Abstract
White adipose tissue (WAT) can develop into several phenotypes with different pathophysiological impact on type 2 diabetes. To better understand the adipogenic process, the transcriptional events that occur during in vitro differentiation of human adipocytes were investigated and the findings linked to WAT phenotypes. Single-molecule transcriptional profiling provided a detailed map of the expressional changes of genes, enhancers, and long noncoding RNAs, where different types of transcripts share common dynamics during differentiation. Common signatures include early downregulated, transient, and late induced transcripts, all of which are linked to distinct developmental processes during adipogenesis. Enhancers expressed during adipogenesis overlap significantly with genetic variants associated with WAT distribution. Transiently expressed and late induced genes are associated with hypertrophic WAT (few but large fat cells), a phenotype closely linked to insulin resistance and type 2 diabetes. Transcription factors that are expressed early or transiently affect differentiation and adipocyte function and are controlled by several well-known upstream regulators such as glucocorticosteroids, insulin, cAMP, and thyroid hormones. Taken together, our results suggest a complex but highly coordinated regulation of adipogenesis.
Introduction
Although obesity is a major risk factor for type 2 diabetes (T2D), not all forms of excess body fat are equally dangerous (1,2). White adipose tissue (WAT) expands by increasing the size (hypertrophy) and/or number (hyperplasia) of fat cells. There is a constant turnover of adipocytes in adult humans (3), and the formation of human adipocytes is linked to WAT morphology (i.e., hyperplasia/hypertrophy) (4). Subjects with adipose hypertrophy, independent of obesity, have increased risk of development of T2D (5,6). Thus, changes in adipogenesis (differentiation of precursor cells to fat cells) are important in distinguishing diabetogenic from less pernicious excess WAT.
Cell differentiation is governed by gene regulatory programs that involve transcription factors (TFs) and coregulators acting on promoters and enhancers, as well as less well-studied regulators such as long noncoding RNAs (lncRNAs). Adipocyte differentiation has been studied extensively, particularly in the murine 3T3-L1 cell line. This has allowed for identification of major regulators like PPARγ (peroxisome proliferator–activated receptor γ [7,8]) and the C/EBPs (CCAAT/enhancer binding proteins [9–12]) (for review, see refs. 13,14). However, a detailed mapping of the complete transcriptome (including enhancer RNA and lncRNA) during human adipocyte differentiation is lacking and essential to better understand the interactions that transform a progenitor cell into an adipocyte.
Recent advances in high-throughput methods enable studies of genome-wide transcriptional events in an unbiased manner. Here, single molecule 5′ cap analysis of gene expression (CAGE) (15) was used to map the transcriptional output at promoters and enhancers during human adipocyte differentiation in vitro. The influence of selected early and transiently expressed TFs on adipocyte differentiation was assessed functionally using small interfering RNA (siRNA). Finally, we overlapped enhancers active during adipogenesis to single nucleotide polymorphisms (SNPs) associated with T2D-related traits, such as obesity and WAT morphology.
This work is part of the FANTOM5 project. Data downloads, genomic tools, and copublished articles are summarized at http://fantom.gsc.riken.jp/5/.
Research Design and Methods
Cell Culture and Isolation
Subcutaneous WAT (scWAT) was obtained as a waste product of cosmetic liposuction from otherwise healthy subjects. Apart from sex, age, and BMI, no clinical details are available from the donors. All studies were approved by the regional ethics board, and all donors gave their written informed consent.
Human Adipose-Derived Stem Cells
Human adipose-derived stem cells (hASCs) were isolated from scWAT of a male donor (aged 16 years, BMI 24 kg/m2). The cells were isolated, propagated, and differentiated to adipocytes using protocols described in refs. 16–18. Briefly, stromal vascular fraction (SVF) cells were isolated by collagenase treatment of scWAT followed by centrifugation. Cells were then selected for their ability to adhere to plastic and proliferated/subcultured in proliferation medium containing 10% FBS and FGF2 (2.5 ng/mL).
Primary Preadipocytes
The SVF cells and mature adipocytes from four donors were isolated using a previously described method (19). Briefly, scWAT was collagenase treated, and SVF and mature fat cells were separated by centrifugation. SVF cells were allowed to adhere to plastic in plating medium containing 10% FBS. To avoid proliferation, cells were kept in plating medium <20 h before inducing differentiation. SVF, unlike hASC cultures, is believed to contain both mesenchymal-like stem cells and committed adipocyte precursor cells.
RNA Isolation
RNA for CAGE library preparation from primary preadipocytes (PreAds), hASCs, and mature adipocytes was isolated using Qiagen (GmbH, Germany) miRNeasy Mini Kit (cat. no. 217004). RNA for quantitative RT-PCR (RT-qPCR) was isolated using the Macherey-Nagel (GmbH & Co., Germany) NucleoSpin RNA kit (no. 740955). Both kits were used according to the manufacturer’s instructions.
CAGE Library Preparation, Sequencing, and Mapping
CAGE libraries were prepared, sequenced, mapped, and clustered into transcription start site regions as previously described (20). Briefly, libraries were prepared using 5 μg of total RNA by manual and automated protocols followed by sequencing on the HeliScope Single Molecule Sequencer platform. Three (hASC) or four (PreAd, mature adipocyte) replicates were used per time point, where the hASCs were derived from the same donor and the PreAds and mature adipocytes from four (one donor per replicate). hASC replicates were defined as cells differentiated in separate wells and treated separately throughout the sample processes afterward. Reads corresponding to ribosomal RNA were removed using the rRNAdust program (http://fantom.gsc.riken.jp/5/suppl/rRNAdust/). Remaining CAGE reads were mapped to the genome (hg19) using Delve (http://fantom.gsc.riken.jp/software/). Reads mapping with a quality of <20 (<99% chance of being true) were discarded. Furthermore, all reads that mapped to the genome with a sequence identity of <85% were discarded.
RNA integrity measurements were made using an Agilent Bioanalyser for samples with more than 1 μg of RNA available. An RNA integrity number score of at least 7.5, a mapped tag count of 250,000, and at least 0.8 Pearson correlation of log10(promoter expression) to other replicates for the same time point was required for a library to be included in the analysis. In 16 out of 21 time points, all replicates passed quality control after sequencing and mapping, whereas five had one replicate discarded because of low tag counts.
FANTOM5 CAGE data, including the libraries analyzed here (Supplementary Table 1), is available at the DNA Data Bank of Japan under accession numbers DRA000991, DRA002711, DRA002747, and DRA002748.
CAGE Promoter Analysis
To identify peaks/promoters in the CAGE profiles, we used decomposition peak identification as previously described (21). We used the relative log expression method (22) to calculate normalization factors for the expression of promoters used in this study. For normalization of the libraries, we used the same method but with calculation of the geometric mean taken from a previous study (21).
Differential Expression of Promoters
For each pair of time points, differential expression was calculated using edgeR (22), using default parameters and common dispersion, on promoters having at least five CAGE tags in total across all samples. Comparisons with a q value (false discovery rate [FDR]) <0.05 were defined as differentially expressed.
CAGE Enhancer Analysis
Candidate active enhancers were predicted using all FANTOM5 phase 1 and 2 samples, as described previously (20,23). A minimum of three CAGE tags was required for each enhancer to be considered expressed in that CAGE library. Pairwise differential enhancer expression between time points was determined using edgeR (22), using common and tagwise dispersion. Only enhancers expressed in at least half of CAGE library replicates for at least one of the time points in each time point pair were considered for differential expression. Enhancers with a Benjamini–Hochberg adjusted FDR ≤0.05 were considered differentially expressed. Enhancers enriched in adipocytes, PreAds, and adipose tissue in a previous study (23) and enhancers expressed in PreAds in this study were compared with enhancers expressed in at least one sample during the hASC time course, and statistical significance of overlaps was computed using Fisher exact test. Histone modification data from ref. 24 was converted from human genome assembly hg18 to hg19 using the UCSC liftOver tool (25), where the absolute majority (>99.9%) of the histone modification reads could be unambiguously converted between the genome versions. Histone modification enrichment was calculated in a 2-kb window around the center point of all enhancers identified in the FANTOM5 project and separately in enhancers expressed in at least one library in the hASC time course. Finally, enhancers expressed in at least one library were overlapped with SNPs with disease association in the following way: SNPs were obtained from the GWAS catalog (26), and an additional two recent GWAS studies where traits with high relevance to adipocytes were studied (27,28). This set of 14,816 GWAS SNPs was extended by including SNPs in linkage disequilibrium (r2 > 0.8) to GWAS SNPs (within 250 kb) in the 1000 Genomes Project pilot data, using the SNAP search tool (29), yielding a total of 207,431 SNPs with disease association according to the above definitions.
Annotation of Promoters
Promoters were associated to gene symbols and TFs as described in ref. 21. Any promoter not associated to a gene symbol was annotated as a putative lncRNA promoter.
Expression Clustering
Clustering using the k-means method was applied to promoters and enhancers differentially expressed in at least one time point compared with time zero. For each data type, expression was averaged in each time point and normalized to range between 0 and 1 across the time course prior to clustering. Manual inspection after trial of different choices of the k parameter suggested that k = 3 captured the major patterns in enhancers and k = 5 captured major patterns in promoters.
Gene Ontology Analysis
Genes in k-means expression clusters were analyzed for Gene Ontology (GO) enrichment using the DAVID web resource (30) with all genes expressed during the hASC time course as background. In cases where the same gene was represented by several promoters, only the most highly expressed promoter was used. Gene groups were subjected to Functional Annotation Clustering using the GOTERM_BP_FAT, BIOCARTA, and KEGG_PATHWAY categories, and annotation clusters with enrichment score at least 1.3, corresponding to P ≤ 0.05 (30), were considered enriched. A separate analysis on the same gene groups was also performed with the ReactomePA package (31) using default parameters.
Principal Component Analysis of k-means Cluster Gene Expression Extracted From WAT Data
Gene expression data for the genes in each k-means cluster were extracted from a previously published microarray experiment from human WAT (32). Each subject was classified according to obesity status (nonobese/obese, according to BMI) and morphology (hyperplastic/hypertrophic). The gene expression levels of genes in each k-means cluster were subjected to principal component analysis in R (http://www.R-project.org/). Principal components were plotted against each other and subjects annotated with color/shape according to their corresponding BMI/obesity status or obesity/morphology status.
siRNA
One day before differentiation induction, hASCs were reverse transfected with 50 nmol/L siRNA (GE Healthcare Life Sciences, Dharmacon ON-TARGETplus) using 0.35 µL DharmaFECT 3 (GE Healthcare Life Sciences, Dharmacon) and 10,000 cells per well in a 96-well plate. TFs with effects on adipogenesis were reinvestigated using electroporation as transfection method. The NEON system (Invitrogen, Carlsbad, NM) 10-μL electroporation tips were used with 105 cells per reaction. Two pulses (1,200 V, 20 ms) were used to transfect a final concentration of 40 nmol/L siRNA to cells. All siRNAs were from the ON-TARGETplus series from GE Healthcare Dharmacon, Inc. Order numbers are found in Table 1.
Table 1.
siRNA oligos and qPCR primers
| Gene | siRNA order number, Dharmacon | qPCR primer 1 | qPCR primer 2 |
|---|---|---|---|
| NegC | D-001810-10 | NA | NA |
| PPARG | L-003436-00 | TCATAATGCCATCAGGTTTG | CTGGTCGATATCACTGGAG |
| BCL6 | L-011591-01 | GTGTGATCAATCTAGATCCTG | CACTGGCCTTAATAAACTTCC |
| CREB3L1 | L-008579-02 | CAAATTACAGGGGACATCAG | CACATACTCCTTCTTCTTACG |
| PRRX1 | L-012402-00 | CAGGTGTGGTTTCAGAAC | CTGAGTAGGATTTGAGGAGG |
| SOX4 | L-011779-00 | AGTGACAGTATCCCTTAACC | GTGTTTGGCTAATTCTCCTC |
| ZFP36L2 | L-013605-01 | ACAGAAATCGATCTTGATGC | TGATTGCAAAGTATGTGGTC |
NA, not available.
RT-qPCR
cDNA from a total of 50 ng of RNA was prepared using iScript cDNA Synthesis Kit (no. 170-8891, Bio-Rad Laboratories, Inc.). On a CFX96 RT-qPCR system (Bio-Rad Laboratories, Inc.), 0.1 ng/µL cDNA, 300 nmol/L SYBR primer, and 1xiQ SYBR Green mix (no. 172-5006, Bio-Rad Laboratories, Inc.) were run. Data were analyzed using the ΔΔCt method (33). Primer sequences are found in Table 1.
Lipid Accumulation
Cells were fixed at day 9 of differentiation and neutral lipids stained as described (18).
Lipolysis (Glycerol Release)
Cell culture medium was collected at day 9 of differentiation and assayed for glycerol content as described (18).
Statistical Methods
Values are presented as individual points and as mean ± SD. The level of significance was calculated using statistical tests as indicated.
Results
Study Design and Initial Results
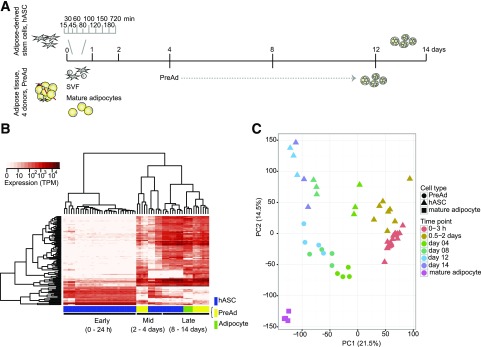
5′ ends of capped RNA from 17 time points during the 14-day-long in vitro differentiation of subcutaneous hASCs from one donor was sequenced using CAGE. In four additional donors we sequenced RNA from three time points of adipocyte precursor cells during differentiation (PreAds) and from mature fat cells isolated by collagenase treatment from WAT (Fig. 1A). Quality control demonstrated high correlation between replicates across all time points (Supplementary Fig. 1) and confirmed induction of typical adipogenic markers (Supplementary Fig. 2). Samples isolated at late time points of adipogenesis (hASC day 8–14, PreAd day 8–12) displayed similar transcriptional profiles as mature adipocytes as observed in the following two analyses. First, we assessed whether an unbiased selection of variable promoters would be sufficient to divide the samples into biologically meaningful groups using an unsupervised clustering method. Hierarchical clustering of the 100 promoters with highest variance across all samples showed that the samples could be divided into three major temporal groups: early (hASC 0–24 h), middle (hASC 48 h to 4 days, PreAd day 4 and one sample day 8) and late (hASC 8–14 days, PreAd 8–12 days, mature adipocytes) (Fig. 1B and Supplementary Fig. 3). Second, principal component analysis (Fig. 1C and Supplementary Fig. 4) revealed similar results in hASCs and PreAds: the trajectory of differentiation was traced mainly along the first principal component, which captured 21.5% of the variance. The second principal component mainly captured cell type differences, whereas the third component distinguished samples from the middle of the hASC time course (12 h to 4 days) as a separate group (Supplementary Fig. 4B). These analyses suggest that hASCs and PreAds display similar dynamics during adipogenesis with low donor bias and that the end results of in vitro differentiation mirror the results of in vivo differentiated adipocytes (i.e., the mature fat cells).
Figure 1.
Experimental set up and model system comparison. A: Model system and sampling schematic. B: Hierarchical clustering of the 100 promoters with the highest variance in all samples that passed quality control. C: Principal component (PC) analysis of promoters with at least 10 tags per million (TPM) expression level in at least one sample. The two components capturing the highest variance are plotted.
Temporal Expression Patterns During Adipogenesis
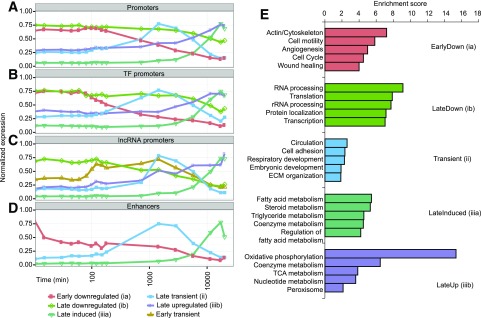
To find global patterns of transcript expression, unsupervised clustering of the CAGE expression data were performed (Fig. 2). We observed transcripts with initial high expression that dropped after addition of adipogenic medium (i); transient transcripts with expression that increased between 12 h and 2 days after adipogenic induction and then dropped again (ii); and late transcripts with expression that started to increase between 1 and 4 days after induction (iii). Furthermore, the downregulated (i) and upregulated (iii) groups were separated into early/late downregulated (ia and ib) and late induced/upregulated (iiia and iiib) clusters (Fig. 2A, genes in clusters listed in Supplementary Table 2A). Separate clustering of functional subclasses such as TFs (Fig. 2B and Supplementary Table 2B), lncRNAs (Fig. 2C), and enhancers (Fig. 2D) resulted in similar subdivisions, although a subgroup of lncRNAs showed an early transient expression pattern absent in the other feature types. For all promoters (including TFs and lncRNAs), there was an initial phase of about 120 min when expression was largely stable on a global scale.
Figure 2.
Temporal expression patterns and functional annotations of gene clusters. k-means clustering of the following: all promoters with a minimum expression over 10 tags per million in any hASC sample and a statistically significant (FDR corrected P value ≤ 0.05), at least twofold, change in expression in any time point compared with time zero (A); the subset of promoters in A annotated as belonging to TFs (B); the subset of promoters in A annotated as belonging to lncRNAs (C); and enhancer RNA, expressed in at least half of all libraries and differentially expressed between any two time points (D). The x-axis in A–D corresponds to log10 (time of sampling in minutes), and the y-axis is the average of the normalized expression [expression/max(expression)] of each gene in each cluster. E: Functional annotation clustering of genes belonging to the clusters/temporal classes identified in A. The five most enriched functional clusters of each k-means class are given in the plot. For complete list of clusters and included GO terms and pathways, see Supplementary Table 3. rRNA, ribosomal RNA; TCA, tricarboxylic acid cycle.
To elucidate the possible functional role of genes associated to promoters in the different clusters, we linked their expression patterns to biological processes and pathways (Fig. 2E). Actin/cytoskeleton processes, angiogenesis, cell cycle, and wound healing were enriched in the early downregulated group (ia), while the late downregulated group (ib) was enriched for RNA processing. The group of transiently expressed transcripts (ii) contained developmental genes and genes involved in tissue formation such as cell adhesion, circulation, and extracellular matrix (ECM) organization. Late induced genes (iiia) were enriched for lipid (fatty acid, triglyceride, and steroid) metabolism, while the late upregulated group (iiib) contained primarily energy metabolism–related pathways such as the respiratory chain and the tricarboxylic acid cycle (Fig. 2E and Supplementary Tables 3 and 4).
Enhancer Dynamics and Overlap With Functional Variants
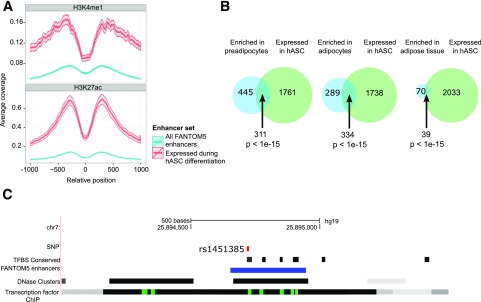
CAGE sequencing captures expression of enhancer RNA (eRNA) transcribed by polymerase II. In total, 2,072 enhancers were expressed during adipogenesis in the hASC data and 2,846 in the PreAd time course. Many of the identified enhancers were common between hASCs and PreAds (1,182 enhancers, P < 1e-15, Fisher exact test), indicating that enhancer usage during differentiation is largely independent of donor and subculturing. Overlapping enhancers with two enhancer histone markers (H3K4me1, H3K27ac) from a previous study of adipogenesis (24) showed substantial enrichment in hASC-expressed enhancers compared with those from all other cells and tissues in the FANTOM5 project (Fig. 3A). We also found significant overlaps (P < 1e-15, Fisher exact test, Fig. 3B) with previously identified enhancers from commercially available PreAds, adipocytes, and adipose tissue (23).
Figure 3.
Enhancers. A: Enrichment of H3K4me1 and H3K27ac ChIP-seq reads from an independent study (24) surrounding enhancers expressed in hASCs (red) compared with all enhancers detected in the FANTOM5 project (blue). B: Overlaps between enhancers expressed during hASC adipogenesis time course (green) and enhancers enriched in primary cell types profiled in FANTOM5 (blue). From left to right: PreAds, adipocytes, and adipose tissue. Statistical significance of overlap calculated by Fisher exact test. C: Genomic context of an SNP (rs1451385) associated to waist-to-hip ratio adjusted for BMI, located in an enhancer expressed during hASC differentiation. The SNP is visualized as a red bar and the enhancer as a blue line. Locations of conserved TF binding sites (TFBS), as well as clusters of DNase hypersensitive sites and TF ChIP-seq signal, are also visualized.
Genetic variation within enhancers can affect traits connected to adipose tissue function (34). We therefore investigated enhancer overlap with adipose-related SNPs identified in genome-wide association studies (GWAS) (26–29). Overall, 80 GWAS-associated SNPs resided within enhancers active during adipogenesis, of which 26% (n = 21, Supplementary Table 5) were associated with traits that could be related to adipogenesis/WAT tissue function such as obesity, body shape, and components of the metabolic syndrome. For example, an SNP (rs1451385) associated with waist-to-hip ratio (a measure of fat distribution) (28) overlapped with an enhancer expressed during adipogenesis (Fig. 3C). This enhancer is located in open chromatin (DNase hypersensitive sites) and has conserved binding sites for the TFs USF1, USF2, BHLHE40, MYC, MAX, and ARNT (confirmed for all but ARNT by ChIP-seq data from ENCODE) (35).
Genes With Transient or Induced Temporal Expression Patterns in Adipogenesis Are Associated With Adipose Morphology
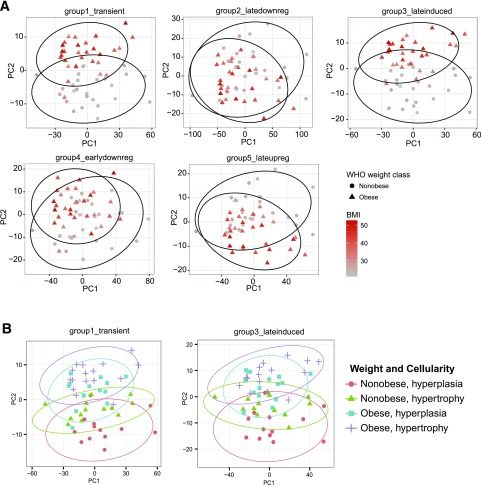
The GO term enrichment analysis of temporal gene clusters suggested that the different differentiation states are associated to specific functions. To elucidate if these gene groups had relevance in vivo, we reanalyzed previously published gene expression data in WAT from a cohort of lean and obese human subjects (32). Adipocyte cell size had been recorded in this cohort and WAT classified as hypertrophic or hyperplastic. Gene expression data for each dynamic cluster from the WAT data set was subjected to principal component analysis. We plotted the first two principal components against each other and labeled subjects according to weight status (obese/nonobese and BMI, Fig. 4A) or weight status in combination with hyperplasia/hypertrophy (Fig. 4B). Genes from all adipogenesis-based expression clusters separated patients according to obesity status, with transient and late upregulated or induced genes showing stronger separation than genes downregulated during adipogenesis (Fig. 4A). Furthermore, only genes in the transient and late induced clusters separated subjects based on morphology/obesity combined, with nonobese hyperplastic subjects being most distant from obese hypertrophic subjects (Fig. 4B and Supplementary Fig. 5).
Figure 4.
Transient and late induced genes can separate subjects based on adipose tissue morphology and obesity status. Gene expression for the genes in each of the dynamic clusters identified in Fig. 2A was extracted from a microarray experiment of WAT from 56 female subjects with differing BMI. These data were subjected to principal component (PC) analysis and the first and second or first and third principal component plotted against each other. Dots representing individual subjects were shaped and colored according to the weight status/BMI of each subject (A) or according to weight status and adipose tissue cellularity (B). Circles represent 95% CI. WHO, World Health Organization.
We next reexamined an analysis from Gao et al. (36) where the subset of nonobese subjects from a study by Arner et al. (32) was analyzed for mRNA expression changes between hypertrophic and hyperplastic scWAT. We focused on the genes with an adjusted P value <0.05 and mapped whether these genes were altered by adipogenesis and, if they were, which k-means cluster they mapped to. We also included the genes CCL2 (MCP-1) (37), DUSP1 (MKP-1) (37), and PLXND1 (38) as they have been implicated in adipose hypertrophy. A total of 54% of the morphology genes were regulated by adipogenesis, and all k-means clusters were represented among the genes regulated based on morphology (Table 2).
Table 2.
Adipose tissue morphology regulated genes and their distribution in the adipogenesis k-means clusters
| GeneID (36) | Log2 fold change, morphology (36) | Adjusted P value, morphology (36) | Regulated in adipogenesis** | In k-means cluster** | |
|---|---|---|---|---|---|
| Probe set ID (36) | |||||
| 8123744 | F13A1 | 0.94 | 0.0263 | No | |
| 8020384 | GREB1L | −0.50 | 0.0263 | Yes | group1_transient |
| 8120428 | FKBP1A | 0.32 | 0.0263 | Yes | group2_latedownreg |
| 8057887 | STK17B | 0.48 | 0.0263 | Yes | group4_earlydownreg |
| 8008263 | PDK2 | −0.50 | 0.0263 | Yes | group5_lateupreg |
| 7950933 | NOX4 | 0.90 | 0.0263 | No | |
| 8014903 | GSDMB | −0.85 | 0.0264 | No | |
| 8072798 | CYTH4 | 0.54 | 0.0289 | No | |
| 7948364 | MPEG1 | 0.77 | 0.0307 | No | |
| 8088745 | FRMD4B | 0.65 | 0.0363 | No | |
| 8003903 | ARRB2 | 0.55 | 0.0405 | Yes | group2_latedownreg |
| 8091511 | P2RY14 | 0.65 | 0.0405 | No | |
| 7968344 | ALOX5AP | 0.54 | 0.0405 | No | |
| 8066117 | SAMHD1 | 0.63 | 0.0405 | Yes | group1_transient |
| 8108217 | TGFBI | 0.61 | 0.0405 | Yes | group4_earlydownreg |
| 8178193 | HLA-DRA | 0.63 | 0.0405 | No | |
| 7916229 | ECHDC2 | −0.31 | 0.0405 | Yes | group3_lateinduced |
| 8049246 | INPP5D | 0.41 | 0.0405 | No | |
| 7946579 | LYVE1 | 0.82 | 0.0405 | No | |
| 8137264 | TMEM176A | 0.62 | 0.0412 | No | |
| 7960794 | CD163 | 0.98 | 0.0454 | No | |
| 8115147 | CD74 | 0.52 | 0.0454 | Yes | group4_earlydownreg |
| 8180100 | HLA-DPA1 | 0.63 | 0.0455 | Yes | group4_earlydownreg |
| 8064790 | RASSF2 | 0.51 | 0.0481 | Yes | group4_earlydownreg |
| Other candidates | |||||
| (36) | EBF1 | Yes | group1_transient | ||
| (37) | DUSP1 (MKP1) | Yes | group2_latedownreg | ||
| (38) | PLXND1 | Yes | group1_transient | ||
| (37) | CCL2/MCP1 | Yes | group4_earlydownreg |
**Data from k-means clustering, see Supplementary Table 2.
BCL6, PRRX1, ZFP36L2, and CREB3L1 Are Early or Transient TFs With Effects on Adipogenesis
On the basis of expression patterns (Supplementary Table 2B) and expression levels (above 50 tags per million in at least one time point during the hASC time course and expressed in PreAd and/or mature adipocyte samples), we manually curated a list of 20 TFs with either recognized (e.g., PPARG and CEBPA as positive controls) or less-established roles in adipocyte differentiation, mainly selecting TFs from the transient and early clusters.
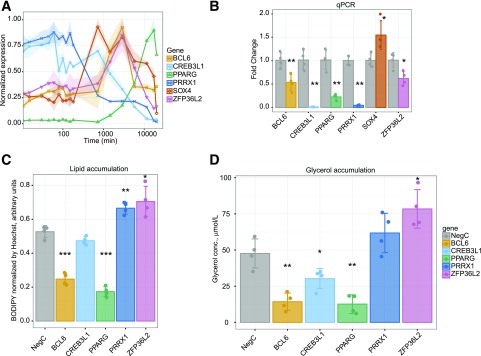
Genes were knocked down using siRNA transfected 1 day before induction of differentiation. Lipid accumulation and lipolysis (glycerol release) were measured at day 9 as proxies of adipogenesis. PPARG and CEBPA knockdown resulted in the expected reduction in both parameters. An initial screen identified five candidates (BCL6, CREB3L1, PRRX1, SOX4, and ZFP36L2) with significant effects on lipid accumulation and/or lipolysis (Supplementary Fig. 6A and B and Supplementary Table 6, with Bonferroni corrected Student t test P value <0.05). The expression of these five TFs (and PPARG) during adipogenesis is depicted in Fig. 5A. We assessed the knockdown efficiency on the five by qPCR and confirmed significant knockdown at the mRNA level for all but SOX4, which was excluded from further analyses (Fig. 5B). We confirmed the knockdown at the protein level for BCL6 and PRRX1 using Western blotting (Supplementary Fig. 7A and B), but for CREB3L1 and ZFP36L2 we had technical issues with the antibodies tested and were not able to detect specific bands at the correct molecular weight. The four TFs were again tested for effects on lipid accumulation (Fig. 5C) and glycerol release (Fig. 5D). Knockdown of BCL6 reduced lipid accumulation and glycerol release, whereas CREB3L1 knockdown reduced glycerol release. Knockdown of PRRX1 increased lipid accumulation, and ZFP36L2 knockdown increased lipid accumulation and glycerol release. Thus, BCL6 and CREB3L1 knockdown reduced, while PRRX1 and ZFP36L2 increased, measures of adipogenesis, indicating that they may be inducers (BCL6, CREB3L1) and repressors (PRRX1, ZFP36L2) of adipogenesis.
Figure 5.
TFs BCL6, CREB3L1, PRRX1, and ZFP36L2 have effects on adipogenesis. A: Temporal expression during adipogenesis from CAGE data of four TFs identified in a screen for early/transient TFs that affect adipogenesis when knocked down using siRNA. The x-axis is log-scaled time in minutes. B: RT-qPCR data showing efficiency of siRNA knockdown for each candidate TF and the positive control PPARG. Cells were electroporated with siRNA day −1 of differentiation and RNA harvested at day 1 (48 h after transfection) of differentiation. C: Intracellular lipid accumulation as measured by neutral lipid staining (BODIPY) normalized to cell number (Hoechst staining of nuclei) of cells with candidate TFs knocked down at day −1 of differentiation. D: Glycerol concentration in cell culture medium as a measure of lipolysis. Concentrations measured at day 9 of differentiation of cells with candidate TFs knocked down at day −1 of differentiation. Each point represents an individual measurement, each bar the mean value, and each error bar SD. Four independent experiments; significance compared with negative control calculated by Student t test with Benjamini–Hochberg corrected *P < 0.05; **P < 0.01; ***P < 0.001.
We next explored whether these factors could be connected to adipose morphology by reexamining data from a cohort (32) used for the analysis in Fig. 4. We correlated the mRNA expression of BCL6, CREB3L1, PRRX1, and ZFP36L2 with the morphology value (delta) (4): a measure of fat cell volume at the prevailing total fat mass (4). PRRX1 expression correlated with delta (P = 0.02, r = 0.31) and also with fat cell volume after adjusting for BMI (standardized coefficient 0.5, P = 0.007). No such correlations were observed with the other three genes. In addition, we plotted the mRNA expression of all four genes when grouped according to the subjects’ BMI status and adipose morphology. There was a significant difference between PRRX1 mRNA expression in nonobese subjects with hyperplasia compared with the other groups (Supplementary Fig. 8).
Glucocorticoids, Insulin, and cAMP Affect the Expression of BCL6, PRRX1, ZFP36L2, and CREB3L1 During Adipogenesis
The differentiation medium that induces hASC transformation to adipocytes includes five compounds of special importance: dexamethasone, acting through the glucocorticoid receptor; the phosphodiesterase inhibitor IBMX, increasing intracellular cAMP levels; insulin, acting through the insulin receptors; thyroid hormone (T3), acting on the thyroid hormone receptor; and rosiglitazone, acting through PPARG. We excluded one inducer at the time and measured the effects on the candidate TF mRNA expression 1, 4, and 12 days after differentiation induction.
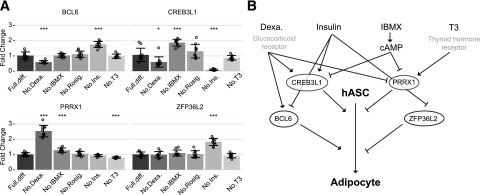
At day 1 (Fig. 6A), omission of dexamethasone had opposing effects on BCL6 and PRRX1 expression, just as BCL6 and PRRX1 had opposing effects on adipogenesis, and also decreased CREB3L1. Insulin omission increased BCL6 and ZFP36L2 expression while decreasing CREB3L1. IBMX removal increased the expression of CREB3L1 and PRRX1, while removing T3 slightly decreased PRRX1. Omission of rosiglitazone had no significant effect on the expression of the candidate TFs at this early time point.
Figure 6.
Glucocorticoid, insulin, and cAMP but not PPARG agonist affect BCL6, CREB3L1, PRRX1, and ZFP36L2 expression. A: Expression of candidate TFs after 24 h of differentiation when dexamethasone (Dexa.), 3-isobutyl-1-methylxanthine (IBMX), rosigiltazone (Rosig.), insulin (Ins.), or triiodothyrine (T3) had been removed from the complete differentiation medium (Full.diff.). Each point represents an individual measurement, each bar the mean value, and each error bar SD. A total of 10 experiments; significance calculated by Student t test with Benjamini–Hochberg corrected *P < 0.05; **P < 0.01; ***P < 0.001. B: Summarization of effects in A.
At days 4 and 12 (Supplementary Fig. 9A and B), expression of CREB3L1 had dropped and was increasingly hard to detect, and no significant effects of any of the omitted compounds were detected. Opposing effects of dexamethasone were again seen for BCL6 compared with PRRX1. Exclusion of rosiglitazone significantly increased PRRX1 and ZFP36L2 mRNA levels, while exclusion of insulin increased BCL6 and ZFP36L2 expression.
At day 12 (Supplementary Fig. 9B), CREB3L1 mRNA was no longer detectable. Exclusion of rosiglitazone increased PRRX1 and ZFP36L2 while decreasing BCL6 expression, thus showing the same opposing effects as omission of dexamethasone. Omission of T3 had the same opposing effects as dexamethasone and rosiglitazone, while insulin increased PRRX1 and ZFP36L2.
Discussion
We have generated detailed transcriptomic data from hASCs during their differentiation into adipocytes. Unbiased clustering identified at least three general expression patterns during the 14-day adipogenesis time course: downregulated, transient, and induced transcripts. These temporal patterns agree with other studies of adipogenesis that show that cells undergo epigenomic transitions during differentiation resulting in three states: undifferentiated, transient, and differentiated (39). Importantly, we also show that protein-coding RNA, enhancer RNAs, and lncRNAs all follow the same dynamic expression patterns. This suggests a complex but highly coordinated chain of regulatory events for human adipogenesis.
The three general transcription patterns were characterized by specific gene expression signatures enriched for different functional classes of genes. A transition from the undifferentiated to the transient state results in altered expression of cell cycle, cytoskeletal, and angiogenic genes, possibly reflecting the change from a dividing to a differentiating cell. The transient state is enriched for ECM structure, adhesion, and morphology, perhaps representing the changes necessary to form and make space for the maturing adipocyte. Lastly, the differentiated cell expresses genes that define adipocyte function, such as those involved in lipid metabolism and energy consumption. Thus, each cluster has a specific role in adipogenesis.
By intersecting regulatory regions for gene expression with disease-associated gene variants identified in GWAS studies of relevant traits, it is possible to functionally assign SNPs with previously unknown functions (34). CAGE-defined enhancers are more likely to represent functional enhancers than predictions based on chromatin marks (23). This notion is supported by our observation that a number of SNPs associated with T2D-related traits such as obesity and adipose tissue function were localized to enhancers expressed during adipogenesis. In the highlighted example (Fig. 3C), a variant associated with waist-to-hip ratio overlapped predicted and experimentally determined TF binding sites within the enhancer region, raising the possibility that the variant affects enhancer function by altering binding efficacy of TFs bound to the enhancer region. Future studies of these enhancers will give a more complete understanding of how genetic variation can contribute to development of T2D. Compared with studies in other cell types using the same approach (20,23), we find few dynamically transcribed enhancers. This could be due to the depth of sequencing achieved, as eRNA expression is much lower than gene expression. The observation that the eRNAs identified herein overlap with enhancer chromatin marks from previous studies of adipogenesis (24) suggests that they are indeed relevant for human fat cell differentiation and that deeper CAGE sequencing in future studies of adipocytes and related cell types may identify additional enhancers contributing to the regulation of fat cell formation and maintenance.
Changes in the regulation of adipogenesis are likely to be important for WAT expansion and different WAT morphologies. For example, lack of the TF EBF1 has been linked to the pernicious hypertrophic obesity in humans and mice (36). The current study suggests that genes in all clusters are related to obesity. However, those expressed transiently or late during adipogenesis distinguish obese hypertrophic WAT from nonobese hyperplastic WAT to a higher degree than early downregulated genes. Late induced genes should be expected to be involved in WAT morphology, as they likely influence final differentiation and fine tune specific aspects of adipocyte function. Nevertheless, impaired function in fat cell progenitors may also contribute to hypertrophy by limiting the ability to recruit new adipocytes (40), thereby directing WAT expansion through increased fat cell size. This is corroborated by the observation that obese hypertrophy was also reflected in the transiently expressed genes (Fig. 4B).
To shed light on the control and the regulatory role of the early and transient gene group we functionally assessed selected early and transient TFs with a less-established role in adipocyte biology. Four were found to be involved in the early steps of converting a hASC into an adipocyte. Two were expressed primarily in the undifferentiated hASCs (early downregulated cluster) and two in the transiently expressed cluster. PRRX1 (early downregulated) siRNA-mediated knockdown increased measures of adipogenesis, suggesting that it is antiadipogenic, in agreement with data from murine 3T3-L1 cells (41), as did that for ZFP36L2 (transiently expressed), in agreement with previous data from murine 3T3-L1 cells (42). CREB3L1 (early downregulated) and BCL6 (transient) had proadipogenic effects. CREB3L1 is part of the CREB3-like family which also contains CREB3L4, a protein shown to have a negative effect on adipogenesis in mice (43). CREB3L1 mRNA was expressed at low levels in our system, and we were unable to detect the protein via Western blotting; further studies are required to determine its importance in adipogenesis. For BCL6, our data from human cells was in line with previous murine studies: BCL6−/+ heterozygote mice (44) demonstrated reduced adipose mass and disturbed lipid metabolism; the lack of fat mass is in agreement with a role for BCL6 in lipolysis and lipid filling. In addition, Hu et al. (45) recently showed that BCL6 is involved in early commitment of a murine pluripotent cell line (C3H10T1/2) to adipocytes. It thus appears that TFs expressed early and transiently during adipogenesis may have opposing effects on fat cell formation despite sharing similar expression patterns. Out of the four TFs relevant to adipogenesis studied herein, only PRRX1 showed any correlation to measures of in vivo adipose morphology. It will be interesting to see whether future studies determine the correlation to be an effect of disturbed adipogenic activity or a direct regulatory effect of this early downregulated TF on adipose morphology.
To place our TF candidates into an established network of events, we studied how omission of selected proadipogenic compounds affected their expression 24 h after adipogenic induction. Individual removal of a corticosteroid (dexamethasone), insulin, a cAMP elevator (IBMX) or T3 enabled us to identify the TF response to each compound (Fig. 6A and B). In contrast, although PPARγ is a key regulator of adipogenesis, removal of a PPARγ agonist (rosiglitazone) did not alter the early expression levels of any of the four TFs (Fig. 6). This is probably due to the fact that PPARγ exerts its major effects at later stages of differentiation, a notion supported by its expression pattern during adipogenesis (low levels at day 1 compared with day 14). At later stages (day 4 and 12), omission of these compounds generally increased the levels of antiadipogenic factors while decreasing the adipogenic, which was especially apparent for rosiglitazone at day 12 (Supplementary Fig. 9).
In summary, the transcriptional dynamics during human adipogenesis display a complex but highly coordinated chain of events. The different regulatory factors (genes, enhancers, lncRNAs) can be divided into similar temporal expression patterns indicating at least three distinct steps involved in the birth of a fat cell, where the gene clusters appear to have distinct functions in adipogenesis. Several enhancers are linked to genetic variation previously shown to be important for adiposity-related traits. The transiently expressed and late induced genes are linked to obese hypertrophic WAT, which is closely associated with development of insulin resistance and T2D. TFs expressed before or within this cluster have profound effects on late differentiation and function of fat cells and are regulated by key upstream factors. We have identified four, in this context, either novel or not extensively studied TFs that should be further investigated regarding their contributions to adipogenesis. The transcriptional map is available as part of the FANTOM5 project, and we encourage further studies based on this resource.
Supplementary Material
Article Information
Funding. FANTOM5 was made possible by a research grant for RIKEN Omics Science Center from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) (to Y.H.) and a MEXT grant for the Research Program of Innovative Cell Biology by Innovative Technology (Cell Innovation Program) (to Y.H.). FANTOM5 was also supported by MEXT research grants for the RIKEN Preventive Medicine & Diagnosis Innovation Program (to Y.H.) and the RIKEN Center for Life Science Technologies Division of Genomic Technologies. R.A. and A.S. were supported by the Novo Nordisk Foundation and the Lundbeck Foundation. The studies at the Karolinska Institutet were supported by grants from the Swedish Research Council, the Diabetes Research Program at Karolinska Institutet, CIMED, and the Novo Nordisk Foundation. All data within this article are easily searchable through the FANTOM5 gateways (46), http://fantom.gsc.riken.jp/5/.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Samples were prepared by A.E., N.M., C.B., A.K., G.Å., M.I., and J.L. Analyses were carried out by A.E., R.A., H.K., T.L., A.S., E.I., M.R., P.A., and E.A. The experiments were designed and managed by C.O.D., P.C., A.R.R.F., Y.H., P.A., and E.A. The manuscript was written by A.E., M.R., P.A., and E.A. with contributions, edits, and comments from all authors. E.A. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-0631/-/DC1.
References
- 1.Blüher M. Adipose tissue dysfunction contributes to obesity related metabolic diseases. Best Pract Res Clin Endocrinol Metab 2013;27:163–177 [DOI] [PubMed] [Google Scholar]
- 2.Wronkowitz N, Romacho T, Sell H, Eckel J. Adipose tissue dysfunction and inflammation in cardiovascular disease. Front Horm Res 2014;43:79–92 [DOI] [PubMed] [Google Scholar]
- 3.Spalding KL, Arner E, Westermark PO, et al. . Dynamics of fat cell turnover in humans. Nature 2008;453:783–787 [DOI] [PubMed] [Google Scholar]
- 4.Arner E, Westermark PO, Spalding KL, et al. . Adipocyte turnover: relevance to human adipose tissue morphology. Diabetes 2010;59:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weyer C, Foley JE, Bogardus C, Tataranni PA, Pratley RE. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia 2000;43:1498–1506 [DOI] [PubMed] [Google Scholar]
- 6.Lönn M, Mehlig K, Bengtsson C, Lissner L. Adipocyte size predicts incidence of type 2 diabetes in women. FASEB J 2010;24:326–331 [DOI] [PubMed] [Google Scholar]
- 7.Rosen ED, Sarraf P, Troy AE, et al. . PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 1999;4:611–617 [DOI] [PubMed] [Google Scholar]
- 8.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994;79:1147–1156 [DOI] [PubMed] [Google Scholar]
- 9.Wang ND, Finegold MJ, Bradley A, et al. . Impaired energy homeostasis in C/EBP alpha knockout mice. Science 1995;269:1108–1112 [DOI] [PubMed] [Google Scholar]
- 10.Umek RM, Friedman AD, McKnight SL. CCAAT-enhancer binding protein: a component of a differentiation switch. Science 1991;251:288–292 [DOI] [PubMed] [Google Scholar]
- 11.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J 1997;16:7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z, Rosen ED, Brun R, et al. . Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 1999;3:151–158 [DOI] [PubMed] [Google Scholar]
- 13.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell 2014;156:20–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siersbæk R, Nielsen R, Mandrup S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab 2012;23:56–64 [DOI] [PubMed] [Google Scholar]
- 15.Kanamori-Katayama M, Itoh M, Kawaji H, et al. . Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res 2011;21:1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez A-M, Pisani D, Dechesne CA, et al. . Transplantation of a multipotent cell population from human adipose tissue induces dystrophin expression in the immunocompetent mdx mouse. J Exp Med 2005;201:1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaragosi L-E, Ailhaud G, Dani C. Autocrine fibroblast growth factor 2 signaling is critical for self-renewal of human multipotent adipose-derived stem cells. Stem Cells 2006;24:2412–2419 [DOI] [PubMed] [Google Scholar]
- 18.Pettersson AML, Acosta JR, Björk C, et al. . MAFB as a novel regulator of human adipose tissue inflammation. Diabetologia 2015;58:2115–2123 [DOI] [PubMed] [Google Scholar]
- 19.van Harmelen V, Skurk T, Hauner H. Primary culture and differentiation of human adipocyte precursor cells. Methods Mol Med 2005;107:125–135 [DOI] [PubMed] [Google Scholar]
- 20.Arner E, Daub CO, Vitting-Seerup K, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 2015;347:1010–1014 [DOI] [PMC free article] [PubMed]
- 21.Forrest ARR, Kawaji H, Rehli M, et al.; FANTOM Consortium and the RIKEN PMI and CLST (DGT) . A promoter-level mammalian expression atlas. Nature 2014;507:462–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson R, Gebhard C, Miguel-Escalada I, et al.; FANTOM Consortium . An atlas of active enhancers across human cell types and tissues. Nature 2014;507:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, Xu Z, Zhang X, et al. . Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010;143:156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, et al. . The human genome browser at UCSC. Genome Res 2002;12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welter D, MacArthur J, Morales J, et al. . The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001–D1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locke AE, Kahali B, Berndt SI, et al.; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shungin D, Winkler TW, Croteau-Chonka DC, et al.; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGC; ICBP; International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium . New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015;518:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008;24:2938–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 31.Yu G, He Q-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst 2016;12:477–479 [DOI] [PubMed] [Google Scholar]
- 32.Arner E, Mejhert N, Kulyté A, et al. . Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes 2012;61:1986–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 34.Claussnitzer M, Dankel SN, Kim K-H, et al. . FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med 2015;373:895–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M; ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao H, Mejhert N, Fretz JA, et al. . Early B cell factor 1 regulates adipocyte morphology and lipolysis in white adipose tissue. Cell Metab 2014;19:981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito A, Suganami T, Miyamoto Y, et al. . Role of MAPK phosphatase-1 in the induction of monocyte chemoattractant protein-1 during the course of adipocyte hypertrophy. J Biol Chem 2007;282:25445–25452 [DOI] [PubMed] [Google Scholar]
- 38.Minchin JEN, Dahlman I, Harvey CJ, et al. . Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proc Natl Acad Sci U S A 2015;112:4363–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steger DJ, Grant GR, Schupp M, et al. . Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 2010;24:1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gustafson B, Gogg S, Hedjazifar S, Jenndahl L, Hammarstedt A, Smith U. Inflammation and impaired adipogenesis in hypertrophic obesity in man. Am J Physiol Endocrinol Metab 2009;297:E999–E1003 [DOI] [PubMed] [Google Scholar]
- 41.Du B, Cawthorn WP, Su A, et al. . The transcription factor paired-related homeobox 1 (Prrx1) inhibits adipogenesis by activating transforming growth factor-β (TGFβ) signaling. J Biol Chem 2013;288:3036–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin N-Y, Lin T-Y, Yang W-H, et al. . Differential expression and functional analysis of the tristetraprolin family during early differentiation of 3T3-L1 preadipocytes. Int J Biol Sci 2012;8:761–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim T-H, Jo S-H, Choi H, et al. . Identification of Creb3l4 as an essential negative regulator of adipogenesis. Cell Death Dis 2014;5:e1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaPensee CR, Lin G, Dent AL, Schwartz J. Deficiency of the transcriptional repressor B cell lymphoma 6 (Bcl6) is accompanied by dysregulated lipid metabolism. PLoS One 2014;9:e97090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu X, Zhou Y, Yang Y, et al. . Identification of zinc finger protein Bcl6 as a novel regulator of early adipose commitment. Open Biol 2016;6:160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lizio M, Harshbarger J, Shimoji H, et al.; FANTOM Consortium . Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 2015;16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.