Abstract
Summary
MIRMMR predicts microsatellite instability status in cancer samples using methylation and mutation information, in contrast to existing methods that rely on observed microsatellites. Additionally, MIRMMR highlights those genetic alterations contributing to microsatellite instability.
Availability and implementation
Source code is freely available at https://github.com/ding-lab/MIRMMR under the MIT license, implemented in R and supported on Unix/OS X operating systems.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Microsatellites consist of short DNA sequence repeats and may change in size due to errors in DNA replication, in particular because of strand slippage (Vilar et al., 2010). Normally, such errors are caught and repaired through mechanisms of the mismatch repair (MMR) pathway. However, changes in the methylation level of gene promoters and deleterious mutations in MMR pathway genes such as MLH1 may be responsible for dysregulation of the MMR pathway and increases in microsatellite instability (MSI) (Kim et al., 2013). MSI is strongly associated with inherited cancer syndromes such as Lynch syndrome and is an important diagnostic indicator that may influence treatment options.
Experimental and computational methods exist to detect MSI in patient samples. Experimentally, the length of known microsatellites is measured using gel electrophoresis and compared between normal and tumor samples. Computational methods such as MSIsensor (Niu et al., 2014) and mSINGS (Salipante et al., 2014) measure the prevalence of unstable microsatellites by examining sequence data from normal and tumor samples. MSIseq (Ni Huang et al., 2015) and MOSAIC (Hause et al., 2016) use machine learning classifiers based on microsatellite variants and other microsatellite features.
The experimental measurement process is time consuming and only assays a limited number of markers. Measuring microsatellite length in DNA-seq data requires computational resources to store and process sequencing data. MOSAIC and MSIseq mitigate these issues by working on smaller files but still focus on microsatellite features such as the number of microindels observed in simple repeat regions per megabase.
Instead of observing microsatellites directly to evaluate MSI status, we created an orthogonal prediction method using methylation levels and mutations in MMR pathway genes. Here we present MIRMMR (pronounced ‘murmur’): Microsatellite Instability Regression using Methylation and Mutations in R. MIRMMR trains logistic regression models using DNA methylation and mutation information from MMR pathway genes to classify MSI status. Once a prediction model has been trained, MIRMMR quickly reports the likely MSI status of new samples.
2 Materials and methods
MIRMMR consists of several independent modules to build logistic regression models, compare method outcomes, and classify MSI status in new samples. Users may select penalized, stepwise, or univariate modules to perform logistic regression modeling. Given a binary measure of MSI status, MIRMMR trains logistic regression models based on predictors such as MMR pathway gene methylation levels or mutation severity indicators, like Combined Annotation Dependent Depletion (CADD) scores (Kircher et al., 2014).
Penalized regression can perform variable selection by setting the coefficients of unimportant predictors to zero, which is vital to finding an informative and relevant model. MIRMMR’s penalized module performs elastic net regression based on R’s glmnet package (Friedman et al., 2010), which lets users balance the penalty term’s L1 and L2 norms.
A vital task in penalized regression is selecting an appropriate weight (lambda) to give the entire penalty term. Minimizing cross validation (CV) error is one way to find the optimal lambda value. However, due to the randomness of fold selection, the best lambda value may not be consistent between successive CV runs. After many independent CV runs, MIRMMR selects the lambda value with minimal average CV error. It fits a penalized logistic regression model using that lambda value and reports a logistic model based on the automatically selected variables.
See Supplementary Information for a description of all MIRMMR parameters, including options to train and test models on subsets of data.
3 Results
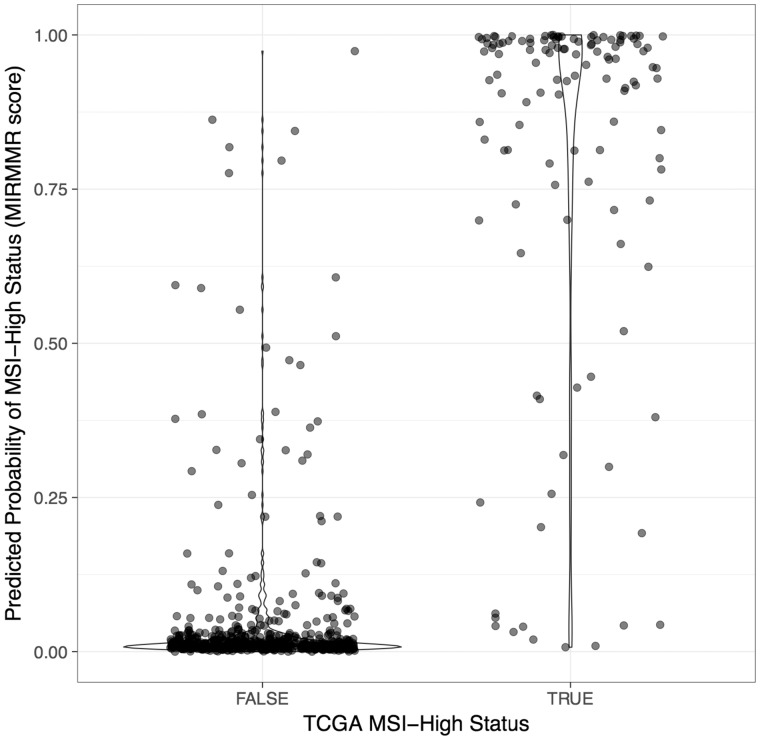
We used MIRMMR’s penalized module to train a model on colorectal (COADREAD), stomach (STAD) and uterine (UCEC) tumor samples from The Cancer Genome Atlas (TCGA) (TCGA Network et al., 2013). Of 676 total samples, 123 (123/676, 18.2%) were called MSI-High by TCGA. We trained the model using 10-fold CV with no samples withheld for testing. Model predictors included point mutation rate, methylation beta levels at MMR genes, and CADD scores for mutations found in MMR genes. See Supplementary Information for a full list of MMR pathway genes included and a summary of the final model produced, which highlights predictors important for MSI status prediction. Figure 1 illustrates the distribution of MIRMMR scores and shows a clear separation between TCGA MSI-High and Not-MSI-High groups.
Fig. 1.
MIRMMR scores. MIRMMR scores (y-axis) indicate a sample‘s predicted probability of having MSI-High status. Higher scores indicate higher probability of being MSI-High. The x-axis indicates MSI-High status reported by TCGA. The prediction model was built using 676 COADREAD, STAD and UCEC samples from TCGA
MIRMMR reports a score between zero and one, so a suitable cutoff to separate MSI-High samples from Not-MSI-High samples is necessary. Individual users may decide on a cutoff to balance their own needs for sensitivity and specificity. We selected a cutoff score of 0.1922 to maximize the sum of sensitivity (0.9187) and specificity (0.9421). With this cutoff, we found 634 samples (634/676, 93.8%) for which the original TCGA experimental MSI status call matched the MIRMMR call. Missed calls could be due to incomplete or inaccurate mutation and methylation reporting. We found similar areas under the curve when comparing the ROC curves of MIRMMR (0.9727), mSINGS (0.9799) and MSIsensor (0.9977), indicating that MIRMMR offers a promising new option for integrated MSI diagnosis that does not rely on measuring microsatellites. Given the high accuracy of existing, sequence-based methods, MIRMMR also offers an orthogonal measurement to reinforce concordant calls and flag potentially misclassified samples for further review.
4 Conclusion
MIRMMR provides a new dimension in MSI diagnosis and modeling. Although previous studies (Hause et al., 2016) have used regression to infer relationships between certain gene mutations and MSI, only MIRMMR performs full logistic regression model building for the purpose of MSI status prediction via binary classification. Building a prediction model highlights genes contributing to the MSI phenotype, and users can set intuitive classification thresholds based on probabilities.
We trained a logistic regression model to predict MSI status based only on mutation and methylation data using samples from COADREAD, STAD and UCEC cancer types. MIRMMR’s classification performance was on par with methods that rely on measuring microsatellites in BAM files, providing an additional, accurate tool for MSI diagnosis.
Supplementary Material
Acknowledgements
We thank the TCGA Research Network for providing the primary data and Michael C. Wendl for critical reading and discussion.
Funding
This work was supported by the National Cancer Institute [RO1CA180006, R01CA178383 and U24CA2110006 to L.D.].
Conflict of Interest: none declared.
References
- Friedman J. et al. (2010) Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw., 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- Hause R.J. et al. (2016) Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med., 22, 1342–1350. [DOI] [PubMed] [Google Scholar]
- Kim T.-M. et al. (2013) The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell, 155, 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M. et al. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Huang M. et al. (2015) MSIseq: software for assessing microsatellite instability from catalogs of somatic mutations. Sci. Rep., 5, 13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B. et al. (2014) MSIsensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics, 30, 1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- Salipante S.J. et al. (2014) Microsatellite instability detection by next generation sequencing. Clin. Chem., 60, 1192–1199. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network et al. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar E. et al. (2010) Microsatellite instability in colorectal cancer—the stable evidence. Nat. Rev. Clin. Oncol., 7, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.