Abstract
Motivation
The collection, management and visualization of clinical pedigree (family history) data is a core activity in clinical genetics centres. However, clinical pedigree datasets can be difficult to manage, as they are time consuming to capture, and can be difficult to build, manipulate and visualize graphically. Several standalone graphical pedigree editors and drawing applications exist but there are no freely available lightweight graphical pedigree editors that can be easily configured and incorporated into web applications.
Results
We developed ‘pedigreejs’, an interactive graphical pedigree editor written in JavaScript, which uses standard pedigree nomenclature. Pedigreejs provides an easily configurable, extensible and lightweight pedigree editor. It makes use of an open-source Javascript library to define a hierarchical layout and to produce images in scalable vector graphics (SVG) format that can be viewed and edited in web browsers.
Availability and implementation
The software is freely available under GPL licence (https://ccge-boadicea.github.io/pedigreejs/).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The collection, management and visualization of pedigree (family history) data is a core activity in clinical settings, such as genetic centres and primary care, where healthcare professionals manage patients at risk of disease. Pedigree data are used to identify individuals at risk of familial disease and to inform their clinical management, including referral of individuals at elevated risk for mutation screening and/or recommending enhanced screening. For example, generating a pedigree is a requirement for calculating BRCA1 and BRCA2 mutation carrier probabilities and breast cancer risks in the BOADICEA (Lee et al., 2014) and IBIS (Tyrer et al., 2004) programs recommended in the National Institute for Health and Care Excellence guidelines (NICE, 2013, https://www.nice.org.uk/guidance/cg164).
Clinical pedigree datasets can be difficult to manage for several reasons: (i) they are supplied by humans not machines, and are therefore time consuming to capture in digital form; (ii) they include both family structure (which can be difficult to visualize in the presence of complex structure and still remain clear and comprehensible) and separate information for each family member (which must be linked); (iii) they often require updating over time; (iv) they can include data errors (e.g. broken family structures) and internal inconsistencies; and (v) they must be stored securely in order to fulfill data privacy requirements.
Many existing software applications capture and display pedigree data. Some open-source tools (e.g. Kinship2: Sinnwell et al., 2014) generate a vector image file from an input data file, whereas others (e.g. Pelican: Dudbridge et al., 2004 and PhenoTips: Girdea et al., 2013) provide a graphical pedigree editor. There are also commercial subscription pedigree drawing services (http://www.progenygenetics.com, http://pedigreedraw.com/, http://www.pedigreexp.com). However, after evaluating available tools (see Supplementary Data) we found no freely available lightweight pedigree drawing tools (components implemented solely in JavaScript) that could be easily incorporated into other web applications, and configured to provide a means of capturing and visualising pedigree data. Pedigreejs has been developed to fulfil these requirements as part of the ongoing BOADICEA project (http://ccge.medschl.cam.ac.uk/boadicea/, Lee et al., 2014) (Fig. 1).
Fig. 1.
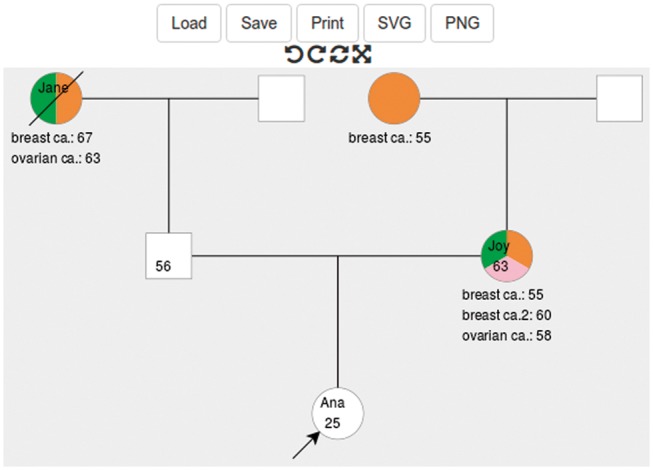
Screenshot showing a pedigree extending to second degree built using pedigreejs. The pedigree is annotated in the conventional manner: the proband is identified with an arrow; family members who have developed cancer are shaded; and unaffected family members are unshaded. Users can add or delete family members, and change their properties using widgets, which appear when the individual is selected. Buttons directly above the pedigree are used to ‘undo’ and ‘redo’ actions, ‘reset’ the initial structure, and switch to ‘fullscreen’ mode. The buttons at the top are used to ‘Load’ and ‘Save’ pedigree data files, to print the pedigree, and to export it as a SVG/PNG image
2 Materials and methods
Pedigreejs has been implemented as a configurable JavaScript library to provide a graphical pedigree editor in a web-based environment. The pedigreejs algorithm uses the d3-hierarchy JavaScript module (https://d3js.org) to assist the layout of individuals’ nodes in the pedigree representation. Pedigrees are rendered as SVG images in the browser window which results in a fast and responsive user experience to facilitate clinical data entry. The standard nomenclature for representing pedigrees (Bennett et al., 1995, 2008) is used. Pedigrees are built by adding parents, partners and siblings (including twins) using widgets that appear on mouseover of an individual.
Local browser storage is used to store the pedigree data (including family structure and linked data for each family member, e.g. sex, age). This means that confidential patient data can be stored on the client computer and not transmitted over the Internet. For reasons of data privacy the default is to hold these data for the session so that they do not persist after the browser or tab is closed. Previous versions of the pedigree dataset are also stored so that when a change is made it can be undone. Basic validation of the dataset is provided by default (e.g. check all mothers are female) and this can be overridden to define specific validations.
The pedigree data are stored in the lightweight JSON format (https://www.w3schools.com/js/js_json_intro.asp) to facilitate data interchange with other applications. JSON is easily extendable, so that the individual level data that are gathered are not limited. Once constructed, the pedigree dataset can be saved to a JSON format data file and loaded back into pedigreejs at a later date, so that pedigrees can be easily updated. In addition, pedigree data files in BOADICEA Web Application (v4) pedigree format and PED format (used by PLINK http://www.cog-genomics.org/plink/1.9, Chang et al., 2015) can be loaded into pedigreejs.
Pedigreejs has a variety of configuration options to assist integration with other web applications. For example, it can be configured to define the size of the SVG image that is rendered on the web-page, and define whether the image can zoom in and out. To allow for different property editors, an edit function can be provided in the configuration. In addition diseases can be configured and colours defined to identify affected individuals.
3 Results
Pedigreejs is a freely available, configurable, interactive, fast and responsive JavaScript pedigree editor. It is lightweight (implemented solely in JavaScript) and uses D3.js to assist in the rendering and does not send requests to a backend server. Creating and deleting individuals is made easy by clicking on widgets that appear on mouseover of a family member. ‘Undo’, ‘redo’ and ‘reset’ options and the ‘fullscreen’ option add to this being a user friendly editing tool.
Future developments will include further support for other commonly used pedigree data file formats (e.g. Madeline, Progeny). Pedigreejs will also be updated to support more complex family structures including consanguineous partners beyond those at the same level in the tree such as cousins. Pedigreejs will be used in future releases of the BOADICEA Web Application.
Supplementary Material
Acknowledgements
We would like to thank Beverley Speight and Aoife O'Shaughnessy-Kirwan from the East Anglian Medical Genetics Service for providing feedback and suggestions on the usability of the software.
Funding
This work was supported by Cancer Research UK grants: C12292/A20861 & C1287/A16563, the European Union’s Horizon 2020 research and innovation programme under grant agreements No 633784 (B-CAST) and 634935 (BRIDGES), and a Wellcome Trust Collaborative Award (203477/B/16/Z).
Conflict of Interest: none declared.
References
- Bennett R.L. et al. (1995) Recommendations for standardized human pedigree nomenclature. Pedigree Standardization Task Force of the National Society of Genetic Counselors. Am. J. Hum. Genet., 56, 745–752. [PMC free article] [PubMed] [Google Scholar]
- Bennett R.L. et al. (2008) Standardized human pedigree nomenclature: update and assessment of the recommendations of the National Society of Genetic Counselors. J. Genet. Couns., 17, 424–433. [DOI] [PubMed] [Google Scholar]
- Chang C.C. et al. (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 4, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. et al. (2004) Pelican: pedigree editor for linkage computer analysis. Bioinformatics, 20, 2327–2328. [DOI] [PubMed] [Google Scholar]
- Girdea M. et al. (2013) PhenoTips: patient phenotyping software for clinical and research use. Hum. Mutat., 34, 1057–1065. [DOI] [PubMed] [Google Scholar]
- Lee A.J. et al. (2014) BOADICEA breast cancer risk prediction model: updates to cancer incidences, tumour pathology and web interface. Br. J. Cancer, 110, 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICE (2013) Familial breast cancer: classification, care and managing breast cancer and related risks in people with a family history of breast cancer. Guidance and guidelines. [PubMed]
- Sinnwell J.P. et al. (2014) The kinship2 R package for pedigree data. Hum. Hered., 78, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrer J. et al. (2004) A breast cancer prediction model incorporating familial and personal risk factors. Stat. Med., 23, 1111–1130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


