Abstract
Motivation
In an effort to better understand the molecular drivers of synaptic and neurophysiologic dysfunction in Alzheimer’s disease (AD), we analyzed neuronal gene expression data from human AD brain tissue to identify master regulators of synaptic gene expression.
Results
Master regulator analysis identifies ZCCHC17 as normally supporting the expression of a network of synaptic genes, and predicts that ZCCHC17 dysfunction in AD leads to lower expression of these genes. We demonstrate that ZCCHC17 is normally expressed in neurons and is reduced early in the course of AD pathology. We show that ZCCHC17 loss in rat neurons leads to lower expression of the majority of the predicted synaptic targets and that ZCCHC17 drives the expression of a similar gene network in humans and rats. These findings support a conserved function for ZCCHC17 between species and identify ZCCHC17 loss as an important early driver of lower synaptic gene expression in AD.
Availability and implementation
Matlab and R scripts used in this paper are available at https://github.com/afteich/AD_ZCC
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
One of the earliest events in Alzheimer’s disease (AD) is synaptic dysfunction (Scheff et al., 2007). In microscopic post-mortem studies, synaptic loss correlates strongly with pre-mortem cognitive status (DeKosky and Scheff, 1990), and serves as a better predictor of pre-mortem cognitive status than either plaque or tangle pathology (Terry et al., 1991). In animal models of AD, early aberrations in synaptic physiology have been widely reported, and AD-associated synaptic structural abnormalities (such as altered dendritic spine morphology) have also been linked to neurophysiologic defects in AD animal models (see Pozueta et al., 2013 for a recent review). Faced with this evidence, multiple groups have proposed that synaptic dysfunction is central to the pathophysiology of AD (Gonatas et al., 1967; Teich et al., 2015).
In an effort to identify master regulators of synaptic and neurophysiologic dysfunction in AD, we have applied novel data mining techniques (Lefebvre et al., 2010; Margolin et al., 2006) to neuronal RNA expression data from AD brain tissue. This computational effort identified a protein (ZCCHC17, also known as pNO40), which is hypothesized to normally support the expression of a network of synaptic genes, and whose dysfunction in AD is predicted to cause impaired expression of these genes. Motivated by this theoretical work, we experimentally demonstrate that ZCCH17 is expressed in neurons and is lost early in the course of AD. We knock-down ZCCHC17 in rat neurons and demonstrate that a majority of the synaptic genes from our computational work show decreased expression, and that the ZCCCH17 regulon from our human data is predictive of the gene expression changes in our rat data, which suggests that ZCCHC17 has a conserved function across species at a systems level. This work establishes ZCCHC17 loss as an early driver of lower synaptic gene expression in AD.
2 Materials and methods
We identified ZCCHC17 through a computational screen of previously published neuronal RNA expression data (Liang et al., 2007, 2008, 2010). Initial work from this analysis identified master regulators (MRs) that were predicted to drive changes in gene expression in AD (Aubry et al., 2015). Here, we use this initial work as the starting point to identify MRs that drive synaptic dysfunction in AD. Aubry et al. identified MRs by region in six regions of brain, and each region had a separate list of MRs that are predicted to drive gene expression in AD brain tissue in that region (see Supplementary Methods for full details). With this initial list from Aubry et al., we first assembled a list of MRs that were predicted to have differential activity in three or more of these six regions, as we are interested in finding MRs whose dysfunction may be generalizable to neurons throughout the brain in AD. For each MR that passed this threshold, we then calculated the average change in gene expression for each ‘synaptic’ gene across all regions where this MR is predicted to have differential activity. We defined ‘synaptic’ genes using a pre-defined list of ‘synaptic’ genes from the Ingenuity database (Kramer et al., 2014). We curated this list using a broad definition that tagged any gene that was related to synaptic function (according to Ingenuity‘s filter), even if its protein product is not technically localized to the synapse. This relatively broad definition fit our purpose, as our goal was to identify MRs that drive synaptic and neurophysiologic dysfunction. Although a narrower focus limited to proteins actually localized to the synapse would be interesting, it was not the goal of this project. We then calculated the synaptic score for a given MR (SMR) with the following equation:
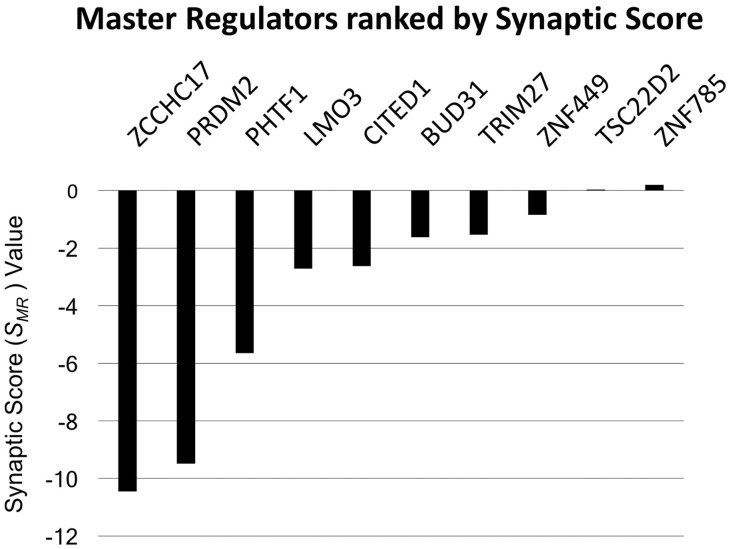
where n is the number of synaptic genes that correlate with a given MR, ri is the Spearman‘s rank correlation between the MR and ith synaptic gene, and di is the difference in the expression values of the ith synaptic gene. Thus, MRs with low synaptic scores in Figure 1 are predicted to normally support the upregulation of synaptic genes, and dysfunction of these MRs is predicted to lead to lower expression levels of these genes in AD.
Fig. 1.
ZCCHC17 loss is predicted to contribute to synaptic dysfunction in AD. We used novel data mining techniques to identify putative MRs of synaptic dysfunction in AD (see Supplementary Methods for details). Each MR was assigned a synaptic score (SMR), which is essentially a sum of the change in a MR’s synaptic targets in AD, weighted by the correlations between the MR and each target. MRs with large negative SMR values have large positive correlations with synaptic genes with large decreases in expression in AD. ZCCHC17 has the most negative value in our analysis, which suggests that ZCCHC17 (1) Normally supports the expression of a large number of synaptic genes, and (2) ZCCHC17 dysfunction contributes to loss of support for these synaptic genes in AD
Immunohistochemistry and western blotting was performed with adult human brain tissue acquired from the Columbia University ADRC brain bank, and qPCR and RNA-seq was performed on 14 day-old rat cortical cultures. Detailed Methods, including full computational details of our MR analysis, can be found in Supplementary Information.
3 Results
3.1 ZCCHC17 loss correlates with loss of synaptic genes in AD
We identified ZCCHC17 through a computational screen of prior work that had identified master regulators (MRs) that are predicted to drive gene expression in AD (Aubry et al., 2015) (see Materials and methods and Supplementary Methods for details). Here, we use this initial work to identify MRs that 1) Normally support the expression of a large number of synaptic genes, and 2) Whose impairment in AD is predicted to lead to lower levels of expression in these genes. As seen in Figure 1, the top ranked MR from this analysis is ZCCHC17. Note that the ‘synaptic score’ value in Figure 1 is low for MRs that (i) have large numbers of synaptic targets, (ii) positively correlate with these targets, and (iii) these targets are decreasing in AD. Using more conventional ontology analysis, ZCCHC17’s regulon also has a high representation of synaptic ontology groups (Supplementary Table S1).
ZCCHC17 is a poorly understood protein that has been shown to interact with transcription factors (Chang et al., 2003; Joo et al., 2010) as well as splicing factors (Lin et al., 2017; Ouyang, 2009). ZCCHC17 contains a zinc-finger (CCHC) domain and a nuclear localization signal, and microscopy has localized ZCCHC17 to the nucleus and nucleolus (Chang et al., 2003; Gueydan et al., 2002; Lin et al., 2017; Ouyang, 2009). Although there is no published experimental study of ZCCHC17 protein in brain tissue, a recent meta-analysis of RNA expression data from six previous gene expression studies of AD brain tissue included ZCCHC17 as one of the top 30 most reliably downregulated genes in AD (Li et al., 2015).
3.2 ZCCHC17 is expressed in neurons and is lost early in the course of AD pathology
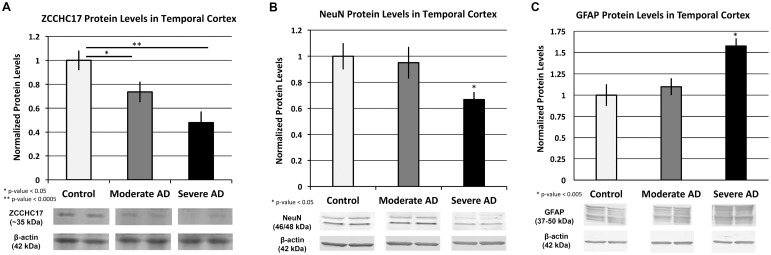
ZCCHC17 protein has not previously been studied in brain tissue, and so we examined which cell types express ZCCHC17. Immunohistochemistry at higher antibody concentrations stain mainly nucleus but also cytoplasm of cells throughout the grey matter (Supplementary Fig. S1A) and to a lesser extent white matter neuropil (Supplementary Fig. S1B). Although ZCCHC17 does not appear to be exclusively neuronal, it does stain neurons strongest. This is seen at lower antibody concentrations, where staining is limited to the nucleolus of large pyramidal neurons (Supplementary Fig. S1C and D). Western blot analysis with AD brain tissue shows that ZCCHC17 protein levels decrease with increasing severity of AD pathology (Fig. 2A). In addition, ZCCH17 loss occurs early in AD, before significant neuronal loss or gliosis (Fig. 2B and C), which supports an early role for ZCCHC17 loss in AD pathophysiology.
Fig. 2.
ZCCHC17 is lost at early stages of AD, before significant gliosis and neuronal loss. (A) ZCCHC17 shows statistically significant changes in both moderate AD and severe AD in temporal cortex. This is before significant neuronal loss (as measured by NeuN, B) or astrogliosis (as measured by GFAP, C). All bands are normalized by beta-actin; see Supplementary Figures S2–S4 for full blots. n = 9 for control, 10 for moderate AD and 10 for severe AD (P-values for ZCCHC17 blots: Ctl versus Mod = 0.02, Mod versus Sev = 0.03, Ctl versus Sev = 0.0003). In (B) we see the classic NeuN doublet (Dredge et al., 2011); the difference between control and severe is significant (P-value = 0.007), as is the difference between moderate and severe (P-value = 0.03). In (C), the GFAP signal shows the characteristic banding pattern from 50 kDa to 37 kDa (Zoltewicz et al., 2012). The difference between control and severe is also significant (P-value = 0.001), as is the difference between moderate and severe (P-value = 0.001). All P-values generated with one-sided t-test; error bars are standard error
3.3 ZCCHC17 supports the expression of synaptic genes in rats and has a conserved function across species
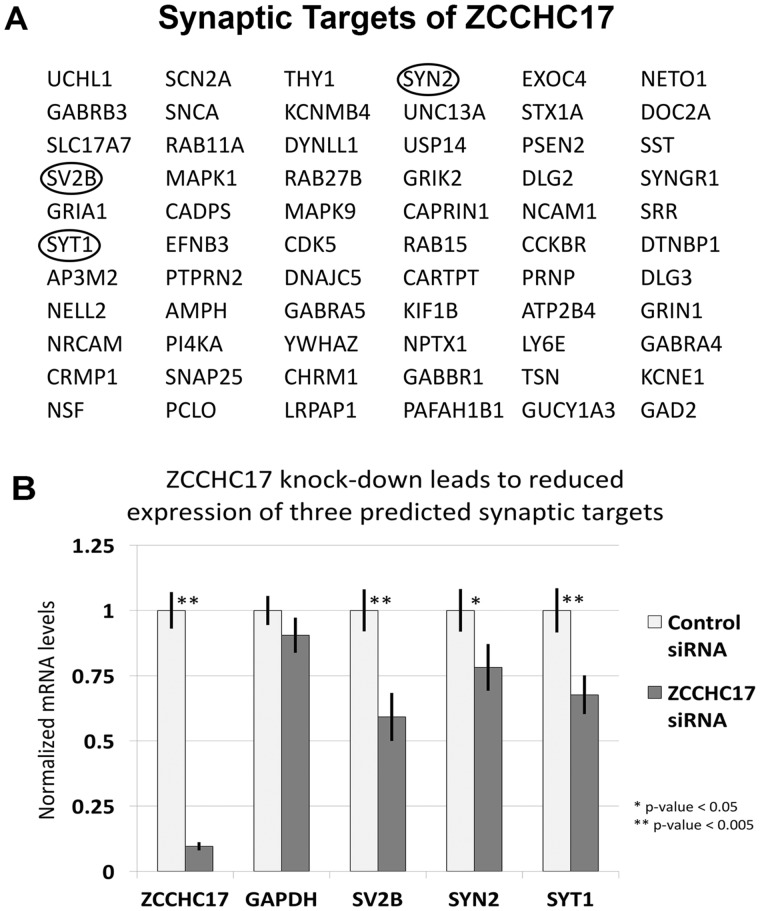
We next tested our hypothesis that ZCCHC17 influences expression of synaptic genes. To test this hypothesis, we knocked-down ZCCHC17 in rat neuronal cultures (Supplementary Fig. S5). As expected from our human immunohistochemistry and prior literature (Chang et al., 2003; Lin et al., 2017; Ouyang, 2009), ZCCHC17 is localized primarily to the nucleus of rat cortical neuronal cultures. Introducing ZCCHC17 siRNA led to a reduction of approximately 50% of ZCCHC17 protein. Our computational work defines an interactome, which allows us to hypothesize which genes are in the regulon of a given MR. We tested the hypothesis that lowering ZCCHC17 protein levels will lead to reductions in mRNA transcripts of the subset of synaptic genes in the ZCCHC17 regulon. We first performed qPCR on three synaptic genes from our dataset that are predicted to have particularly strong positive correlations with ZCCHC17: SYT1, SYN2 and SV2B. qPCR for these three genes demonstrated statistically significant reductions in mRNA levels after ZCCHC17 knock-down (Fig. 3A and B). We then performed RNA-seq and discovered that a majority of the synaptic genes predicted to positively correlate with ZCCHC17 have reduced expression after ZCCHC17 knock-down. Of the 66 synaptic genes that positively correlate with ZCCHC17 in our interactome analysis with human expression data, 37 are significantly decreased in rat cortical cultures after ZCCHC17 knock-down (Supplementary Table S2).
Fig. 3.
ZCCHC17 has a conserved function across species. (A) Our interactome work identified target genes whose expression ZCCHC17 supports. Shown here are the synaptic genes whose expression is predicted to be driven by ZCCHC17. After ZCCHC17 knock-down, three of ZCCHC17’s predicted synaptic targets were selected based on particularly strong positive Spearman‘s rank correlation coefficients in our computational work (circled). (B) All three targets (SYT1, SYN2 and SV2B) showed a statistically significant reduction using qPCR after ZCCHC17 knock-down. All qPCR values were normalized by RPL13, a ribosomal protein previously used as a housekeeping gene control in qPCR of AD brain tissue (Gebhardt et al., 2010); RPL13 is not predicted to be affected by ZCCHC17. qPCR was also performed on GAPDH as an additional positive control. n = 11 biological replicates in each group (P-values: ZCCHC17 = 3.4 × 10-8, GAPDH = 0.14, SV2B = 0.002, SYN2 = 0.04, SYT1 = 0.0049). All of the P-values were generated with a one-sided t-test. The displayed error bars are standard error
The above analysis suggests that ZCCHC17 has a relatively conserved function in neurons across species. To test this hypothesis further, we analyzed the relationship between ZCCHC17 and its full regulon between rats and humans. Although ZCCHC17 is predicted to regulate 66 synaptic genes, this is a fraction of the 775 total number of genes predicted to be regulated by ZCCHC17 in our human interactome analysis. We found that 668 of these 775 genes have a single unambiguous rat homologue that was also expressed at detectable levels in our rat cortical cultures (Supplementary Table S3). Each ZCCHC17-target interaction from our interactome work has an associated Spearman‘s rank correlation value (Aubry et al., 2015). We tested the hypothesis that the Spearman‘s rank correlation between ZCCHC17 and each target gene in our human interactome is predictive of the identical Spearman‘s rank correlation in our rat data. Our human interactome was built using 193 samples (see Supplementary Methods). Using our 12 rat RNA-seq profiles, we found that 233 of the 668 ZCCHC17 human regulon genes with an expressed rat homologue reached a significant correlation with ZCCHC17 in our rat data. Further analysis demonstrates that 81% of these identified genes in our rat data show the same sign of Spearman‘s rank correlation with ZCCHC17 in our human interactome (P-value < 2.2 × 10−16, binomial test; Supplementary Fig. S6A). In addition, across all of these 668 rat homologue genes, the correlation value of each ZCCHC17-target gene pair in our human data is mildly predictive of the equivalent value in our rat data (whether or not it reached significance in the rat data), with a Spearman‘s rank correlation of 0.28 between these two values across all 668 genes (P-value = 1.41 × 10−13 using a one-sided t-test). Note that our human interactome was generated with RNA expression data from aged human brain tissue, while our experimental data is derived from embryonal rat neuronal cultures. In addition, the experimental protocols used to generate the two datasets are completely different; laser-captured neurons were profiled using Affymetrix U133 Plus 2.0 Array for our human data, while rat cortical cultures were profiled using RNA-seq analysis. Taken together, we believe this implies a high degree of robustness to our findings, and supports a relatively conserved function for ZCCHC17 between species and across multiple targets.
Although the above findings are encouraging for our interactome analysis, one may still ask whether genes related to synaptic function are the most relevant net effect of ZCCHC17 knock-down. In other words, our computationally derived ZCCHC17 regulon is a fraction of the total number of genes that have changes in expression after ZCCHC17 knock-down. The ARACNe algorithm we used to establish the interactome is designed to detect only direct interactions between a transcriptional regulator and a target gene. This is accomplished by eliminating correlations between an MR and a secondary effect gene using the data processing inequality (Basso et al., 2005; Margolin et al., 2006; Palomero et al., 2006). However, secondary effects in gene expression will be seen in the RNA-seq profile after knocking-down an MR. We performed ontology analysis on the RNA-seq profile of our rat cortical cultures after ZCCHC17 knock-down (Supplementary Table S4) in order to see if synaptic gene categories are represented amongst all changes in gene expression. As seen in Supplementary Figure S6B, six of the top ten most significant ontology categories relate to synaptic function. Of note, all of these ‘synaptic’ categories show a net decrease in expression. This further validates our hypothesis that ZCCHC17 normally supports the expression of genes that facilitate synaptic function, and that decreased ZCCHC17 levels in AD contributes to decreased expression of these genes.
4 Discussion
In this study, we computationally identify ZCCHC17 loss as a potential driver of synaptic and neurophysiologic dysfunction in AD. We show that ZCCHC17 is expressed in neurons and decreases early in AD, before significant neuronal loss or gliosis. We then knock-down ZCCHC17 in rat cortical cultures and show that the majority of ZCCHC17’s target synaptic genes show the hypothesized decrease in expression. In addition, the ZCCHC17 human regulon is predictive of equivalent interactions between ZCCHC17 and homologous genes in rat neurons, which supports a conserved function. Finally, the net effect on gene expression after ZCCHC17 knock-down preferentially affects ontology groups related to synaptic function, even when secondary changes in gene expression are included. Taken together, these data support the view that ZCCHC17 is a conserved nuclear protein whose loss early in AD contributes to loss of support for synaptic gene expression.
ZCCHC17 is still a relatively unstudied protein, and the work detailed here elevates the importance of understating its normal function. As noted earlier, ZCCHC17 has a nuclear localization signal and has been shown to interact with pinin (Chang et al., 2003) [which has several roles in the nucleus, including transcriptional regulation (Joo et al., 2010) and modulating mRNA splicing (Wang et al., 2002)], as well as the splicing factors SRrp37 (Ouyang, 2009), SRSF1 and SRSF2 (Lin et al., 2017). This suggests that ZCCHC17 may function as a co-factor to transcription and/or play a role in regulating mRNA splicing. A recent study (Lin et al., 2017) demonstrated that overexpression of ZCCHC17 in U2OS cells is associated with aberrant mRNA splicing. A role for ZCCHC17 in mRNA splicing would be interesting, and not mutually exclusive from its role in supporting gene expression. Pre-mRNA synthesis and splicing are increasingly recognized as occurring in tandem (Saldi et al., 2016), and a possible coordinating role for ZCCHC17 could lead to disruptions in both processes. In addition to ZCCHC17’s nuclear role, early work also found that cytoplasmic ZCCHC17 co-fractionates with ribosomes (Gueydan et al., 2002). This led some to initially propose that ZCCHC17 was part of the ribosome complex (Gueydan et al., 2002). At higher antibody concentrations, we also find non-nuclear staining (Supplementary Fig. S1A). Although recent work has focused on the role of ZCCHC17 in the nucleus, a ribosomal role would be very interesting. In light of ZCCHC17’s nuclear function, one might speculate that this ribosomal role could involve guidance or stabilization of mRNA transcripts during translation.
The relationship between ZCCHC17 loss and the other stigmata of AD pathology, and what ultimately causes ZCCHC17 loss, should be addressed in future work. Although amyloid and/or tau may play a role, additional toxic sequelae of mitochondrial dysfunction, disruptions in autophagy and neuro-inflammation should all be considered. Considering the large-scale role that ZCCHC17 function has on genome expression generally, and on synaptic gene expression specifically, ZCCHC17 is clearly vital for maintaining normal synaptic gene expression, and could be a target for future therapies aimed at alleviating synaptic dysfunction in AD.
Funding
This work was supported by National Institutes of Health [R03-AG048077 to A.F.T. and K08-AG049938 to A.F.T.]; and Alzheimer’s Association [NIRG-13-283742 to A.F.T.].
Conflict of Interest: none declared.
Supplementary Material
References
- Aubry S. et al. (2015) Assembly and interrogation of Alzheimer's disease genetic networks reveal novel regulators of progression. PLoS One, 10, e0120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K. et al. (2005) Reverse engineering of regulatory networks in human B cells. Nat. Genet., 37, 382–390. [DOI] [PubMed] [Google Scholar]
- Chang W.L. et al. (2003) Molecular characterization of a novel nucleolar protein, pNO40. Biochem. Biophys. Res. Commun., 307, 569–577. [DOI] [PubMed] [Google Scholar]
- DeKosky S.T., Scheff S.W. (1990) Synapse loss in frontal cortex biopsies in Alzheimer's disease: correlation with cognitive severity. Ann Neurol, 27, 457–464. [DOI] [PubMed] [Google Scholar]
- Dredge B.K. et al. (2011) NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PLoS One, 6, e21585.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt F.M. et al. (2010) Housekeepers for accurate transcript expression analysis in Alzheimer's disease autopsy brain tissue. Alzheimers Dement., 6, 465–474. [DOI] [PubMed] [Google Scholar]
- Gonatas N.K. et al. (1967) The contribution of altered synapses in the senile plaque: an electron microscopic study in Alzheimer's dementia. J. Neuropathol. Exp. Neurol., 26, 25–39. [DOI] [PubMed] [Google Scholar]
- Gueydan C. et al. (2002) Identification of ribosomal proteins specific to higher eukaryotic organisms. J. Biol. Chem., 277, 45034–45040. [DOI] [PubMed] [Google Scholar]
- Joo J.H. et al. (2010) Pinin modulates expression of an intestinal homeobox gene, Cdx2, and plays an essential role for small intestinal morphogenesis. Dev Biol., 345, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A. et al. (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics, 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre C. et al. (2010) A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol., 6, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. (2015) Integrated genomic approaches identify major pathways and upstream regulators in late onset Alzheimer's disease. Sci. Rep., 5, 12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.S. et al. (2008) Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set. Physiol. Genomics, 33, 240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.S. et al. (2010) Neuronal gene expression in non-demented individuals with intermediate Alzheimer's Disease neuropathology. Neurobiol. Aging, 31, 549–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.S. et al. (2007) Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol. Genomics, 28, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.M. et al. (2017) Ribosomal protein pNO40 mediates nucleolar sequestration of SR family splicing factors and its overexpression impairs mRNA metabolism. Cell Signal, 32, 12–23. [DOI] [PubMed] [Google Scholar]
- Margolin A.A. et al. (2006) Reverse engineering cellular networks. Nat. Protoc., 1, 662–671. [DOI] [PubMed] [Google Scholar]
- Ouyang P. (2009) SRrp37, a novel splicing regulator located in the nuclear speckles and nucleoli, interacts with SC35 and modulates alternative pre-mRNA splicing in vivo. J. Cell Biochem., 108, 304–314. [DOI] [PubMed] [Google Scholar]
- Palomero T. et al. (2006) NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA, 103, 18261–18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozueta J. et al. (2013) Synaptic changes in Alzheimer's disease and its models. Neuroscience, 251, 51–65. [DOI] [PubMed] [Google Scholar]
- Saldi T. et al. (2016) Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol., 428, 2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheff S.W. et al. (2007) Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology, 68, 1501–1508. [DOI] [PubMed] [Google Scholar]
- Teich A.F. et al. (2015) Synaptic therapy in Alzheimer's disease: a CREB-centric approach. Neurotherapeutics, 12, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry R.D. et al. (1991) Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann. Neurol., 30, 572–580. [DOI] [PubMed] [Google Scholar]
- Wang P. et al. (2002) Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem. Biophys. Res. Commun., 294, 448–455. [DOI] [PubMed] [Google Scholar]
- Zoltewicz J.S. et al. (2012) Characterization of antibodies that detect human GFAP after traumatic brain injury. Biomark Insights, 7, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.