Abstract
Protein families are often represented by profile hidden Markov models (pHMMs). Homology between two distant protein families can be determined by comparing the pHMMs. Here we explored the idea of building a phylogeny of protein families using the distance matrix of their pHMMs. We developed a new software and web server (pHMM-tree) to allow four major types of inputs: (i) multiple pHMM files, (ii) multiple aligned protein sequence files, (iii) mixture of pHMM and aligned sequence files and (iv) unaligned protein sequences in a single file. The output will be a pHMM phylogeny of different protein families delineating their relationships. We have applied pHMM-tree to build phylogenies for CAZyme (carbohydrate active enzyme) classes and Pfam clans, which attested its usefulness in the phylogenetic representation of the evolutionary relationship among distant protein families.
Availability and Implementation
This software is implemented in C/C ++ and is available at http://cys.bios.niu.edu/pHMM-Tree/source/
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Numerous protein family/domain databases are available to provide multiple sequence alignment-based statistical models (e.g. profile hidden Markov models or pHMM (Eddy, 1998)) and literature-derived function descriptions, which are the foundation for computational function annotations of any newly sequenced genomes (Radivojac et al., 2013). Many of these databases also offer hierarchical classification of proteins at different levels, often defined by protein structural (e.g. SCOP (Lo Conte et al., 2000)) or evolutionary similarity (e.g. PANTHER (Thomas et al., 2003)). For example, Pfam defines a clan as a group of families that may share the same evolutionary origin or structural similarity (Finn et al., 2006); PANTHER classifies protein family into subfamilies; and CDD uses superfamily cluster to group a set of evolutionarily related protein domain models (Marchler-Bauer et al., 2007).
Although the parent-child information is known for these databases, it would be more insightful to know how different children (e.g. families of a Pfam clan) are evolutionarily related. Pfam attempted to address the issue using a JavaScript clanviewer to present the families of a clan as a network graph. However, it has never been explored before to represent families of a clan in the form of phylogeny. Here we developed an algorithm and software (pHMM-tree) to build a phylogeny of pHMMs, each pHMM representing a child (e.g. a Pfam family), to depict the evolutionary relationship of these pHMMs (e.g. Pfam families of a clan).
In addition, classification of a large protein superfamily or family into subfamilies has been viewed as a promising way to infer more specific functions for proteins. Tools such as SCI-PHY (Brown et al., 2007) were published for automated subfamily classification. Although not designed for subfamily classification, if given a set of protein sequences, pHMM-tree can also call a clustering program to automatically classify proteins into subgroups and then builds a pHMM phylogeny.
Here, we present pHMM-tree and its web server, and use CAZyme and Pfam families to demonstrate its applications. To our knowledge pHMM-tree is the first pHMM-based phylogeny-building tool.
2 Algorithm
The algorithm of pHMM-tree is depicted in Figure 1A. It can flexibly handle four kinds of inputs: (1) multiple files each in pHMM format; (2) multiple files each with a multiple sequence alignment (MSA) in FASTA format; (3) mixture of (1) and (2), and (4) a file with unaligned protein sequences in FASTA format.
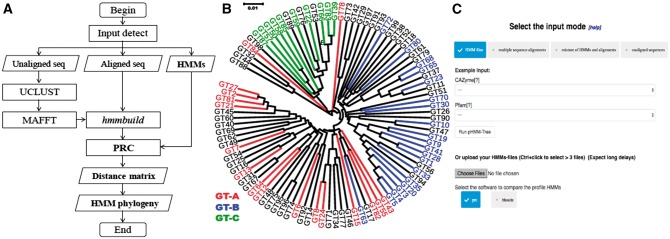
Fig. 1.
(A) Flow chart of the algorithm. (B) Phylogeny of 97 GT family pHMMs, 47 of which have been previously classified into three GT clans (see Supplementary Data). Black colors are families not previously classified. (C) The pHMM-tree web server job submission page. Under upload HMMs option, the CAZyme classes and Pfam clans are provided as example input
If the input is (1), all the pHMMs will be compared with each other using the pHMM comparison tool PRC (Madera, 2008) or HHsuite (Remmert et al., 2012) to calculate a pair-wise distance. These distances will then be used to build a pHMM distance matrix, which will be used to build a distance-based phylogeny. If the input is (2) and (3), each MSA will be used to build a pHMM and then build the phylogeny. If the input is (4), the unaligned sequences will be clustered into subfamilies, and then each cluster will be aligned and all aligned clusters together will be used for building a pHMM phylogeny.
3 Implementation
Standalone program : The pHMM-tree program was written in C/C ++. Users should ensure that enough sequences, MSAs or pHMMs are provided as the inputs, because it needs at least three pHMMs to build a phylogeny. In addition to the parameter specifying the input data type, there are also parameters to (i) select one of the two different methods for pHMM comparison: PRC or HHsuite; (ii) change sequence identity thresholds for clustering. The output is written to a folder with at least three subfolders: (i) phylogeny; (ii) distance matrix; (iii) pHMM. Running time: the time use of pHMM-tree will go exponentially when the number of pHMMs increases (Supplementary data). Web Server: In addition to the standalone program, we also developed a web server for users who do not have programming experience (Fig. 1C).
4 Evaluation
To assess pHMM-tree’s performance, we have run it on the six classes of CAZymes. Each class contains between 13 to 129 protein families. Figure 1B shows a pHMM phylogeny of 97 glycosyltransferase (GT) families. We have also built pHMM phylogenies for 254 Pfam clans having at least five families. Phylogenies and discussions of all the CAZyme classes and Pfam clans are provided in the pHMM-tree website and Supplementary data. Overall, the phylogenies revealed clustering patterns that are largely consistent with previous knowledge about the closeness of different families within a CAZyme class. Furthermore, to assess the effectiveness of the elements used in the matrix building step, we randomly extracted 1000 Pfam clan pairs, and carried out a Wilcoxon rank test (see Supplementary data). The results indicated that in over 95% clan pairs, inter-clan distances were higher than intra-clan distances, and that in over 85% clan pairs, the Wilcoxon rank test supported the larger inter-clan distances with a P-value < 0.05.
Supplementary Material
Acknowledgements
We acknowledge the Department of Computer Science of NIU for providing free access to the Linux computing cluster Gaea.
Funding
This work has been supported by the National Institutes of Health (1R15GM114706) to YY and the Natural Science Foundation of Tianjin (15JCYBJC18900) to HZ.
Conflict of Interest: none declared.
References
- Brown D.P. et al. (2007) Automated protein subfamily identification and classification. PLoS Computat. Biol., 3, e160.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- Finn R.D. et al. (2006) Pfam: clans, web tools and services. Nucleic Acids Res., 34, D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte L. et al. (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res., 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera M. (2008) Profile Comparer: a program for scoring and aligning profile hidden Markov models. Bioinformatics, 24, 2630–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. (2007) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res., 35, D237–D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P. et al. (2013) A large-scale evaluation of computational protein function prediction. Nat. Methods, 10, 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert M. et al. (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods, 9, 173–175. [DOI] [PubMed] [Google Scholar]
- Thomas P.D. et al. (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res., 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.