Abstract
Summary
Precision oncology is an approach that accounts for individual differences to guide cancer management. Omics signatures have been shown to predict clinical traits for cancer patients. However, the vast amount of omics information poses an informatics challenge in systematically identifying patterns associated with health outcomes, and no general purpose data mining tool exists for physicians, medical researchers and citizen scientists without significant training in programming and bioinformatics. To bridge this gap, we built the Omics AnalySIs System for PRecision Oncology (OASISPRO), a web-based system to mine the quantitative omics information from The Cancer Genome Atlas (TCGA). This system effectively visualizes patients’ clinical profiles, executes machine-learning algorithms of choice on the omics data and evaluates the prediction performance using held-out test sets. With this tool, we successfully identified genes strongly associated with tumor stage, and accurately predicted patients’ survival outcomes in many cancer types, including adrenocortical carcinoma. By identifying the links between omics and clinical phenotypes, this system will facilitate omics studies on precision cancer medicine and contribute to establishing personalized cancer treatment plans.
Availability and implementation
This web-based tool is available at http://tinyurl.com/oasispro; source codes are available at http://tinyurl.com/oasisproSourceCode.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Oncology is a targeted field for precision medicine (National Research Council Committee on A Framework for Developing a New Taxonomy of Disease, 2011). Cancers cause a significant number of deaths and loss of function worldwide (Ashley, 2015; Collins and Varmus, 2015; Siegel et al., 2016), and cancer patients exhibit diverse clinical traits and prognoses (National Research Council Committee on A Framework for Developing a New Taxonomy of Disease, 2011). Previous research has shown strong correlations between molecular profiles and clinical outcomes in various cancers (Ashley, 2015; Collins and Varmus, 2015). A number of machine learning algorithms proved to be useful in revealing previously unrecognized molecular patterns associated with clinical phenotypes (Collins and Varmus, 2015), and the development of cloud-based computing has facilitated the implementation of machine learning models.
However, most data mining tools are not tailored for predicting clinically important variables nor are they accessible to physicians or researchers without significant training in data science (Mirnezami et al., 2012). Although many state-of-the-art data mining methods can provide novel insights into the omics data (Han et al., 2011; Snyder, 2016; Yu et al., 2016; Yu and Snyder, 2016), such tools are generally inaccessible to clinicians. In addition, the size of raw human omics data is on the order of terabytes (Larranaga et al., 2006), making it difficult to manipulate without a cloud-based system. The absence of a user-friendly web-based omics data analysis system prevents timely discovery of cancer biomarkers and hinders the study of biological mechanisms leading to different disease phenotypes.
To address this gap, we built the Omics Analysis System for Precision Oncology (OASISPRO), a cloud-based omics analysis tool for clinical prediction. We established data visualization modules that effectively summarized the clinical characteristics of cancer patients and built a data mining interface that allows users to discover associations between omics profiles and clinical traits, such as the extent of tumor and survival outcomes. Our system enables clinicians, medical researchers, as well as citizen scientists to explore the omics data with ease, provides biological insights into cancer development and generates robust clinical prediction models that guide clinical cancer managements. This system facilitates the development of precision oncology, which will improve the quality of care for cancer patients.
2 Methods and results
We built the OASISPRO system to visualize and analyze transcriptomics, proteomics, microRNA, DNA methylation and clinical data for all available cancer types in TCGA (Larranaga et al., 2006; Taylor, 2010; Yu and Snyder, 2016). This system automatically obtains the updated omics and clinical data of all cancer patients from the TCGA Data Portal, processes them, formats the results into data matrices, visualizes the clinical as well as quantitative omics data through interactive web-based modules, compares the difference of quantitative omics profiles in patients with different phenotypes, and analyzes the data using machine learning methods with desired input and outcome of interest specified by the users. The features were selected and the models were built with the training set, and the performance of the models was evaluated by a held-out test set or by cross-validation. A combination of omics inputs and parallel processing was implemented for machine learning tasks. The user interface is shown in Figure 1. A list of all available tumor types from TCGA is shown in Supplementary Table S1, and the features of OASISPRO are summarized in Supplementary Table S2. Implementation details and three example analyses that generate novel biomedical insights are included in Supplementary Methods and Results (Supplementary Figs S1–S3 and Table S3).
Fig. 1.
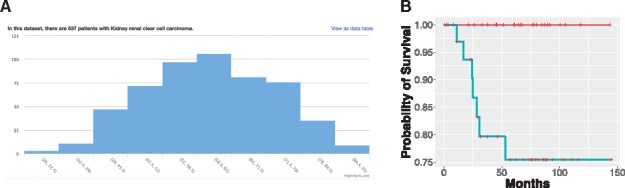
The OASISPRO system. We built several modules for data visualization and data analysis, including clinical and omics data visualization, classification of clinical phenotypes and survival analysis. Users can select any clinical variables of interest, and the system will generate the descriptive statistics and visualize the distribution of the selected variable. The distribution of continuous clinical data is summarized by a histogram. Categorical variables are presented by pie charts. Examples that (A) illustrate the distribution of age at initial pathological diagnosis of the TCGA renal clear cell carcinoma cohort and (B) survival prediction of the TCGA chromophobe renal cell carcinoma patients using their proteomics data are shown
3 Conclusion
Our system is the first cloud-based interface that integrates quantitative omics data and builds prediction models for cancer phenotypes, making the state-of-the-art machine-learning methods accessible to researchers of all backgrounds and citizen scientists. OASISPRO enables quantitative omics analysis, provides insights into the biology of cancer and empowers accurate clinical predictions. This data mining system is extensible to other diseases and health conditions. By building accurate prediction models for clinical phenotypes, this system will contribute to establishing personalized cancer treatment plans, thereby increasing the quality of care and reducing the cost of cancer management.
Supplementary Material
Acknowledgements
The authors thank Mr. Chung Yu Wang, Drs Jim Notwell, Sandeep Chinchali, Wei Wang, Christopher Ré, Gill Bejerano and Serafim Batzoglou for their helpful comments and suggestions. The authors thank the AWS Cloud Credits for Research, the Microsoft Azure Research Award and the NVIDIA GPU Grant Program for their support.
Funding
This work was supported by the National Human Genome Research Institute and the National Cancer Institute [5P50HG00773502 and 5U24CA16003605]; Harvard Data Science Postdoctoral Fellowship [to K.-H.Y.]; Howard Hughes Medical Institute International Student Research Fellowship [to K.-H.Y.]; and Winston Chen Stanford Graduate Fellowship [to K.-H.Y.].
Conflict of Interest: none declared.
References
- Ashley E.A. (2015) The precision medicine initiative: a new national effort. JAMA, 313, 2119–2120. [DOI] [PubMed] [Google Scholar]
- Collins F.S., Varmus H. (2015) A new initiative on precision medicine. N. Engl. J. Med., 372, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C.L. et al. (2011) An informatics-assisted label-free approach for personalized tissue membrane proteomics: case study on colorectal cancer. Mol. Cell. Proteomics, 10, M110 003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larranaga P. et al. (2006) Machine learning in bioinformatics. Brief. Bioinform., 7, 86–112. [DOI] [PubMed] [Google Scholar]
- Mirnezami R. et al. (2012) Preparing for precision medicine. N. Engl. J. Med., 366, 489–491. [DOI] [PubMed] [Google Scholar]
- National Research Council Committee on A Framework for Developing a New Taxonomy of Disease. ( 2011) Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. National Academies Press (US; ), Washington, D.C., USA. [PubMed] [Google Scholar]
- Siegel R.L. et al. (2016) Cancer statistics. CA Cancer J. Clin., 66, 7–30. [DOI] [PubMed] [Google Scholar]
- Snyder M. (2016) Genomics and Personalized Medicine: What Everyone Needs to Know. Oxford University Press, Oxford, UK. [Google Scholar]
- Taylor R.C. (2010) An overview of the Hadoop/MapReduce/HBase framework and its current applications in bioinformatics. BMC Bioinformatics, 11 (Suppl. 12), S1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K.H., Snyder M. (2016) Omics profiling in precision oncology. Mol. Cell. Proteomics, 15, 2525–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K.H. et al. (2016) Predicting ovarian cancer patients’ clinical response to platinum-based chemotherapy by their tumor proteomic signatures. J. Proteome Res., 15, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


