCF is a text format usually stored in compressed files that supports rich annotations and relatively efficient data retrieval. However, VCF files are large and the computational burden associated with large-scale data retrieval from text files can be significant for a WGS study with thousands of samples. As the number of individuals sequenced increases from thousands to tens of thousands and even more in studies like the Precision Medicine Initiative program (Collins and Varmus, 2015), the need for an efficient and flexible and high-performance environment to store and analyze WGS variant data becomes essential.
The binary variant call format (BCF) was developed as a complementary format to extract information from VCF files without having to parse text (Danecek et al., 2011; Li, 2011). VCF and BCF files are commonly accessed using either VCFtools (https://vcftools.github.io/index.html) or BCFtools (https://samtools.github.io/bcftools). VCFtools consists of a perl module with APIs for manipulating files and a binary executable providing general analysis routines including the calculations of allele frequency, linkage disequilibrium statistics and fixation index population statistics. BCFtools is designed as a faster replacement for most of the perl VCFtools commands.
Other file formats have been developed to improve upon VCF and BCF. For example, BGT and GQT were developed for fast genotype queries (Layer et al., 2016; Li, 2016). In either VCF or BCF, a single genotype requires at least a full byte of storage and genotypic data are usually compressed by the zlib algorithm (http://www.zlib.net). Li (2016) developed a new binary format, BGT, specific to genotypes, which represents diploid genotypes as a 2-bit integer matrix and utilizes the positional Burrows-Wheeler transform (PBWT) to store bit matrices (Durbin, 2014). In general, PBWT has a higher compression ratio and faster access capability than BCF. However, BGT format only allows three different alleles at a site.
Complementary to the variant-centric indexing strategy implemented in VCF, BCF and BGT, Layer et al. (2016) proposed a sample-centric storage approach (GQT) for variant data by transposing the genotypes originally stored in VCF files. GQT represents sample genotypes as compressed bitmap indices and provides efficient algorithms for genotype queries, allele counting and other calculations. However, GQT query is inefficient when analyzing a small genomic region and does not compress well compared with other new formats (Li, 2016).
GenomicsDB is used by the Broad Institute (https://github.com/Intel-HLS/GenomicsDB) for storing and processing variant data, and it is built on top of the Intel TileDB system (http://istc-bigdata.org/tiledb/) which is optimized for both dense and sparse multi-dimensional arrays (Papadopoulos et al., 2016). However, none of these binary formats are integrated with the R programming environment.
To provide an efficient alternative to VCF and BCF for WGS variant data, we developed a new data format and accompanying Bioconductor package, ‘SeqArray’. Key features of the SeqArray package are efficient storage including multiple high compression options, data retrieval by variant or sample subsets, support for parallel access and computing, and C ++ integration in the R programming environment. The SeqArray package provides R functions for efficient block-wise computations, and enables scientists to develop custom R scripts for exploratory data analysis. In addition, an R ecosystem built on top of the SeqArray package is available in the Bioconductor repository, including the SeqVarTools, SNPRelate and GENESIS packages which provide a wide range of tools for WGS variant analysis.
2 Materials and methods
2.1 Implementation
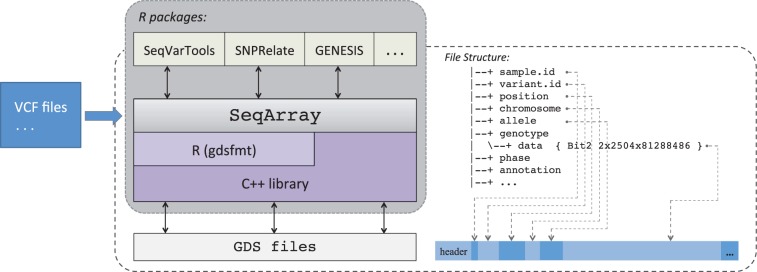
The SeqArray file format is built on top of the Genomic Data Structure (GDS) format (Zheng et al., 2012), a flexible and scalable data container with a hierarchical structure that can store multiple array-oriented data sets. GDS supports large-scale genomic datasets, especially those which are much larger than the available main memory, by providing memory- and performance-efficient operations specifically designed for integers of less than 8 bits, since a diploid genotype usually occupies fewer bits than a byte. Data compression and decompression are available with relatively efficient random access. GDS is implemented using an optimized C ++ library (CoreArray, http://corearray.sourceforge.net) and a high-level R interface is provided by the platform-independent R package gdsfmt (http://bioconductor.org/packages/gdsfmt). Figure 1 shows the relationship between a SeqArray file and the underlying infrastructure upon which it is built. All of the information represented in a VCF file can be captured by a SeqArray file. At a minimum, the data fields required are sample and variant identifiers, chromosome, position and reference and alternate alleles. The SeqArray package is available at Bioconductor (Gentleman et al., 2004; R Core Team, 2016) under the GNU General Public License v3: http://www.bioconductor.org/packages/SeqArray.
Fig. 1.
SeqArray framework and ecosystem. The SeqArray file format is built on top of the GDS format, a generic data container with hierarchical structure for storing multiple array-oriented data sets. Access to GDS is either through an efficient C ++ library or a high-level R interface. The SeqArray package creates GDS files and offers functionality specific to WGS variant data. At a minimum a SeqArray file contains sample and variant identifiers, position, chromosome and reference and alternate alleles for each variant. The functionality of the SeqArray package is extended by other R/Bioconductor packages such as SeqVarTools, SNPRelate and GENESIS that provide an ecosystem for WGS analyses on top of the SeqArray file format
The SeqArray file format stores genotypes in a 2-bit array with ploidy, sample and variant dimensions. If two bits cannot represent all alleles at a site, a 2-bit matrix is appended to store additional bits according to the variant coordinate; hence any number of alleles can be stored including a missing value. An index vector associated with genotypes is used to indicate how many bits are used for each variant. An example of genotype decoding with graphical illustrations is shown in Supplementary Figure S1.
The annotations in a SeqArray file can be integers, floating point numbers or characters. For a variant, variable-length vectors in an annotation are padded with missing values, and all vectors are packed together in an array. If each variant has different length, an extra vector is used to store the length information. Per-sample read depth (DP), allelic depths, conditional genotype quality (GQ) and Phred-scaled genotype likelihoods (PLs) are stored as integers according to the VCF Specification (http://samtools.github.io/hts-specs/). Variable-length integer encoding is used to store these variables to reduce the file size, e.g. a 32-bit integer between −64 and 63 is saved in a byte instead of a double word. The encoding details are shown in Supplementary Figure S2.
Currently, two primary lossless compression methods are available for the SeqArray file format: either the zlib or Lempel-Ziv Markov chain (LZMA) algorithm. LZMA has a higher compression ratio than zlib, although it takes more random-access memory and time to compress. To enable efficient random access of compressed data, a storage strategy using independently compressed data blocks is employed with an indexing strategy (see Supplementary Fig. S3). Independent data blocks allow for fast performance for data filtering since not all data have to be decompressed.
2.2 Parallel computing
The SeqArray package utilizes the framework implemented in the R package ‘parallel’ (Rossini et al., 2007; R Core Team, 2016) to support multiple forms of parallelism. First, the majority of WGS variant data consists of bi-allelic SNVs (1000 Genomes Project Consortium, 2010), four of which can be encoded in a single byte and operated on simultaneously. Second, multi-core parallelism on symmetric multiprocessing (SMP) computer architectures is supported via POSIX forking on Unix-like systems. Third, job-level parallelism on loosely coupled compute clusters is supported via communication over sockets or by the Message Passing Interface. Most importantly, all of these forms of parallelism can be combined together allowing high-speed access and operations on hundreds of different parts of the genome simultaneously.
When multiple processes are writing data, the SeqArray package stores the data in a separate local file for each process and merges these files together when the processes finish. Since data are stored in independent blocks, merging files is easily done by copying data directly without having to uncompress and re-compress it.
Streaming SIMD Extensions 2 (SSE2) is a programming technique supported on Intel architectures to expedite calculations when an algorithm can be expressed using vector operations. Each SSE2 register has 128 bits and can operate on 16 bytes or four 32-bit integers. In the situation that an allele is stored in a 32-bit integer for operations, four alleles can be unpacked with SSE2 instructions simultaneously from a 2-bit array. Bit unpacking can be further accelerated if a byte is used for storing an allele and an SSE register saves 16 alleles together. Other operations such as allele counting can be accelerated with SSE2 instructions by scanning multiple genotypes at the same time. Advanced Vector Extensions 2 (AVX2) instructions extend SSE2 registers from 128 bits to 256 bits and are available in newer Intel microprocessors. The SeqArray package also provides an AVX2 implementation.
3 Performances
We used data from the 1000 Genomes Project Phase 3 (1000G) to compare the SeqArray package with other popular file formats and toolsets. The 1000G data consists of 2504 individuals and 81 271 745 autosomal variants distributed in 22 compressed VCF files. Benchmarks were run on an Amazon Web Services instance (c4.8xlarge − 36 cores, 60 Gb main memory, 2.9 GHz Intel Xeon Haswell) with a 200 Gb solid-state disk drive. The software used were zlib v1.2.8, liblzma v5.2.2, R v3.3.2, htslib v1.3.1, bcftools v1.3.1, vcftools v0.1.15, bgt v1.0-r282, PLINK v1.9 (stable beta 3.44), gdsfmt v1.10.1 and SeqArray v1.14.1. All C/C ++ codes were compiled with GCC v4.8.3 with ‘-O2’ optimization. Since SSE2 instructions are well supported by current x86-64 processors while AVX2 instructions are only available on new computers, we report wall-clock times in an SSE2-enabled and AVX2-disabled situation.
Table 1 shows the time and file size to convert 22 autosomal compressed VCF files to a single compressed output file in BCF, BGT and SeqArray file formats. The R scripts used for testing are shown in Supplementary Figure S4. The first row of the table shows the time to merge the 22 compressed VCF files into a single compressed VCF file. This provides a baseline for comparison since no actual format conversion is conducted, only recompression. The VCF to VCF and VCF to BCF conversion were both performed using the bcftools software (https://samtools.github.io/bcftools). The encoded genotype sizes of the VCF and BCF files are not shown since neither file format stores genotypes and annotations separately. The information density is 5.5 genotypes per bit for the SeqArray file with zlib compression, 8.0 for BGT and 14.1 for SeqArray/lzma, respectively. The SeqArray files are smaller than the VCF and BCF files and the SeqArray file with lzma compression is smaller than the BGT file. The conversion times for the SeqArray package are similar to those for the implememtation in bcftools and the BGT utilities, and they can be further reduced when multiple cores are used. The formats VCF, BCF and SeqArray zlib utilize the same compression algorithm to encode data, and the smaller file size for SeqArray (5.7 versus 14.4 and 12.3 Gb) may have an additional performance advantage over the others since less data is written to the file system.
Table 1.
Format conversion and compression. Shown are the time (in hours) and file size (in gigabytes) to convert 22 autosomal compressed genotype VCF files from the 1000 Genomes Project Phase 3 (2504 samples, 81M variants) to a single compressed output file
| Software | Output file format | Compression algorithm | Total file size | Encoded genotype size | Information densitya | Time: 1 core | 4 cores |
|---|---|---|---|---|---|---|---|
| bcftools | VCF.gzb | zlib | 14.4 Gb | — | — | 2.6 h | 0.9 h |
| bcftools | BCF | zlib | 12.3 Gb | — | — | 4.5 h | 3.3 h |
| BGT | BGTc | pbwt | 3.5 Gb | 3.0 Gb | 8.02 | 2.4 h | — |
| SeqArray | SeqArray | zlib | 5.7 Gb | 4.3 Gb | 5.47 | 2.3 h | 0.9 h |
| SeqArray | SeqArray | lzma | 2.6 Gb | 1.7 Gb | 14.08 | 6.3 h | 2.2 h |
the number of genotypes per encoded bit.
merging 22 VCF files into a single file with no format conversion.
pbwt, positional Burrows-Wheeler transform; BGT, does not store the phasing states and annotations.
The comparison of storage usage with genotypes and per-sample annotations from the 1000G is shown in Table 2. The annotations are per-sample DP and PLs, and the R scripts for re-formatting VCF files are shown in Supplementary Figure S10. The SeqArray format with zlib compression reduces disk space 16% compared with the VCF.gz files and further 33% using lzma compression.
Table 2.
Comparison of storage usage with genotypes, per-sample read DP and PLs
| File format | Compression algorithm | Total file size | File size ratio |
|---|---|---|---|
| VCF.gz | zlib | 338.2 Gb | 1.00 |
| BCF | zlib | 309.1 Gb | 0.91 |
| SeqArray | zlib | 282.5 Gb | 0.84 |
| SeqArray | lzma | 227.6 Gb | 0.67 |
Note: Shown is the total file size (in gigabytes) of 22 autosomes from the 1000 Genomes Project Phase 3 (2504 samples and 77M variants that have the DP and PL annotations).
Table 3 measures the time to retrieve and decompress the genotype data and calculate the allele frequencies. The C and R scripts used for benchmarking are shown in Supplementary Figures S5 and S6. The SeqArray package and BGT are both more efficient than the htslib library and SeqArray is more efficient than BGT. In the tests of VCF and BCF files, we estimate the decompression speed via a C implementation with the htslib library (https://github.com/samtools/htslib). Parsing the VCF text file is almost 12-fold slower than extracting genotypes from the BCF file for 1000G data, when the htslib implementation is used. The SeqArray package utilizes the zlib algorithm to encode and decode compressed data as well as VCF and BCF, and the 3-fold speed improvement relative to BCF could be explained by the decrease in file size. BGT decodes up to 455 million genotypes per second for 1000G data which is consistent with the speed reported in Li (2016), while the SeqArray package is 1.4–1.7 times faster than BGT. When multiple cores were used, the running time of the SeqArray package decreases almost linearly with the number of cores. In addition, calculating allele frequencies with the SeqArray package is more than 16 times faster than VCFtools using VCF and BCF files. The VCF/BCF implementation in PLINK v1.9 (Chang et al., 2015) is significantly faster than VCFtools, but the calculation is still slower than the SeqArray package. Parallel access to VCF, BCF and BGT files is not currently provided in their utilities, so their multi-core running times are not shown here.
Table 3.
Genotype decompression and allele frequency calculation using the 1000 Genomes data (in minutes)
| Software | Input file format | Compression algorithm | Decompression ratea | Time: 1 core | 4 cores | 8 cores | 16 cores |
|---|---|---|---|---|---|---|---|
| Genotype decompression | |||||||
| htslib | VCF.gz | zlib | 18.6 | 182.0 m | |||
| htslib | BCF | zlib | 213.5 | 15.87 m | |||
| BGT | BGT | pbwt | 455.3 | 7.45 m | |||
| SeqArray | SeqArray | zlib | 759.7 | 4.46 m | 1.16 m | 0.59 m | 0.30 m |
| SeqArray | SeqArray | lzma | 629.1 | 5.39 m | 1.42 m | 0.73 m | 0.37 m |
| Allele frequency calculation | |||||||
| vcftools | VCF.gz | zlib | — | 446.1 m | |||
| vcftools | BCF | zlib | — | 125.3 m | |||
| PLINK v1.9 | VCF.gz | zlib | — | 61.9 m | |||
| PLINK v1.9 | BCF | zlib | — | 37.7 m | |||
| SeqArray | SeqArray | zlib | — | 6.65 m | 2.45 m | 1.02 m | 0.66 m |
| SeqArray | SeqArray | lzma | — | 7.56 m | 2.79 m | 1.15 m | 0.75 m |
million genotypes per second on one core.
The performance of the SeqArray package using SSE2 and AVX2 instructions is shown in Supplementary Table S1. We measured the elapsed time for genotype decompression using the same R code (Supplementary Fig. S5b), but different compiler settings. The compression libraries zlib and lzma return a 2-bit array, and we manually optimize the performance of bit unpacking from this 2-bit array to a 8-bit or 32-bit integer array. The SSE2 instructions are almost twice as fast as the implementation without SSE2, and AVX2 shows a slight improvement compared with SSE2.
To evaluate random access performance we randomly selected 250 regions of one million base pairs each for 20 randomly selected populations and exported the genotypes to a VCF file. We used the SeqArray package, bcftools and the BGT utilities for these queries. As shown in Supplementary Table S2, the SeqArray package with zlib compression performs similarly to BGT, and both are faster than bcftools. The SeqArray package using lzma compression is slower for random indexing compared with SeqArray/zlib. Note that random access performance depends on the internal size of the independently compressed data blocks. The running time could be reduced if a smaller block size is used, but it would also decrease the compression ratio. The default block size in the SeqArray file format is tuned for gigabyte-scale data sets. The random-access results suggest the SeqArray package performs at least as well as other tools when real-time response to complex genotype queries is needed.
4 R integration
The R/Bioconductor package SeqArray provides functions for efficient file manipulation and parallel execution. Importantly, the SeqArray package has been extended by other R packages for WGS variant data analyses and integrates smoothly with core Bioconductor data structures. Below we give several examples, and a full tutorial is available in the package vignette ‘R Integration’.
4.1 SeqArray features
4.1.1 Key R functions
Genotype data and annotations are stored in an array-oriented manner, providing efficient data access using the R programming language. Table 4 lists five key functions provided in the SeqArray package and many data analyses can be done using just these functions.
Table 4.
Key R functions in the SeqArray package
| Function | Description |
|---|---|
| seqVCF2GDS | Reformat a VCF file |
| seqSetFilter | Define a subset of samples or variants |
| seqGetData | Get data with a defined filter |
| seqApply | Apply a user-defined function over array margins |
| seqParallel | Apply a function in parallel |
seqVCF2GDS() converts VCF files to the SeqArray file format. Multiple cores in an SMP architecture within one or more compute nodes in a compute cluster can be used simultaneously to reformat the data. seqVCF2GDS() utilizes R’s connection interface to read VCF files incrementally. This allows importing uncompressed VCF data without the need to first save the entire file such as from an http/ftp URL or the standard output of a command-line tool. The LZMA compression algorithm is the default option in seqVCF2GDS().
seqSetFilter() and seqGetData() can be used together to retrieve data for a selected set of samples from a defined genomic region. GRanges and GRangesList objects defined in the Bioconductor core packages are supported via seqSetFilter() (Gentleman et al., 2004; Lawrence et al., 2013).
seqApply() applies a user-defined function to the array margins of genotypes and annotations. The function that is applied can be defined in R as is typical, or via C/C ++ code using the Rcpp package (Eddelbuettel et al., 2011). seqParallel() utilizes the facilities in the packages parallel and BiocParallel (Gentleman et al., 2004; Rossini et al., 2007; R Core Team, 2016) to perform calculations on a SeqArray file in parallel.
4.1.2 Calculating allele frequencies
We illustrate the function seqApply() by implementing an example to calculate the frequency of the reference allele across all chromosomes. An optimized version of this calculation is natively provided in the SeqArray package function seqAlleleFreq().
# open a SeqArray file
file <- seqOpen(seqExampleFileName(“KG_Phase1”))
# user-defined allele frequency function
CalcFreq <- function(x) mean(x==0, na.rm = TRUE)
# apply the function to a genotype margin
seqApply(file, “genotype”, as.is=“double”,
margin=“by.variant”, FUN = CalcFreq)
Here, file is a SeqArray file, ‘genotype’ is the variable in the SeqArray file to act on, as.is indicates the result type, margin specifies that the user function should be applied to each variant. The variable x in the user-defined function is an allele-by-sample integer matrix at a single variant site and 0 denotes the reference allele.
The preceding code can be optimized by writing the allele frequency calculation function in C ++. The Rcpp package simplifies integration of compiled C ++ code with R and allows C ++ functions to be dynamically defined inline and the C ++ function name passed to seqApply(), as shown in Supplementary Figure S7. Using C ++ is several times faster than the R implementation in this situation.
The calculations can be run in parallel. Below, the parameter parallel specifies the use of four cores. The genotypes of a SeqArray file are automatically split into non-overlapping parts according to different variants or samples, and the results from client processes collected internally.
seqApply(file, “genotype”, as.is=“double”,
margin=“by.variant”, FUN = CalcFreq, parallel = 4)
4.1.3 Principal component analysis implementation
Principal component analysis (PCA) is a common tool used in exploratory data analysis for high-dimensional data. PCA often involves the calculation of covariance matrix, and the following R code implements the calculation proposed in Patterson et al. (2006). The user-defined function computes the covariance matrix for each variant and adds up to a total matrix s. The operator ‘%o%’ calculates the outer product of normalized genotypes, and the argument ‘.progress’ enables the display of progress information during the calculation.
# covariance variable with an initial value
s <- 0
seqApply(file, “$dosage”, function(x) {
p <- 0.5 * mean(x, na.rm = TRUE)# allele freq
g <- (x - 2*p)/sqrt(p*(1-p))# normalization
g[is.na(g)] <- 0 # missing values
s <<- s + (g %o% g) # update the cov matrix s
# in the parent envir.
}, margin=“by.variant”,.progress = TRUE)
# scaled by the number of samples over the trace
s <- s * (nrow(s)/sum(diag(s)))
# eigen-decomposition
eig <- eigen(s)
The algorithmic efficiency of the preceding R code can be significantly improved by blocking the computations. seqBlockApply() applies the user-defined function on a data block instead of a single variant, and the corresponding implementation is shown in Supplementary Figure S8.
seqParallel() utilizes the facilities offered by the R parallel or BiocParallel package to perform calculations within a cluster or SMP environment, and the genotypes are automatically split into non-overlapping parts. The R parallel implementation is shown in Supplementary Figure S9, and a C-optimized and memory-efficient function is also available in the SNPRelate package (see Section 4.4).
4.2 Integration with bioconductor data structures
The example below uses Bioconductor core packages to perform common queries and retrieve data from a SeqArray file. The GRanges and GRangesList classes in the GenomicRanges package manipulate genomic range data and can be used in the function seqSetFilter() to define a data subset. For example, the annotation information of each exon, the coding range and transcript ID are stored in the TxDb.Hsapiens.UCSC.hg19.knownGene object for the UCSC known gene annotations defined for hg19. The code fragment below loads this object and uses the GenomicFeatures package function exonsBy() to return a GRangesList object for all known exons within each gene. seqSetFilter() takes this object as an argument and only exports to a VCF file exonic variants.
library(TxDb.Hsapiens.UCSC.hg19.knownGene)
# get the exons grouped by gene
txdb <- TxDb.Hsapiens.UCSC.hg19.knownGene
txs <- exonsBy(txdb, “gene”)
seqSetFilter(file, txs)
seqGDS2VCF(file, “exons.vcf.gz”)
The SeqArray package can also export data with selected variants and samples as a VCF object for use with the VariantAnnotation package (Obenchain et al., 2014).
4.3 Integration with SeqVarTools
The SeqVarTools package (http://www.bioconductor.org/packages/SeqVarTools) extends the SeqArray package by providing methods for many tasks common to quality control and analysis of WGS variant data. Methods include: transition/transversion ratio, heterozygosity and homozygosity rates, singleton counts, Hardy-Weinberg equilibrium, Mendelian error checking, and linear and logistic regression. In the example below SeqVarTools defines a new class (SeqVarData) to link the information present in a SeqArray file with additional sample annotation and runs a regression analysis on a subset of the samples. KG_P1_SampData is an AnnotatedDataFrame with columns sample.id, sex, age, and phenotype, where the identifiers in sample.id match those in the SeqArray file.
library(SeqVarTools)
# link sample data to SeqArray file
data(KG_P1_SampData)
seqData <- SeqVarData(file, KG_P1_SampData)
# select female samples
female <- sampleData(seqData)$sex == “female”
seqSetFilter(seqData, sample.sel = female)
# run linear regression
res <- regression(seqData, outcome=“phenotype”,
covar=“age”)
4.4 Integration with SNPRelate
The SNPRelate package (http://www.bioconductor.org/packages/SNPRelate) provides parallel implementations of relatedness and PCA to detect and estimate population structure and cryptic relatedness. SNPRelate was originally designed for analysis of bi-allelic SNPs in GWAS studies where the implementation was optimized with SIMD instructions and multithreading (Zheng et al., 2012). In order to analyze SNVs from sequence data, SNPRelate has been rewritten to take the dosages of the reference alleles as an input genotype matrix from SeqArray files. Therefore, no format conversion is required for WGS analyses.
PCA is implemented in the SNPRelate function snpgdsPCA() using both exact and randomized algorithms (Galinsky et al., 2016; Patterson et al., 2006). The randomized matrix algorithm is designed to reduce the running time for a large number of study individuals (i.e. >10 000 samples). Relatedness analyses include PLINK’s method of moment, KING kinship methods, GCTA genetic relationship matrix and individual-perspective beta estimator (Manichaikul et al., 2010; Purcell et al., 2007; Weir and Zheng, 2015; Yang et al., 2011; Zheng and Weir, 2016). These algorithms are all computationally efficient and optimized with SIMD instructions. In addition, the calculations of fixation index (Fst), a widely used statistic measuring the genetic difference between populations, is available in SNPRelate using either all variants or sliding windows (Weir and Cockerham, 1984; Weir and Hill, 2002; Weir et al., 2005).
4.5 Integration with GENESIS
The GENESIS package offers methodology for estimating and accounting for population and pedigree structure in genetic analyses. It provides functions to perform the PC-AiR and PC-Relate methods (Conomos et al., 2015, 2016). PC-AiR performs PCA on genome-wide genotypes taking into account known or cryptic relatedness in the study samples. PC-Relate uses ancestry representative principal components to estimate measures of recent genetic relatedness. GENESIS also includes support for SeqArray files in mixed model association testing and aggregate burden and SKAT tests of rare variants.
5 Discussion
In this article, we have presented a new file format, SeqArray, and associated toolset to provide an efficient alternative to VCF and BCF files for storing WGS variant data. The SeqArray file format is built on top of the GDS format which can store multiple array-oriented data sets in a hierarchical structure. It stores genotypes in two bits and supports multiple lossless compression options. Benchmarks show file sizes ∼5× smaller than VCF and BCF and 35% smaller than the most storage efficient alternative (BGT). The SeqArray package uses an optimized C ++ library and supports parallel access by both multi-core SMP computers and loosely coupled compute clusters. In our testing simple operations such as fetching genotypes were two to three times faster compared with the htslib library with BCF files. For the allele frequency calculation, the SeqArray implementation is over 5 times faster than PLINK v1.9 with VCF and BCF files, and over 16 times faster than vcftools. The SeqArray package is available on the Bioconductor website and enables analysts to develop custom R scripts for exploratory data analysis. Wide functionality for WGS analysis is provided by the SeqVarTools, SNPRelate and GENESIS packages which extend the functionality of SeqArray.
Other file formats have been developed to improve upon VCF and BCF. BGT and GQT were developed for fast genotype queries (Layer et al., 2016; Li, 2016). GenomicsDB is used by the Broad Institute for storing and processing sequence data, and it is built on top of the TileDB system. However, none of these are integrated with the R programming environment. The HDF5 format (https://www.hdfgroup.org/HDF5) provides similar utilities to the GDS format upon which the SeqArray format is built.
The computational efficiency of file formats rely on both the design and implementation. When compared with the implementations of other formats, the gdsfmt and SeqArray packages have been highly optimized and offer performance-efficient operations specifically designed for integers of less than 8 bits typically found in genomic data.
The SeqArray package provides R functions for convenient block-wise computations and enables users to easily extend the functionality with R codes or C/C ++ codes integrated with the Rcpp package. The Bioconductor integration enables scientists to reuse the functionality of exploratory data analysis and statistical modeling in existing R packages. It also encourages new developers to write custom R packages to extend analyses of sequencing data.
Supplementary Material
Acknowledgements
We thank Roy Kuraisa and other members of the Genetic Analysis Center in the Department of Biostatistics at University of Washington for using and testing the utilities in the SeqArray package.
Funding
This work was supported, in part, by NIH Grant [GM099568].
Conflict of Interest: none declared.
References
- 1000 Genomes Project Consortium. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.C. et al. (2015) Second-generation plink: rising to the challenge of larger and richer datasets. GigaScience, 4, 7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F.S., Varmus H. (2015) A new initiative on precision medicine. N. Engl. J. Med., 372, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos M.P. et al. (2015) Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol., 39, 276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos M.P. et al. (2016) Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet., 98, 127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. (2011) The variant call format and vcftools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R. (2014) Efficient haplotype matching and storage using the positional burrows-wheeler transform (pbwt). Bioinformatics, 30, 1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddelbuettel D. et al. (2011) Rcpp: Seamless R and C ++ integration. J. Stat. Softw., 40, 1–18. [Google Scholar]
- Galinsky K.J. et al. (2016) Fast principal-component analysis reveals convergent evolution of ADH1B in europe and east asia. Am. J. Hum. Genet., 98, 456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C. et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol., 5, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S. et al. (2016) Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet., 17, 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. et al. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol., 9, e1003118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer R.M., Exome Aggregation Consortium, and Quinlan, A. R. et al. (2016) Efficient genotype compression and analysis of large genetic-variation data sets. Nat. Methods, 13, 63–65., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011) A statistical framework for snp calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2016) BGT: efficient and flexible genotype query across many samples. Bioinformatics, 32, 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A. et al. (2010) Robust relationship inference in genome-wide association studies. Bioinformatics, 26, 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker M.L. (2010) Sequencing technologies – the next generation. Nat. Rev. Genet., 11, 31–46. [DOI] [PubMed] [Google Scholar]
- Obenchain V. et al. (2014) VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics, 30, 2076–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos S. et al. (2016) The tiledb array data storage manager. Proc. VLDB Endow, 10, 349–360. [Google Scholar]
- Patterson N. et al. (2006) Population structure and eigenanalysis. PLoS Genet., 2, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2016). R: A Language and Environment for Statistical Computing.
- Rossini A.J. et al. (2007) Simple parallel statistical computing in R. J. Comput. Graph. Stat., 16, 399–420. [Google Scholar]
- Weir B.S., Cockerham C.C. (1984) Estimating F-statistics for the analysis of population structure. Evolution, 38, p 1358–1370. [DOI] [PubMed] [Google Scholar]
- Weir B.S., Hill W.G. (2002) Estimating F-statistics. Annu. Rev. Genet., 36, 721–750. [DOI] [PubMed] [Google Scholar]
- Weir B.S., Zheng X. (2015) SNPs and SNVs in forensic science. Forensic Sci. Int., 5, e267–e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B.S. et al. (2005) Measures of human population structure show heterogeneity among genomic regions. Genome Res., 15, 1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Weir B.S. (2016) Eigenanalysis of SNP data with an identity by descent interpretation. Theor. Popul. Biol., 107, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. et al. (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics, 28, 3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.