Abstract
Environmental or social challenges can stimulate a cascade of coordinated physiological changes in stress response systems. Unfortunately, chronic activation of these adaptations under conditions such as low socioeconomic status (SES) can have negative consequences for long-term health. While there is substantial evidence tying low SES to increased disease risk and reduced life expectancy, the underlying biology remains poorly understood. Using pilot data on 120 older adults from the Health and Retirement Study (United States, 2002–2010), we examined the associations between SES and gene expression levels in adulthood, with particular focus on a gene expression program known as the conserved transcriptional response to adversity. We also used a bioinformatics-based approach to assess the activity of specific gene regulation pathways involved in inflammation, antiviral responses, and stress-related neuroendocrine signaling. We found that low SES was related to increased expression of conserved transcriptional response to adversity genes and distinct patterns of proinflammatory, antiviral, and stress signaling (e.g., sympathetic nervous system and hypothalamic-pituitary-adrenal axis) transcription factor activation.
Keywords: aging, gene expression, inflammation, social genomics, socioeconomic status, stress
Editor's note: An invited commentary on this article appears on page 510.
Low socioeconomic status (SES) has been linked to elevated chronic disease incidence, accelerated physical and cognitive decline, faster rates of aging, and lower life expectancy (1–4). Many of these processes are mediated by differential expression of the human genome (5), and current research seeks to identify the underlying molecular pathways mediating socioeconomic gradients in health. Previous work has shown that measurement noise, tissue-specific differences in gene expression, and variability in the cellular composition of tissue samples (e.g., differential prevalence of various leukocyte subsets) can complicate the detection of consistent SES-related differences in gene expression when examined at the level of individual gene transcripts (i.e., RNA encoding a single protein) (6). Fortunately, many of these obstacles can be overcome by adopting an “abstractionist” or set-based analytical approach that incorporates higher-order biological themes based on biological characteristics shared in common across sets of genes (7). Thus, instead of examining SES-related differences in gene expression on a specific gene-by-gene basis, we instead examine differential activity of specific a priori–defined sets of gene sets that were selected to tap generalized physiological processes that have previously been hypothesized to underlie the link between chronic stress and disease risk.
One biological pathway through which stressful life circumstances may produce health differentials involves modulation of gene expression in immune cells that contribute to the development of chronic illnesses such as cardiovascular disease, cancer, and neurodegenerative diseases (5). For instance, genome-wide transcriptional profiling studies have found that sets of genes that are up-regulated in response to chronic stress tend to be enriched with genes involved in inflammation, whereas those that are down-regulated in chronic stress are enriched with genes involved in immunoglobulin G antibody production and interferon-mediated antiviral responses (8, 9). This profile—termed the conserved transcriptional response to adversity (CTRA)—is thought to have evolved to facilitate wound-healing and prevent bacterial infection in the face of threatening conditions (including social conflict), while down-regulating resource allocation for combating viral infection (5, 9, 10). When chronically activated, however, the CTRA may contribute to an array of chronic neurodegenerative, metabolic, cardiovascular, and neoplastic diseases that share a common proinflammatory pathology (5).
Population studies have also begun to examine the upstream regulatory pathways that shape SES-related differences in gene expression. Gene expression is fundamentally regulated by protein transcription factors (TFs) that are activated by cellular receptors (e.g., detecting stress-related neuroendocrine factors) and subsequently bind to stereotyped DNA sequences in the human genome to coordinately alter the transcription of multiple genes involved in a common physiological process (e.g., inflammation) (11). One method for investigating the transcriptional consequences of low SES is to identify transcription factor binding motifs (TFBMs) that are overrepresented (enriched) in the promotor regions of genes that are differentially expressed as a function of SES (6, 12). As a result, further insight into the links between low SES and CTRA can be gained by examining whether promoter regions of over- or underexpressed genes are enriched for specific TFBMs that serve as targets for known biological mediators of stress or immune responses (12). For instance, there is evidence that social adversity contributes to inflammation through increased activity of the proinflammatory nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) family of TFs and through reduced activity of the antiinflammatory glucocorticoid receptor (GR). Other TFs implicated in CTRA gene expression include reduced activity of interferon response factors (IRFs) and increased activity of the cyclic adenosine monophosphate (cAMP) response element binding protein (CREB)/activating transcription factor TFs, which mediate signaling from “fight-or-flight” stress responses of the sympathetic nervous system (13).
The present study examines the leukocyte transcriptional alterations associated with major socioeconomic influences that have been found to affect health outcomes in adults. We hypothesized that low SES would be associated with a significant increase in expression of previously defined CTRA indicator genes. Further, genes that are empirically up-regulated in association with low SES would be characterized by high prevalence of TFBMs linked to proinflammatory activation (NF-κB family) and sympathetic nervous system signaling (CREB family) and low prevalence of TFBMs for the antiinflammatory GR and interferon-stimulated response elements (ISRE).
METHODS
Sample description
Participants were a subsample of individuals enrolled in the Health and Retirement Study (HRS) who provided venous blood samples, including a PAXgene RNA Tube sample (Qiagen, Valencia, California) that was subsequently analyzed as part of a transcriptomics pilot study. The HRS is a longitudinal prospective study of members of the US population over age 50 years and their spouses. It is conducted by the University of Michigan, under the sponsorship of the National Institute on Aging. The pilot sample consisted of about 200 participants randomly selected from a group of 1,000 respondents living in 13 areas who had completed a face-to-face interview in either 2006 or 2008 and who responded to the 2010 interview. Selected respondents were asked to provide a blood sample in the near future and at the time of the interview; 16% declined to participate, and an additional 15% did not complete the blood draw. The final completion rate was 69% (122 respondents). Of these 122, our final analytical sample included 120 individuals. One participant was excluded based on missing data for SES, and the other was excluded for not yielding suitable quantities of high-quality (intact) RNA (described below).
Low socioeconomic status
SES in adulthood was estimated based on self-reports of current educational and financial status. Low SES was classified from educational attainment and poverty status assessed over 5 waves. From wave 6 (2002) on, measures of current family poverty status were available via the RAND HRS files, based on threshold levels from the US Census Bureau. Participants were given a score of 0, 1, or 2 for poverty status. Scores of 2 corresponded to participants who were classified as being below poverty at any of the 5 waves. A score of 1 was assigned to participants who were never below the poverty level but who were at less than twice the poverty level for at least 1 wave. Participants who were always classified as being at least twice the poverty level were given a score of 0. Next, 1 point was added to these scores if participants also had less than a high school education (reported having less than 12 years of schooling). These scores (ranging from 0–3) were then used to create 2 categories—low SES in adulthood (a score of 2 or 3) versus middle to high SES in adulthood (a score of 1 or less).
Transcriptional profiles
Blood samples were collected into PAXgene RNA Tubes and shipped overnight to a central lab for storage at −80°C. RNA was extracted in parallel for all samples using an automated nucleic acid processing system (Qiagen QIAcube), and all samples were tested for suitable mass and purity (by spectrophotometry on an ND-1000 instrument; NanoDrop Technologies, Wilmington Delaware) and for suitable RNA integrity (by capillary electrophoresis on a TapeStation instrument; Agilent, Santa Clara, California). Overall, 121 of the total 122 PAXgene samples yielded suitable quantities of high-quality (intact) RNA, and these samples were subject to genome-wide transcriptional profiling using Illumina HT-12 v4 bead arrays (Illumina Inc., San Diego, California) in the UCLA Neuroscience Core Laboratory using standard target synthesis (TotalPrep; Ambion Inc., Austin, Texas) and hybridization/scanning protocols (Illumina). Transcript abundance values were quantile normalized and log2-transformed for analysis. Data were posted as GEO GSE68526.
Statistical analysis
In an initial estimation analysis, the relationship between low SES and expression of individual genes was quantified. This was done using standard linear statistical models adjusting for age, sex, race/ethnicity, body mass index, disease prevalence (diabetes, cardiovascular disease, cancer, or stroke), smoking status, weekly alcohol consumption, and the relative prevalence of 5 major leukocyte subsets, estimated using abundance of RNA for cardinal markers of monocytes (CD14), natural killer cells (CD16/FCGR3A, CD56/NCAM1), CD4+ and CD8+ T-lymphocyte subsets (CD3D, CD3E, CD4, CD8A), and B lymphocytes (CD19). Next, our 2 primary substantive hypotheses were tested using bioinformatics analyses based on inputs from the estimation analysis. To test associations between SES and the a priori–specified set of 53 CTRA indicator genes, we computed a contrast score (weighted +1 for 19 proinflammatory genes and −1 for 31 antiviral and 3 antibody-related genes) and assessed its sampling variability by bootstrap resampling of residuals from initial linear-model estimation analyses as previously described (14). To assess potential activity of 4 a priori–specified proinflammatory, antiviral, and neuroendocrine-related transcription control pathways, we identified all gene transcripts showing >1.2-fold differential expression between low-SES and middle/high-SES individuals and quantified the prevalence of transcription factor–binding DNA motifs within the upstream regulatory region (promoter) of genes that were up-regulated versus down-regulated based on data from the Transcription Element Listening System (TELiS) database (www.telis.ucla.edu), as previously described (11). Effect-size thresholding was used for consistency with previous research in this area (14) and has been shown to yield more replicable gene lists than does thresholding based on P values (15–18). Analyses focused specifically on hypotheses involving SES-related up-regulation of the proinflammatory NF-κB family (detected by TRANSFAC DNA motif V$CREL_01), the antiviral interferon response factor family (V$ISRE_01), and the neuroendocrine-related CREB family (V$CREB_02) and GR (V$GR_Q6). Statistical testing of (log-transformed) ratios of DNA motif prevalence in up- versus down-regulated genes was based on bootstrap standard errors as described above. To assess confounding by prevalent disease and leukocyte subset distributions, ancillary analyses omitted each of these covariates and reestimated SES associations with CTRA gene expression and TF activity. This sample had 80% power to detect small/moderate-sized associations (f2 ≥ 0.65) at P < 0.05, assuming a multiple regression analysis with control for 12 weakly correlated covariates with variance inflation factor of <2 (Web Figure 1, available at https://academic.oup.com/aje).
RESULTS
Sample characteristics
As shown in Table 1, the final pilot sample (n = 120) ranged in age from 48 to 95 years, with a mean of 73.3 (standard deviation, 9.8) years, and just over half the sample (56%) was female. Approximately 90% of respondents self-identified their race as non-Hispanic white, 5% (n = 6) self-identified as non-Hispanic black, and 5% (n = 6) self-identified as Hispanic. On average, participants had a body mass index of 27.9 (standard deviation, 7.1). About 9% (n = 11) of participants had ever smoked, and on average participants drank 2.37 alcoholic beverages per week. Prevalence rates for diabetes, cardiovascular disease, cancer, and stroke in our sample were 28%, 33%, 17%, and 16%, respectively. Finally, 16% (n = 19) were classified as having low SES in adulthood.
Table 1.
Characteristics From the Pilot Sample (n = 120), Health and Retirement Study, United States, 2002–2010
| Variable | No. of Participants | % | Mean (SD) |
|---|---|---|---|
| Age, years | 73.3 (9.8) | ||
| Sex (female) | 67 | 55.8 | |
| Race/ethnicity | |||
| Non-Hispanic white | 108 | 90.0 | |
| Non-Hispanic black | 6 | 5.0 | |
| Hispanic | 6 | 5.0 | |
| Body mass indexa | 27.9 (7.1) | ||
| Ever smoked | 11 | 9.2 | |
| Alcohol consumption, no. of beverages/week | 2.3 (5.0) | ||
| Disease prevalence | |||
| Diabetes | 34 | 28.3 | |
| Cardiovascular disease | 39 | 32.5 | |
| Cancer | 20 | 16.7 | |
| Stroke | 19 | 15.8 | |
| Low adulthood SES | 19 | 15.8 |
Abbreviations: SD, standard deviation; SES, socioeconomic status.
a Body mass index was calculated as weight (kg)/height (m)2.
Differential gene expression by SES
SES-related differences in leukocyte gene expression were quantified by genome-wide transcriptional profiling of 34,581 transcripts from venipuncture blood samples. Results identified 141 genes that were differentially expressed at a 1.2-fold difference or greater for individuals with low SES versus those with middle/high SES, after adjusting for age, sex, race/ethnicity, body mass index, disease prevalence, smoking status, weekly alcohol consumption, and the relative prevalence of 5 major leukocyte subsets. Of these, 67 genes were up-regulated in individuals with low SES, and 74 were down-regulated among low-SES individuals. While results from differential expression analysis point to the potential for SES to alter transcription, it doesn't provide insight into the biological pathways for which SES is relevant.
Gene contrast analysis
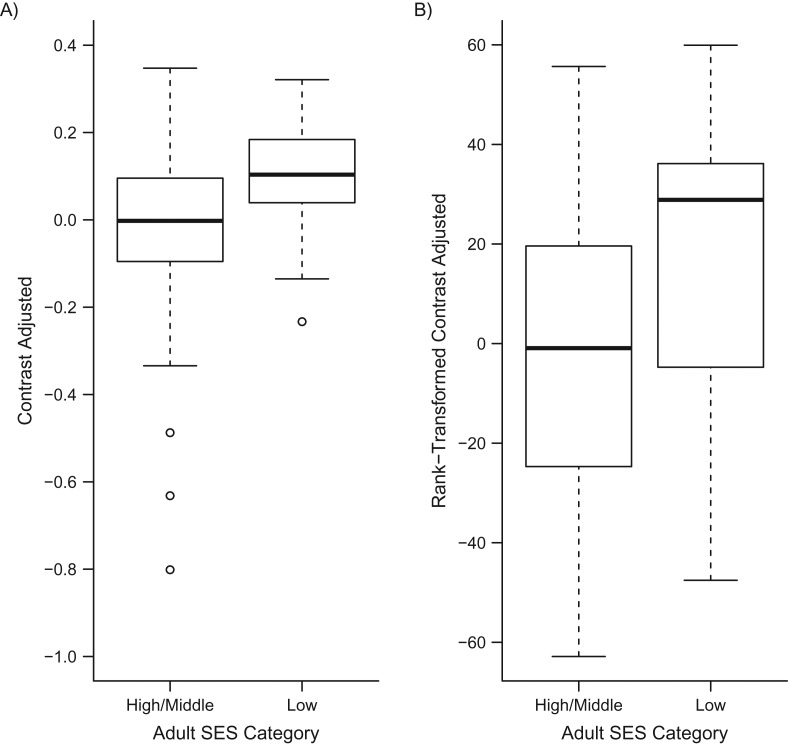
To determine whether low SES was associated with increased expression of the CTRA gene expression profile, we analyzed an a priori–specified contrast of 53 previously identified CTRA indicator genes (19 proinflammatory transcripts weighted +1, 3 antibody-related genes weighted −1, and 31 interferon-related genes weighted −1; transcripts listed in Web Table 1). Results (Figure 1) showed that low SES was associated with significant elevation in expression of the overall CTRA profile—characterized by increased proinflammatory gene expression and decreased antibody and antiviral gene expression (gene contrast = 0.151; standard error, 0.048; P = 0.0029). Similar results emerged in analyses that omitted control for prevalent disease (contrast = 0.194; standard error, 0.051; P = 0.0048) or leukocyte subset distributions (contrast = 0.152; standard error, 0.049; P = 0.0029).
Figure 1.
Conserved transcriptional response to adversity gene expression in 120 older adults as a function of high/middle versus low socioeconomic status (SES), Health and Retirement Study, United States, 2002–2010. Plotted data are residualized on age, sex, race/ethnicity, body mass index, prevalent disease (diabetes, cardiovascular disease, stroke, and cancer), smoking, heavy alcohol consumption, and 8 mRNA indicators of leukocyte subset prevalence. Data represent raw contrast values (log2 RNA expression units) (A) and rank-transformed contrast values (to address outliers) (B).
Transcription control pathways
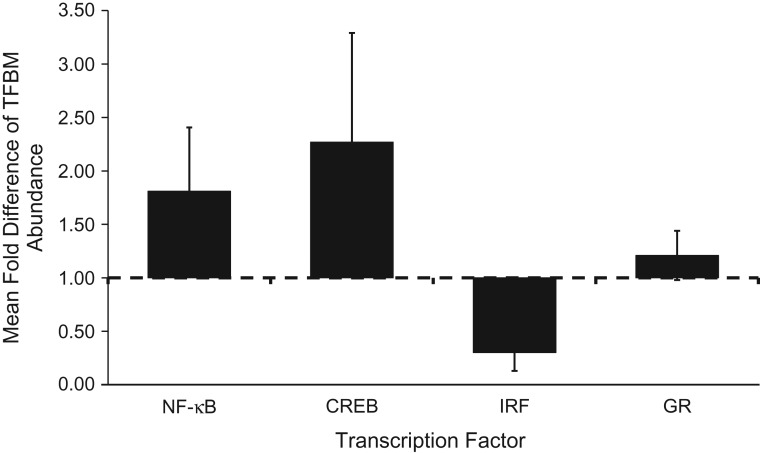
To identify transcription control pathways that may have contributed to the empirically observed differences in gene expression, we conducted Transcription Element Listening System–based bioinformatics analysis of the prevalence of 4 specifically hypothesized TFBMs within the promoters of all genes showing >1.2-fold differential expression as a function of SES (Figure 2). Genes up-regulated in association with low SES showed an overrepresentation of TFBMs for the proinflammatory NF-κB family of transcription factors (fold difference = 1.81; P = 0.038) and the CREB family of transcription factors that mediate sympathetic nervous system signaling (fold difference = 2.27; P = 0.028), and an under-representation of TFBMs for IRFs (fold difference = 0.30; P = 0.009). However, low SES was not linked to any significant asymmetry in GR TFBM distribution (fold-difference = 1.12; P = 0.292). Comparable associations were observed in ancillary analyses that omitted control for prevalent disease (NF-κB fold difference = 1.77, P = 0.033; CREB fold difference = 2.31, P = 0.018; IRF fold difference = 0.30, P = 0.006; and GR fold difference = 1.19, P = 0.291). Absent control for leukocyte subsets, IRF TFBMs continued to show significant asymmetry (fold difference = 0.35; P = 0.007) but NF-κB (fold difference = 1.25; P = 0.397) and CREB (fold difference = 1.23; P = 0.535) were both rendered nonsignificant.
Figure 2.
Transcription Element Listening System (TELiS) bioinformatics results to assess the abundance of transcription factor binding motifs (TFBMs) among the promoters of genes showing >1.2-fold differential expression as a function of socioeconomic status differences, Health and Retirement Study, United States, 2002–2010. CREB, cyclic adenosine monophosphate (cAMP) response element binding protein; IRF, interferon response factor; GR, glucocorticoid receptor; NF-κB, nuclear factor κ-light-chain-enhancer of activated B cells. Error bars represent standard errors.
DISCUSSION
Low SES has been linked to reductions in life expectancy and higher age-specific rates of heart disease, cancer, diabetes, neurodegenerative diseases, and frailty (5, 19, 20). Yet much is still unknown regarding the underlying biology that enables social disparities to “get under the skin.” Our results show that by the time of late adulthood, low-SES individuals—those at or near the poverty limit and/or with less than a high school education—show notable differences in leukocyte gene expression profiles relative to members of higher socioeconomic groups. Specifically, low-SES individuals show greater expression of the CTRA gene expression program previously observed in individuals exposed to extended periods of threat, uncertainty, or adverse life circumstances (8).
CTRA, which is characterized by increased proinflammatory gene expression and decreased expression of antiviral and antibody genes, is hypothesized to be an evolutionarily adaptive response to social threat. Under socially hostile environments, it may have been more advantageous for our ancestors to up-regulate inflammatory defenses to aid in wound-healing via fight-or-flight signaling while deemphasizing resources involved in antiviral defense (9). Unfortunately, when chronically activated in response to the persistent adversity associated with low SES, this functional genomic signature of social stress may contribute to pathogenesis of disease (21). Prolonged inflammatory signaling can damage extracellular molecules and bystander cells (22) and act as a tumor promotor (23). For these reasons, inflammation has been implicated as a central cause of human aging—accelerating the development of atherosclerosis, diabetes, Alzheimer's disease, and a number of cancers (5, 21, 24). Finally, the impaired interferon and antibody components of the CTRA profile may also contribute to infectious disease risk by reducing host resistance to viral pathogens and lowering antibody response after vaccination (25, 26).
Differential gene expression as a function of SES may stem from activation of specific TFs in response to stress signaling. Indeed, results from promoter-based bioinformatics analysis suggest that the differences observed here may stem from increased activity of NF-κB and CREB family TFs, as well as reduced activity of IRF family TFs. These results are consistent with studies from both human and animal models, which have shown that exposure to social stress is often accompanied by increases in circulating levels of proinflammatory cytokines (9, 10). The distinctive patterns of transcription factor activation suggest the association of low SES and physiologic activity may operate through immunoregulatory pathways (NF-κB and IRF) with some additional contribution from the sympathetic nervous system–related CREB pathway. Primary analyses reported here show that these associations occur on a per-cell basis (i.e., after control for variations in the prevalence of specific cell types within the circulating leukocyte pool) and are independent of disease status (e.g., history of diabetes, cardiovascular disease, stroke, or cancer). Ancillary analyses that did not control for disease history found virtually identical results, suggesting that differential pathology is not a major mediator of the relationships observed here. Stress biology can regulate gene expression in the circulating leukocyte pool through per-cell changes in gene transcription and by altering the development and relative prevalence of specific cell subpopulations within the circulating leukocyte pool (6, 13). Primary analyses controlled for individual differences in cell subpopulation prevalence in order to focus on per-cell associations. Ancillary analyses that did not control for cell subpopulation markers found similar up-regulation of the overall CTRA gene expression profile and similar down-regulation of IRF activity. However, no significant SES-related differences in NF-κB or CREB activity emerged (association point estimates regressed partially toward null, and standard errors were not notably affected), suggesting that variations in cell prevalence may partially confound estimates of some SES-related transcription control pathways. Future research using isolated cell populations will be needed to help clarify the respective roles of differential cell development versus per-cell gene regulation in mediating the CTRA correlates of low SES.
There are limitations to this study which need to be acknowledged. Given that this analysis was conducted using a pilot sample from the HRS, our sample size may have limited our statistical power to detect significant differences. Given the limited sample size available, this study focused solely on a priori hypotheses derived from the previous research literature on the CTRA and SES-related differences in inflammatory gene expression (8, 9, 13, 14, 27). As such, the present analyses may have missed some transcriptomic correlates of SES. Discovery of additional genes and gene sets that relate to SES is an important topic for future research in larger samples. Finally, as these genomics-based measures become available in larger samples, it will be important to test causal models hypothesizing mediation of social gradients in health by transcriptional alterations in CTRA-related genes or activation of specific TFs. The associations estimated here derived from an observational study, and this design cannot rule out reverse causation (e.g., wherein the health or behavioral consequences of differential gene expression cause poverty).
Our study was strengthened by the population being examined, which was sampled at random from a large multiethnic, nationally representative study of older adults. We also were able to capitalize on the longitudinal nature of HRS by taking into consideration poverty status over a number of years (2002–2012). Overall, this study provides additional insight into the nature of gene transcriptional dynamics that may contribute to social gradients in health.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California (Morgan E. Levine); Leonard Davis School of Gerontology, University of Southern California, Los Angeles, California (Eileen M. Crimmins); Survey Research Center, Institute for Social Research, University of Michigan, Ann Arbor, Michigan (David R. Weir); and Division of Hematology-Oncology, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California (Steve W. Cole).
This research was supported by National Institute on Aging (grants P30AG017265, K99AG052604, and T32AG0037), the National Institute of Neurological Disorders and Stroke (grant T32NS048004), and the Health and Retirement Study cooperative agreement from the National Institute on Aging (grant U01 AG009740).
Conflict of interest: none declared.
REFERENCES
- 1. Adler NE, Newman K. Socioeconomic disparities in health: pathways and policies. Health Aff. 2002;21(2):60–76. [DOI] [PubMed] [Google Scholar]
- 2. Clark AM, DesMeules M, Luo W, et al. . Socioeconomic status and cardiovascular disease: risks and implications for care. Nat Rev Cardiol. 2009;6(11):712–722. [DOI] [PubMed] [Google Scholar]
- 3. Seeman TE, McEwen BS, Rowe JW, et al. . Allostatic load as a marker of cumulative biological risk: MacArthur studies of successful aging. Proc Natl Acad Sci USA. 2001;98(8):4770–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crimmins EM, Kim JK, Seeman TE. Poverty and biological risk: the earlier “aging” of the poor. J Gerontol A Biol Sci Med Sci. 2009;64(2):286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finch C. The Biology of Human Longevity: Inflammation, Nutrition, and Aging in the Evolution of Life Spans. 1st ed Burlington, MA: Academic Press; 2007. [Google Scholar]
- 6. Cole SW. Elevating the perspective on human stress genomics. Psychoneuroendocrinology. 2010;35(7):955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mellon SH, Wolkowitz OM, Schonemann MD, et al. . Alterations in leukocyte transcriptional control pathway activity associated with major depressive disorder and antidepressant treatment. Transl Psychiatry. 2016;6:e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole SW. Human social genomics. PLoS Genet. 2014;10(8):e1004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cole SW. Social regulation of human gene expression: mechanisms and implications for public health. Am J Public Health. 2013;103(suppl 1):S84–S92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11(9):625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hill CS, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80(2):199–211. [DOI] [PubMed] [Google Scholar]
- 12. Cole SW, Yan W, Galic Z, et al. . Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005;21(6):803–810. [DOI] [PubMed] [Google Scholar]
- 13. Powell ND, Sloan EK, Bailey MT, et al. . Social stress up-regulates inflammatory gene expression in the leukocyte transcriptome via β-adrenergic induction of myelopoiesis. Proc Natl Acad Sci USA. 2013;110(41):16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller GE, Chen E, Fok AK, et al. . Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proc Natl Acad Sci USA. 2009;106(34):14716–14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi L, Jones WD, Jensen RV, et al. . The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics. 2008;9(suppl 9):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Witten D, Tibshirani R. A comparison of fold-change and the t-statistic for microarray data analysis Stanford, CA: Stanford University; 2007.
- 17. Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: a PRIM approach. Bioinformatics. 2003;19(14):1808–1816. [DOI] [PubMed] [Google Scholar]
- 18. Fredrickson BL, Grewen KM, Coffey KA, et al. . A functional genomic perspective on human well-being. Proc Natl Acad Sci USA. 2013;110(33):13684–13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kiecolt-Glaser JK, McGuire L, Robles TF, et al. . Emotions, morbidity, and mortality: new perspectives from psychoneuroimmunology. Annu Rev Psychol. 2002;53:83–107. [DOI] [PubMed] [Google Scholar]
- 20. Glass CK, Saijo K, Winner B, et al. . Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140(6):918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finch CE. Evolution in health and medicine Sackler colloquium: evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci USA. 2010;107(suppl 1):1718–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Finch CE, Morgan TE, Longo VD, et al. . Cell resilience in species life spans: a link to inflammation. Aging cell. 2010;9(4):519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Philip M, Rowley DA, Schreiber H. Inflammation as a tumor promoter in cancer induction. Semin Cancer Biol. 2004;14(6):433–439. [DOI] [PubMed] [Google Scholar]
- 24. Finch CE, Crimmins EM. Inflammatory exposure and historical changes in human life-spans. Science. 2004;305(5691):1736–1739. [DOI] [PubMed] [Google Scholar]
- 25. Sloan EK, Capitanio JP, Tarara RP, et al. . Social stress enhances sympathetic innervation of primate lymph nodes: mechanisms and implications for viral pathogenesis. J Neurosci. 2007;27(33):8857–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weinberger B, Herndler-Brandstetter D, Schwanninger A, et al. . Biology of immune responses to vaccines in elderly persons. Clin Infect Dis. 2008;46(7):1078–1084. [DOI] [PubMed] [Google Scholar]
- 27. Levine ME, Cole SW, Weir DR, et al. . Childhood and later life stressors and increased inflammatory gene expression at older ages. Soc Sci Med. 2015;130:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.