Abstract
Summary
MetExploreViz is an open source web component that can be easily embedded in any web site. It provides features dedicated to the visualization of metabolic networks and pathways and thus offers a flexible solution to analyse omics data in a biochemical context.
Availability and implementation
Documentation and link to GIT code repository (GPL 3.0 license) are available at this URL: http://metexplore.toulouse.inra.fr/metexploreViz/doc/
1 Introduction
Visualization is a much-needed approach in the era of untargeted omics since it enables discovering new insights in large molecular data corpus (Callaway, 2016). For metabolic studies, challenge resides in creating visual representations of all, or part of, the thousands of reactions and metabolites constituting genome scale metabolic networks.
Visualizing metabolic networks necessitates specific graphical features that are not necessarily available in generic network visualization tools (Bastian et al., 2009; Cline et al., 2007) or web based tools (Franz et al., 2016). Resources are available to perform omics data mapping on metabolic pathways (Caspi et al., 2016; Kanehisa et al., 2014) or on overview of metabolism (Noronha et al., 2017), but they mostly rely on manually drawn maps which cannot integrate network updates or be used for new metabolic reconstructions (Karp et al., 2016). Automatic drawing is thus needed to fit current growing research activity in the field.
Current trend in web development is oriented toward modular architectures allowing assembling specialized web components. In this context, we designed MetExploreViz as an easy to install open source web component that can quickly and easily be embedded in any website. Only few lines of code are required to add MetExploreViz to any web page (see installation section at http://metexplore.toulouse.inra.fr/metexploreViz/doc/).
2 Metabolic network visualization features
In order to make MetExploreViz a versatile solution, it is possible to import networks in JSON format. These networks can for instance be retrieved by using MetExplore web services (Cottret et al., 2010). Drawing is achieved using an animated version of force directed layout implemented in D3.js library upon which MetExploreViz is built. It is also possible to manually edit the representation and then save the drawing in JSON format for further reuse.
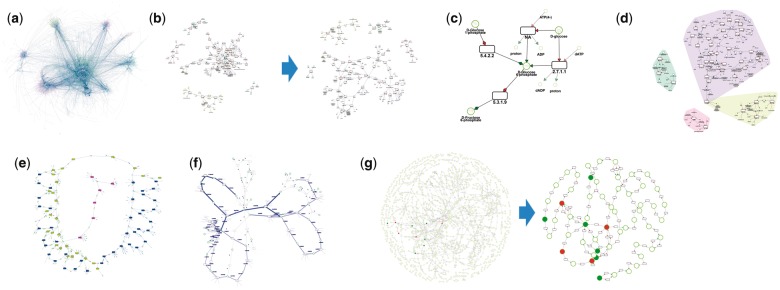
An issue when drawing metabolic networks is the presence of highly connected small molecules like water or CO2 which are not necessarily of biochemical interest for data interpretation. To facilitate the visual inspection, these side compounds can be duplicated or deleted in MetExploreViz (Fig. 1b and c).
Fig. 1.
Main MetExploreViz features. (a) Visualization of Recon2 human metabolic network containing 7440 reactions (Thiele et al., 2013). (b) Sub-network corresponding to glycolysis pathway in cytosol. Highly connected nodes in grey (ATP, ADP, NAD and NADH) on the left network are duplicated to create the sparser representation on the right. (c) Zoom in view on the metabolic pathway, reversible reactions are depicted using double-headed arrows. Smaller nodes are duplicated side compounds. (d) Compartments where glycolysis is taking place are highlighted using colored convex hulls (e.g. cytosol in top right corner). (e) Sub-network with discrete values mapped on reactions. (f) Flux values mapped on edges around reactions. (g) KEGG network of Saccharomyces cerevisiae with metabolomics dataset from (Madalinski et al., 2008) mapped (dark nodes). On the right the sub-network automatically obtained using the algorithm computing the union of lightest paths implemented in MetExploreViz (Color version of this figure is available at Bioinformatics online.)
Each element of a metabolic network can be involved in one or more metabolic pathways and be present in one or several cellular compartments. MetExploreViz offers an original feature to visualize this information using convex hulls (Fig. 1d). Since the force directed algorithm may lead to overlapping hulls, we implemented an algorithm to take these sub-structures into account during the drawing.
MetExploreViz purpose is the visualization of omics data on reactions and metabolites. It is thus possible to import data and map them in a discrete or continuous way using predefined or selected colors (see Fig. 1e and f).
In order to focus on a core sub-network (see Fig. 1g), MetExploreViz implements a sub-network extraction method computing the union of lightest paths between each pairs of mapped (or selected) nodes in the network (Frainay and Jourdan, 2017). Lightest paths aim at avoiding side compounds by weighting nodes according to their degree defined as the number of in and out connections. Classical image exports are available and completed with an SVG export well suited for online representations.
Since MetExploreViz focuses on metabolic networks, it does not provide all the features available in generic network analysis software suites like Cytoscape or Gephi. To allow compatibility with these tools, it is possible in MetExploreViz to export networks in gml and dot formats.
MetExploreViz is provided as an open source web component that only requires few lines of code to be embedded in a web site.
3 Discussion and application
MetExploreViz and metabolic pathways oriented representations like KEGG are complementary visualization solutions. MetExploreViz is focused on integrative and flexible network import and representation, while pathway oriented tools are guiding data interpretation toward a priori defined metabolic functions.
MetExploreViz is embedded in MetExplore web server (Cottret et al., 2010). But its component oriented architecture allows it to be used in various web servers. For instance, it is available in MetaboLights data repository for metabolomics data sets (Kale et al., 2016). We are also currently working on the integration of MetExploreViz in galaxy pipelines (Giacomoni et al., 2015). MetExploreViz provides complementary features to Escher (King et al., 2015) like sub-network extraction and is not focused on network curation, making it a lightweight component.
The tool has been used for several projects as shown on Figure 1g where metabolomics data from (Madalinski et al., 2008) are represented. The extracted sub-network shown on the figure raised similar conclusions as the ones described in Milreu et al. (2014). Tutorial available at http://metexplore.toulouse.inra.fr/metexploreViz/doc/ allows repeating this analysis.
Acknowledgements
Authors would like to thank EBI team and all the users who provided regular feedbacks. We are grateful to the genotoul bioinformatics platform Toulouse Midi-Pyrenees for providing computing and storage resources.
Funding
This work was supported by the French Ministry of Research and National Research Agency as part of the French MetaboHUB [Grant ANR-INBS-0010]; Wellcome Trust [Grant 105614/Z/14/Z]; and PhenoMeNal project, European Commission's Horizon 2020 Program [Grant Agreement Number 654241].
References
- Bastian M. et al. (2009) Gephi: an open source software for exploring and manipulating networks. In: Int. AAAI Conf. WEBLOGS Soc. MEDIA.
- Callaway E. (2016) The visualizations transforming biology. Nature, 535, 187–188. [DOI] [PubMed] [Google Scholar]
- Caspi R. et al. (2016) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res., 44, D471–D480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M.S. et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc., 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottret L. et al. (2010) MetExplore: a web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Res., 38, W132–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frainay C., Jourdan F. (2017) Computational methods to identify metabolic sub-networks based on metabolomic profiles. Brief. Bioinform, 18, 43–56. [DOI] [PubMed] [Google Scholar]
- Franz M. et al. (2016) Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics, 32, 309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomoni F. et al. (2015) Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics, 31, 1493–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale N.S. et al. (2016) MetaboLights: an open-access database repository for metabolomics data. Curr. Protoc. Bioinformatics, 53, 14.13.1–18. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res., 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P.D. et al. (2016) Pathway Tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief. Bioinform., 17, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King Z.A. et al. (2015) Escher: a web application for building, sharing, and embedding data-rich visualizations of biological pathways. PLoS Comput. Biol., 11, e1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madalinski G. et al. (2008) Direct introduction of biological samples into a LTQ-Orbitrap hybrid mass spectrometer as a tool for fast metabolome analysis. Anal. Chem., 80, 3291–3303. [DOI] [PubMed] [Google Scholar]
- Milreu P.V. et al. (2014) Telling metabolic stories to explore metabolomics data: a case study on the yeast response to cadmium exposure. Bioinformatics, 30, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noronha A. et al. (2017) ReconMap: an interactive visualization of human metabolism. Bioinformatics, 31, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I. et al. (2013) A community-driven global reconstruction of human metabolism. Nat. Biotechnol., 31, 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]