Abstract
Motivation
Protein function is directly related to amino acid residue composition and the dynamics of these residues. Centrality analyses based on residue interaction networks permit to identify key residues in a protein that are important for its fold or function. Such central residues and their environment constitute suitable targets for mutagenesis experiments. Predicted flexibility and changes in flexibility upon mutation provide valuable additional information for the design of such experiments.
Results
We combined centrality analyses with DynaMine flexibility predictions in a Cytoscape app called RINspector. The app performs centrality analyses and directly visualizes the results on a graph of predicted residue flexibility. In addition, the effect of mutations on local flexibility can be calculated.
Availability and implementation
The app is publicly available in the Cytoscape app store.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Understanding the function of proteins lies at the heart of fundamental biology. Tools that use either primary (sequence) or tertiary (3D) structure of proteins exist to aid in the identification of their function but few permit to integrate different approaches. Several years ago, a network-based approach was proposed as an alternative representation of a tertiary structure, thus enabling the use of graph analysis tools. This approach consisted of generating a residue interaction network (RIN) from a PDB structure, considering nodes as residues and edges as detected interactions—see e.g. (Amitai et al., 2004; Greene, 2012; Hu et al., 2014). Centrality analyses can then be performed on the graph to evaluate key residues in such a network. These centrality analyses are often based on measures that involve shortest path length calculations like closeness or betweenness, or following the approach of (del Sol et al., 2006), which uses the change of the characteristic path length under removal of individual nodes. They have been shown useful to identify key residues involved in the function of proteins, folding, allostery or long range interactions.
Cytoscape is a tool for the visualization and analysis of networks (Shannon et al., 2003). Recently, the structureViz Cytoscape app (Morris et al., 2007) integrated the possibility of generating RINs from a PDB structure through a connection made with the Chimera structure visualization and analysis program (Pettersen et al., 2004). This simplified the creation and analysis of such networks which can now be done with Cytoscape NetworkAnalyzer tools (Assenov et al., 2008) or i.e. with its RINalyzer app, designed specifically for the purpose of RIN analyses and visualization (Doncheva et al., 2011). While these enable centrality analyses such as closeness or betweenness, based on shortest path length (shortest path centralities), effective electric resistance (current flow centralities) or hitting time (random walk centralities) for distances between two nodes (Doncheva et al., 2012), none permit to perform residue centrality analyses as proposed by (del Sol et al., 2006) and associate a Z-score to each residue.
Besides its sequence and structure, a key ingredient of protein function is its dynamic behavior. Several tools are available to predict residue disorder in a protein, like PrDOS2 or DISOPRED2+3, which were respectively ranked first and second in CASP9 and CASP10 (Atkins et al., 2015), but they require evolutionary information and predictions take several minutes. DynaMine directly predicts backbone dynamics, does not require evolutionary information, with predictions taking only a few seconds and showing good performances in estimating disorder compared to other methods, including PrDOS2 (Cilia et al., 2013, 2014).
Here we present RINspector, a Cytoscape app that combines residue interaction network (RIN) centrality analyses with direct visualization of the predicted local flexibility of the associated sequence. In combination with the structureViz app, it links the three aspects of structure, network and flexibility, making it a useful tool for the investigation of protein structure and subsequent applications such as protein design or the selection of mutants for mutagenesis experiments.
2 Implementation and features
RINspector is written in JAVA as an app for Cytoscape. It incorporates DynaMine predictions from a JSON API. Prior to any RINspector analysis, a RIN needs to be generated, which can be done through the structureViz Cytoscape app (which communicates with Chimera) or any third-party program. RINspector can then perform the following operations on the network (see Supplementary Material for additional details):
Perform one or several centrality analyses: (i) residue centrality, based on the change of average shortest path length under removal of each node; (ii) betweenness centrality; (iii) closeness centrality.
Connect to the DynaMine server to retrieve the prediction of backbone dynamics for a selected protein chain. The backbone flexibility graph is displayed in the result panel.
Nodes and labels sizes and nodes color are adapted following the results of the calculations. Selection of residues highlights them in all three representations (structure, network or flexibility graph).
An on-line tutorial is available through the Cytoscape app store.
3 Example and validation of the approach
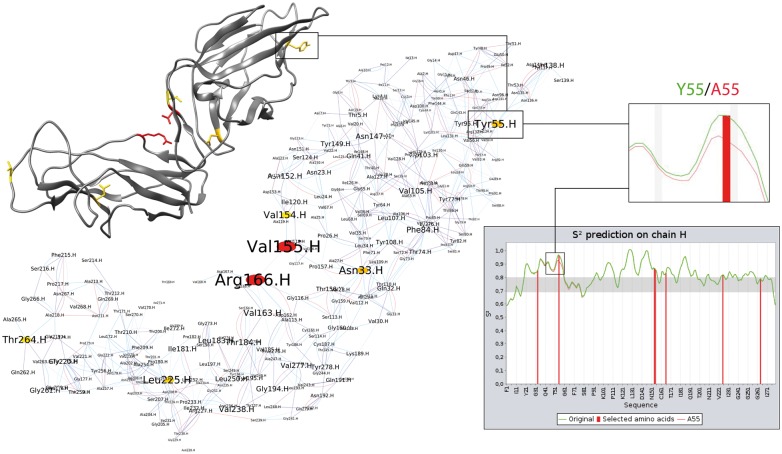
Figure 1 illustrates the use of RINspector on the FimH structure (PDB: 3JWN) of the commensal Escherichia coli F18 strain. FimH is the tip adhesin of the E.coli type 1 fimbriae, which are responsible for adhesion of the bacterium to target cells. FimH consists of two relatively rigid domains that show inter-domain flexibility. We generated the RIN using structureViz (default parameters, hydrogens added) and performed a residue centrality analysis. Seven central residues (V155, R166, N33, T55, L225, T264, V154) are identified on the protein (Z-score ≥ 2). Indeed, R166 has a very high Z-score and is known to be associated to AIEC (Adherent-Invasive Escherichia coli) bacteria when mutated into cysteine, histidine or serine (Dreux et al., 2013). Moreover, mutating Y55 into alanine was shown to have an impact on adhesion (Feenstra et al., 2017) in a close ortholog of FimH originating from the laboratory strain E.coli K12. The lower right-hand side of Figure 1 shows the obtained DynaMine flexibility prediction graph. The central residues are selected and therefore depicted with red bars. The inset zooms on the region surrounding Y55 and shows that a mutation into alanine increases the local flexibility. An investigation of the three-dimensional structure shows Y55 to be located close to the binding pocket (not shown). An increase in flexibility of this region is expected to have an adverse effect on binding efficiency and thus adhesion with respect to wild type FimH. (Feenstra et al., 2017) discussed such an indirect effect for T53, located close to Y55, behind the binding pocket, which when mutated into alanine makes FimH lose adhesion to mannose substrates.
Fig. 1.
Example of RINspector inter-module connectivity. Central residues are colored from yellow (Z-score = 2) to red (Z-score ≥ 4) on structure and network. The central residues are indicated with red bars in the flexibility graph. The zoom shows the increase in local flexibility for the Y55A mutation (green to red curve)
This example shows how RINspector can be used to identify key residues, to mutate these in silico, and to predict the effect of these mutations on local flexibility of the protein.
4 Discussion and perspectives
In addition to investigating single structures, our approach can in principle also be used for an ensemble of conformations like those provided in PDB files from NMR experiments. Calculating centralities on the different conformations in the ensemble identifies those residues that are central in the ensemble. The console dialog provided by Cytoscape makes it possible to run script files but is resource intensive for these calculations. We are currently working on a command line workflow for batch generation of RINs and calculation of centralities.
We also foresee, in a future release of RINspector, the integration of additional analyses like the calculation of Z-scores on eigenvector centralities. This would permit to identify clusters of residues, which could highlight zones of interest as opposed to individual residues.
Funding
This work was supported by Grant ANR-13-BSV8–0002-01 from the Agence Nationale de la Recherche.
Conflict of Interest: none declared.
Supplementary Material
References
- Amitai G. et al. (2004) Network analysis of protein structures identifies functional residues. J. Mol. Biol., 344, 1135–1146. [DOI] [PubMed] [Google Scholar]
- Assenov Y. et al. (2008) Computing topological parameters of biological networks. Bioinformatics, 24, 282–284. [DOI] [PubMed] [Google Scholar]
- Atkins J.D. et al. (2015) Disorder prediction methods, their applicability to different protein targets and their usefulness for guiding experimental studies. Int. J. Mol. Sci., 16, 19040–19054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia E. et al. (2013) From protein sequence to dynamics and disorder with DynaMine. Nat. Commun., 4, 2741.. [DOI] [PubMed] [Google Scholar]
- Cilia E. et al. (2014) The DynaMine webserver: predicting protein dynamics from sequence. Nucleic Acids Res., 42, W264–W270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Sol A. et al. (2006) Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol. Syst. Biol., 2, 2006 0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva N.T. et al. (2012) Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc., 7, 670–685. [DOI] [PubMed] [Google Scholar]
- Doncheva N.T. et al. (2011) Analyzing and visualizing residue networks of protein structures. Trends Biochem. Sci., 36, 179–182. [DOI] [PubMed] [Google Scholar]
- Dreux N. et al. (2013) Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathogens, 9, e1003141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra T. et al. (2017) Adhesion of Escherichia coli under flow conditions reveals potential novel effects of FimH mutations. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol., 36, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene L.H. (2012) Protein structure networks. Brief. Funct. Genomics, 11, 469–478. [DOI] [PubMed] [Google Scholar]
- Hu G. et al. (2014) Residue interaction network analysis of Dronpa and a DNA clamp. J. Theor. Biol., 348, 55–64. [DOI] [PubMed] [Google Scholar]
- Morris J.H. et al. (2007) structureViz: linking Cytoscape and UCSF Chimera. Bioinformatics, 23, 2345–2347. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F. et al. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem., 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Shannon P. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.