Abstract
Motivation: Capabilities in the field of metabolomics have grown tremendously in recent years. Many existing resources contain the chemical properties and classifications of commonly identified metabolites. However, the annotation of small molecules (both endogenous and synthetic) to meaningful biological pathways and concepts still lags behind the analytical capabilities and the chemistry-based annotations. Furthermore, no tools are available to visually explore relationships and networks among functionally related groups of metabolites (biomedical concepts). Such a tool would provide the ability to establish testable hypotheses regarding links among metabolic pathways, cellular processes, phenotypes and diseases.
Results: Here we present ConceptMetab, an interactive web-based tool for mapping and exploring the relationships among 16 069 biologically defined metabolite sets developed from Gene Ontology, KEGG and Medical Subject Headings, using both KEGG and PubChem compound identifiers, and based on statistical tests for association. We demonstrate the utility of ConceptMetab with multiple scenarios, showing it can be used to identify known and potentially novel relationships among metabolic pathways, cellular processes, phenotypes and diseases, and provides an intuitive interface for linking compounds to their molecular functions and higher level biological effects.
Availability and implementation: http://conceptmetab.med.umich.edu
Contacts: akarnovsky@umich.edu or sartorma@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
In recent years, metabolomics has emerged as a new quantitative technique with the ability to characterize large numbers of small molecules in a wide variety of biological samples. Advances in liquid chromatography–mass spectrometry (LC–MS), gas chromatography–mass spectrometry (GC–MS) and nuclear magnetic resonance spectroscopy (NMR), allow rapid and quantitative measurement of several hundreds of metabolites (Jonsson et al., 2004; Wishart, 2011). Untargeted LC–MS based methods have potential to push the number of detected metabolites to several thousands, however securing the identities of the individual features remains challenging and time consuming (Baker, 2011). As experimental detection methods continue to improve, metabolomics has the potential to provide increasingly informative readouts of metabolic changes in complex diseases (Sreekumar et al., 2009; Urayama et al., 2010; Wang et al., 2011; Wisloff et al., 2005; Yap et al., 2010). In contrast to genes and proteins, metabolites have been described as providing direct signatures of biochemical activity and are therefore easier to correlate with phenotype (Patti et al., 2012).
Following these technological advances, a number of pathway databases and tools linking metabolites to biochemical reactions, enzymes, proteins and genes were developed (reviewed in (Sas et al., 2015)). Among these, there are several tools for metabolite set enrichment testing, including MSEA (Xia and Wishart, 2010), MetaboAnalyst 2.0 (Xia et al., 2012) and MBRole (Chagoyen and Pazos, 2011). These programs follow the paradigm of gene set enrichment tools, which test for biological functions or pathways (e.g. Gene Ontology (GO) (Harris et al., 2004) or KEGG Pathways (Kanehisa et al., 2012)) that have significant gene overlap with an experimentally derived set of genes (Khatri et al., 2012).
Biological interpretation of metabolites has unique challenges compared to genes, including a relatively small number of measurable metabolites, low coverage of those by annotation databases, and the presence of ubiquitous metabolites (e.g. co-factors). To improve the annotation of small molecules to their biological contexts, we developed Metab2Mesh (Sartor et al., 2012), which contains 4 646 000 significant associations (P < 0.0001) between 99 871 compounds and 20 683 biomedical terms. Metab2MeSH uses PubChem and Medical Subject Heading (MeSH) terms to identify statistically significant co-occurrences of metabolites and MeSH terms in published manuscripts, thus annotating metabolites to biomedical concepts via the literature.
An additional challenge to working with metabolites is the lack of convenient, standardized identifiers. While IUPAC nomenclature provides a systematic method of naming organic compounds and chemists use the CAS Registry Number, biologists prefer more familiar names that often ignore counter-ions. Consequently, biological databases often contain such names or use their own identifiers. To address these challenges, careful assembly of metabolite sets with synonyms and cross-references is needed.
Due to these challenges, metabolite enrichment testing has not been as widely used as for genes. Enrichment testing among pre-defined biologically relevant metabolite sets can help us better understand and overcome the above challenges, and improve enrichment testing with experimental data. The careful assembly, characterization and exploration of metabolite sets could facilitate the discovery of relationships among metabolic reactions, diseases and other biological phenomena in terms of the metabolites involved. Indeed, several tools for exploring similar relationships based on gene sets exist (Araki et al., 2012; Perez-Llamas and Lopez-Bigas, 2011; Rhodes et al., 2007; Sartor et al., 2010) and have been fundamental in generating novel hypotheses and identifying unexpected associations. However, no comparable tool based on small metabolites yet exists.
We have developed ConceptMetab to explore the relationships among metabolite-based biomedical concepts and generate novel hypotheses. Metabolites were annotated to biomedical concepts using KEGG (Kanehisa et al., 2012), the three branches of GO and Medical Subject headings from the National Library of Medicine (MeSH) (Coletti and Bleich, 2001). Statistically significant associations were identified among all pairs of metabolite sets (concepts), and maintained with additional supporting information. The ConceptMetab website enables searching, browsing, filtering and data exporting capabilities, as well as complementary visualizations (network graphs and heatmaps). We demonstrate the utility of ConceptMetab with example workflows, and by illustrating important relationships identified with metabolites that were not identified with genes. In summary, ConceptMetab assists in understanding links between metabolites, metabolic pathways and biological phenomena, phenotypes, environmental exposures and diseases.
2 Methods
2.1 Mapping small molecules to annotations
Small molecules were annotated to 74 KEGG human metabolic pathways based on the XML pathway representations from the Summer 2011 freeze of KEGG. Metabolites were annotated to GO terms in two stages. First, KEGG Pathways were used to map metabolites to genes through chemical reactions. Second, the Bioconductor package org.Hs.eg.db (R version 3.1.1) was used to map genes to GO terms, providing a complete mapping from metabolites to GO. GO terms are partitioned by their three branches: Biological Process (BP), Cellular Component (CC) and Molecular Function (MF). Enzyme metabolite sets were created by combining all metabolites involved in a reaction with the same enzyme. The metabolite-to-gene mappings from KEGG were used again, and the org.Hs.eg.db package was used to map genes to enzymes.
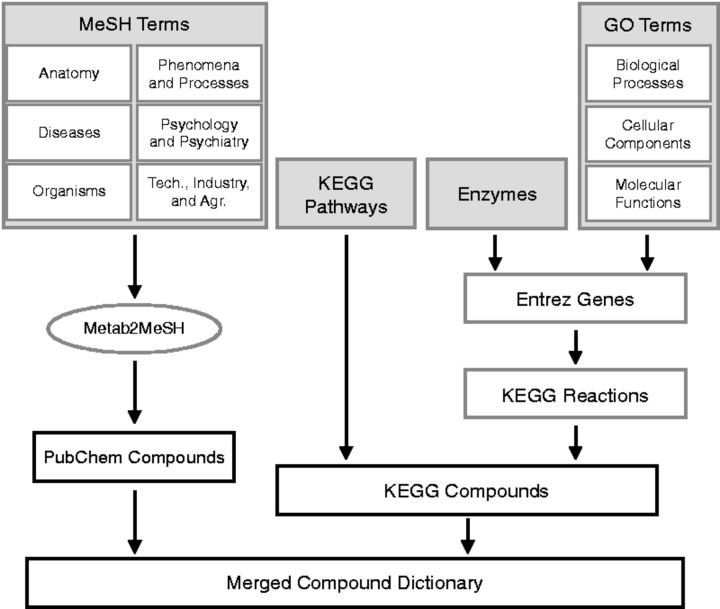
Small molecules were annotated to MeSH terms by their co-occurrences in biomedical literature (PubMed database, updated to version 14 on May 19, 2014) using Metab2MeSH (Sartor et al., 2012). Briefly, Metab2MeSH considers a PubChem compound to be associated with a MeSH term if the number of co-occurring annotations to PubMed articles is significant according to a two-sided Fisher’s exact test. We selected the most relevant top-level MeSH categories for use in ConceptMetab (Fig. 1). Because some MeSH terms occur in more than one top-level category, we assigned membership according to the following priority: (i) Diseases, (ii) Phenomena and Process, (iii) Psychiatry and Psychology, (iv) Anatomy, (v) Organisms and (vi) Technology, Industry, Agriculture. In all cases, we retain only those concepts associated with ≥5 compounds. Figure 1 gives a diagrammatic overview of the compound-to-concept mappings.
Fig. 1.
A diagrammatic view of how small molecules are annotated to concepts in ConceptMetab. PubChem compounds are associated with MeSH Terms via Metab2MeSH. Metabolites with KEGG IDs are associated with KEGG Pathways via their XML representation. Enzymes and GO terms are mapped to KEGG compounds through Entrez genes and KEGG reactions. Finally, PubChem and KEGG small molecules are linked via a dictionary used in Metab2MeSH
The resulting mappings linking compounds to biological concepts are stored in a MySQL database. The same relational database is also used to store all testing results, as described in Section 2.3 below.
2.2 Compound dictionary
To compare MeSH terms (based on PubChem IDs) to the other concept types (based on KEGG IDs), we used the dictionary previously developed for Metab2MeSH (Sartor et al., 2012). In Metab2MeSH, KEGG IDs are linked to PubChem substance IDs (SIDs) via the KEGG REST API (http://www.kegg.jp/kegg/rest/). The SIDs are linked to PubChem compound IDs (CIDs) via PubChem (Wang et al., 2009).
We observed some PubChem compounds having the same name, or differing only by capitalization, that had different CIDs. Therefore, all PubChem compound names were transformed to lower-case, and assigned a new internal ID to each instance where the lower-case names match. Next, any existing CID to KEGG ID links were propagated to all newly equivalent CIDs from the previous step. About 5500 uninformative compounds that had purely numeric or alpha-numeric names in PubChem and were not connected to a KEGG ID were removed. In all, ConceptMetab has 97 104 unique compounds, 68 556 with ≥1 annotation. Among these, 15 231 have both a KEGG ID and a PubChem CID, 1629 have only a KEGG ID, and 80 244 have only a PubChem ID.
2.3 Metabolite set enrichment testing
We tested for associated metabolite sets using a modified one-sided Fisher’s exact test (FET) which tests whether the number of compounds in both concepts exceeds that expected by chance relative to the background set of compounds. Given two concepts, the background set of compounds was the intersection of the sets of compounds in all concepts in the two concept types (e.g. all compounds annotated in both GO and MeSH diseases if testing a GO term versus a disease). We modified FET by subtracting one from the intersection of the two concepts, as has previously been done (Huang da et al., 2009; Sartor et al., 2010). This modification results in a more conservative test for small concepts, which are more likely be affected by chance co-occurrences, while having minimal effect on large concepts. After computing p-values and odds ratios for all pairs of concepts, we applied the False Discovery Rate (FDR) multiple testing correction of Benjamini and Hochberg and stored all results in the database.
2.4 Visualizing relationships among concepts
The main benefits of ConceptMetab are its interactivity, various workflows and visualizations (Supplementary Fig. S1). The web interface was built using Grails and Javascript, which communicate with the MySQL database. Users can either browse or query a compound or concept (disease, biological process, etc.) and choose among the matches to obtain an overview of results. In the case of a concept, users can obtain the significantly overlapping concepts, filter the results, and either output tabular results or visualize the resulting relationships as network graphs or a heatmap created via hierarchical clustering. In the case of a compound, users can retrieve all concepts to which it is annotated and further analyze those concepts.
The Cytoscape Web Javascript API is used to display the two interactive network graphs: the star network and the complete network. In both cases, nodes represent concepts; their size and color represent the number of compounds and the concept type, respectively. Graph edges represent significant enrichment at user-defined levels between concepts (default FDR <0.05 and odds ratio >0). The star network displays only edges connected to the selected concept (Supplementary Fig. S1E) while the complete network shows relationships among all of the concepts enriched relative to the selected concept (Supplementary Fig. S1F). Clicking a node gives concept information, and clicking an edge gives FET results and lists the compounds intersecting the two concepts connected by that edge.
The interactive heatmap, created using the gplots R package, illustrates which compounds are responsible for the enrichment of each concept, and the similarity among those concepts relative to the selected concept (Supplementary Fig. S1G). Rows and columns are hierarchically clustered using the Euclidean distance metric and average-linkage criterion. Heatmaps displayed on the website are interactive, and the underlying, unclustered data is available for download.
For visual clarity, networks may not exceed 200 concepts (nodes). For heatmaps, if the concept of interest has more than 2000 compounds, up to 200 concepts can be selected. When the concept of interest has between 1000 and 2000 compounds, up to 500 concepts can be selected. Users may filter concepts by P-value, q-value, or odds ratio, or may select individual concepts for the network or heatmap in the table view.
2.5 Viewing metabolite sets as networks in Metscape
When viewing the list of metabolites in a concept, users have the option to visualize the KEGG compounds in the MetScape plug-in of Cytoscape (Karnovsky et al., 2012) using the automatic web-start feature. Metscape will construct a network of metabolites and metabolic reactions that allows users to explore the interactions between metabolites, enzymes and genes as determined by metabolic pathways.
3 Results
3.1 Overview of the ConceptMetab database
ConceptMetab annotates 68 556 compounds to 16 069 biomedical concepts including diseases, metabolic pathways, cellular processes and components, phenotypes, environmental exposures and organisms. Concepts originate from 11 concept types: KEGG Pathways, GO, Enzymes (defined by the metabolites involved in their associated chemical reactions) and six of the top level MeSH categories (https://www.nlm.nih.gov/mesh/). Figure 1 shows how compounds were mapped to each of the concepts for the different annotation sources. The number of concepts in a concept type ranges from 74 (KEGG Pathways) to 4089 (MeSH Diseases). Concept sizes vary widely across concept types (Supplementary Fig. S2), with the mean number of compounds in a concept ranging from 11 (Enzymes) to 404 (MeSH Phenomena and Processes; Table 1). The broad range of concept sizes is reflective of the widely differing number of compounds required for very specific chemical tasks compared to much broader biological phenomena.
Table 1.
An overview of the annotation databases in ConceptMetab
| Concept Type | No. of Concepts | Mean Size | Median Size | No. of Compounds |
|---|---|---|---|---|
| Enzyme | 175 | 11 | 7 | 874 |
| GOBP | 3712 | 56 | 20 | 1220 |
| GOCC | 346 | 117 | 19 | 1213 |
| GOMF | 864 | 48 | 14 | 1226 |
| KEGG Pathways | 74 | 42 | 38 | 2427 |
| MeSH Anatomy | 1506 | 357 | 208 | 37 706 |
| MeSH Diseases | 4089 | 182 | 105 | 33 074 |
| MeSH Organisms | 3011 | 150 | 58 | 48 688 |
| MeSH Phen and Proc | 1443 | 404 | 195 | 43 016 |
| MeSH Psy and Psy | 519 | 180 | 79 | 9188 |
| MeSH Tech | 330 | 280 | 198 | 15 721 |
The number of biological concepts, the mean and median number of compounds in them, and the number of unique compounds across all concepts in each concept type are given.
Compounds were annotated to radically different numbers of concepts, ranging from 1 to 1611. A few compounds were annotated to a large number of concepts while the majority occurred in fewer than eight (Supplementary Fig. S3). The compounds annotated to the most concepts were: arachidonate, ATP, AMP, cyclic AMP, GTP, water, cyclic GMP, nitric oxide, glutathione and linoleate (Supplementary Table S1). The distributions for the 3 GO branches were the least skewed among the concept types, with compounds on average belonging to between 20 and 100 concepts (Supplementary Fig. S3).
A dictionary mapping between KEGG and PubChem IDs makes concepts from KEGG Pathways, GO terms and Enzymes comparable to MeSH terms. Upon testing the statistical significance of overlap among all 129 098 346 possible pairs of concepts, a total of 10 334 760 pairs (8%) were statistically significant (FDR < 0.05), providing a rich network of interactions to explore.
3.2 ConceptMetab workflow
The ConceptMetab website (http://conceptmetab.med.umich.edu) allows users to explore biomedical concepts and their relationships to one another in a variety of ways. As specific examples, it can be used to (i) examine the links between an enzyme such as ‘17-beta-estradiol 17-dehydrogenase’ and diseases (70 MeSH Diseases are significant with q-value<0.05) via their common metabolites; (ii) query a disease or phenotype such as ‘confusion’ to identify relationships with processes or other diseases, and the small molecules on which those relationships are based; or (iii) explore the biological annotations of a specific compound of interest, and relationships among them. Users can choose a concept type and then browse the list of concepts in that type, or concepts may be searched directly (Supplementary Fig. S1A and B). Users may also search for a particular compound of interest. Supplementary Table S2 shows how ConceptMetab's features differ from other metabolite analysis tools.
Upon selecting a concept, a summary page provides: (i) a list of the compounds in the concept, (ii) filtering options, (iii) a link to the originating database, (iv) the percent of significant terms in each concept type and (v) links to visualizations including a star network, a complete network, a heatmap and a table view (Supplementary Fig. S1C). The table provides the metabolite enrichment testing results, including P-values, FDR values, odds ratios and numbers of overlapping compounds (Supplementary Fig. S1D). Users may adjust the P-value or FDR cutoff, and also use a cutoff on the odds ratio. Users can link to the list of compounds annotated to both the queried term and any of the enriched terms, and then link to PubChem or KEGG for more information on a compound. Users can also visualize the compounds in metabolic networks using a one-click link to the MetScape Cytoscape plugin (Karnovsky et al., 2012), where there is information about metabolic reactions, enzymes, genes and pathways. The star network shows which concepts have significant overlap with the concept of interest given the selected cutoffs (Supplementary Fig. S1E). The complete network adds significant interactions among all of the associated concepts in the star network (Supplementary Fig. S1F).
The heatmap illustrates the relationships between compounds in the queried metabolite set and the enriched concepts to which they belong. This allows users to find groups of compounds that are closely related functionally, and to determine which compounds were responsible for the enrichment of particular concepts (Supplementary Fig. S1G).
3.3 Significant relationships among metabolite annotation sources
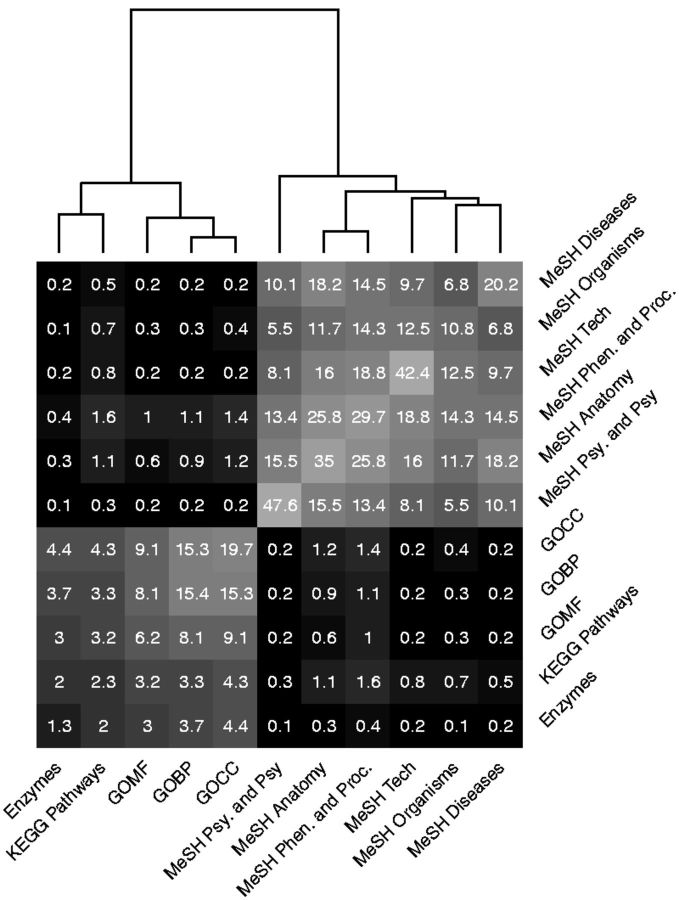
Of the greater than 10 million (8%) significant concept pairs, 4.1 million (39%) were within the same concept type while 6.3 million (61%) were between concept types. Overall, 18% of the tests within a concept type and 6% of tests between two concept types were significant. At the concept type level, KEGG ID-based concept types (Enzyme, GO and KEGG Pathway) have a greater percentage of significant associations with other KEGG-based concepts compared to PubChem-based concepts, and the same is true for PubChem-based concepts (Fig. 2). This is likely because only a subset of the compounds could be mapped between KEGG and PubChem. Certain concept types, most notably MeSH Psychology and Psychiatry, have a large degree of overlap in compounds among their concepts (lighter squares along the diagonal in Fig. 2). Others such as Enzymes, KEGG Pathways and MeSH Organisms, have more unique non-overlapping metabolite sets. Although the percentages are smaller, we found the most interesting associations to be between KEGG ID-based annotation concepts and MeSH-based concepts, as these often link molecular level reactions or cellular processes (KEGG-based) to macro-scale biological phenomena, such as diseases, anatomy, diet, environmental exposure, or other phenotypes (MeSH-based). In any user workflow, one can easily filter to any such subset of results of interest.
Fig. 2.
Percentage enrichments between concept types. Numbers in each cell are the percentage of enrichment tests between the respective concept types which were significant (FDR < 0.05). Observe that KEGG-based concepts tend to be more enriched with other KEGG-based concepts, and similarly for PubChem-based concepts
3.4 Comparing biological associations based on metabolites to those based on genes
Although our development of an interactive tool for exploring relationships among biological metabolite-sets is novel, similar tools for gene-based concepts are relatively well-established. We therefore wanted to assess how well metabolites can predict relationships between various biological phenomena and diseases compared to genes. ConceptMetab annotates 68 556 compounds to 16 069 biological concepts, and includes concepts based on molecular evidence (GO, KEGG and Enzymes) and biomedical literature (MeSH); a similar database based on genes (ConceptGen (Sartor et al., 2010)) annotates 36 393 genes to 21 086 biological concepts, includes many of the same concept types, and uses the same approach for determining significant association between pairs of concepts.
We compared associations identified between MeSH Disease and MeSH Phenomena & Processes. We based our comparison on two MeSH-based concepts (as opposed to MeSH Disease versus GO) because both metabolites and genes are assigned to MeSH terms using the same approach, resulting in a fair comparison of metabolites versus genes. ConceptMetab and ConceptGen tested the association of all such pairs of concepts, identifying 857 378 and 5147 to be significant (FDR < 0.05), respectively. The main reason for the drastically higher number of significant metabolite-based associations is that the majority of MeSH terms did not have ≥5 genes assigned to them, which was a requirement for the test. Overall, 10 515 concept pairs had at least two elements in common and were tested for association based on both metabolites and genes. Among these, 3853 pairs were significant in both approaches, 757 uniquely significant based on genes, and 851 uniquely significant based on metabolites, indicating a high level of agreement when sufficient data exists for both metabolites and genes. However, overall these results point to a strong advantage to using metabolite-based associations, as these result in a > 100-fold increase in the ability to detect associations owing to there being more compounds than genes, and having more compounds annotated to biological functions.
Interestingly, the top 10 types of MeSH Phenomena & Processes terms that are associated with the most diseases in ConceptMetab are in the Organic Chemistry Phenomena, Chemical Processes, Cell Physiological Processes, Metabolism, Biophysical Phenomena and Biochemical Phenomena branches of the MeSH tree. On the other hand, the top 10 types of MeSH Phenomena & Processes terms uniquely significant based on genes are in the Genetic Variation, Phenotype, Gene Frequency, Inheritance Patterns and Genotype branches of the MeSH tree. We found that the terms uniquely enriched in ConceptMetab tended to be more biologically meaningful than those based on genes, for example ‘cell cycle, drug resistance, DNA damage, and platelet aggregation’ as opposed to ‘phenotype, genetic markers, gene frequency, linkage disequilibrium, and genotype’. This illustrates the important (and until now unexplored) contribution that metabolites make to understanding of the relationships among biological concepts.
Doing a similar analysis for MeSH Disease terms, among those uniquely enriched in ConceptMetab we found Nervous and Digestive System Neoplasms, Neurodegenerative Diseases, Neoplastic Processes, Pancreatic and Liver Diseases, Endocrine Gland Neoplasms and Metabolic Diseases within the top 20 types of diseases. In contrast, we find Graft versus Host Disease, Bronchial and Joint Diseases, Connective Tissue and Joint Diseases, RNA Virus Infections and Vascular Diseases uniquely enriched based on genes. Indeed, there are diseases and biological concepts where metabolites play a more important role than genes, and vice versa.
3.5 Using ConceptMetab to understand the molecular and anatomical risks and effects of a disease
Atherosclerosis is an inflammatory disease of the arteries and is characterized by an accumulation of lipids within the artery wall, which can lead to reduced blood flow and infarction. Consequently, atherosclerosis is more than an inflammatory disease; it is also a leading cause of heart attack and stroke (Ross, 1993).
Atherosclerosis is a MeSH Disease concept in ConceptMetab with 755 compounds. It is significantly associated with 203 GO terms, 425 MeSH Phenomena and Processes concepts, 488 MeSH Anatomy concepts, and others at the FDR < 0.05 level. In particular, MeSH Anatomy concepts such as ‘Endothelium’, ‘Macrophages’, ‘Monocytes’, ‘T-lymphocytes’, ‘Blood platelets’ and various specific arteries are significantly associated with atherosclerosis. MeSH Phenomena and Processes that are significantly associated with atherosclerosis include ‘Vasoconstriction’, ‘Platelet adhesiveness’ and ‘Platelet aggregation’.
These terms are expected because atherosclerosis is localized to the inner walls (endothelium) of arteries, wherein monocyte-derived macrophages and subtypes of T-lymphocytes mediate the inflammatory response. The inflammatory response in turn increases adhesiveness of the endothelium, especially with respect to blood platelets, resulting in platelet aggregation. Ultimately, the increased adhesion and aggregation within the artery contributes to vasoconstriction (Ross, 1999).
ConceptMetab also finds a number of GO terms associated with atherosclerosis. Fatty acid catabolism, metabolism and biosynthesis are enriched, along with ‘Smooth muscle contraction’, ‘Foam cell differentiation’ and ‘Prostanoid metabolic process’. The inflammatory response is partly mediated by prostanoids, and foam cell formation, in conjunction with smooth muscle migration, contributes to the growth of the fatty atherosclerotic lesion.
As noted above, atherosclerosis is an inflammatory disease that is the leading cause of heart attack and stroke. ConceptMetab finds MeSH Disease concepts such as ‘Inflammation’, ‘Myocardial infarction’ and ‘Stroke’ to be highly associated with atherosclerosis, thus predicting comorbidities. Risk factors such as ‘Hypercholesterolemia’ and ‘Atherosclerotic plaque’ are also found. Overall, ConceptMetab correctly associates numerous risk factors, molecular mechanisms, anatomical features and observed downstream effects with atherosclerosis, providing a comprehensive overview of related biological concepts and the metabolites that explain these relationships.
3.6 Using ConceptMetab to investigate the diseases associated with an aberrant biological process
The unfolded protein response (UPR) is a well-studied cellular response that occurs under stress and is tightly coupled with endoplasmic reticulum stress. The UPR can be either pro-survival or pro-apoptotic, depending on specific cellular conditions, and leads to the induction of a specific battery of genes while repressing a wealth of genes transcribed under normal growth conditions to allow the cell to regain control. In ConceptMetab, ‘Unfolded Protein Response’ is represented as a MeSH Phenomena and Processes concept, with 189 compounds, and with 36 significantly enriched GO terms and 255 MeSH Diseases at the FDR < 0.05 level and restricting to terms with odds ratio >8.
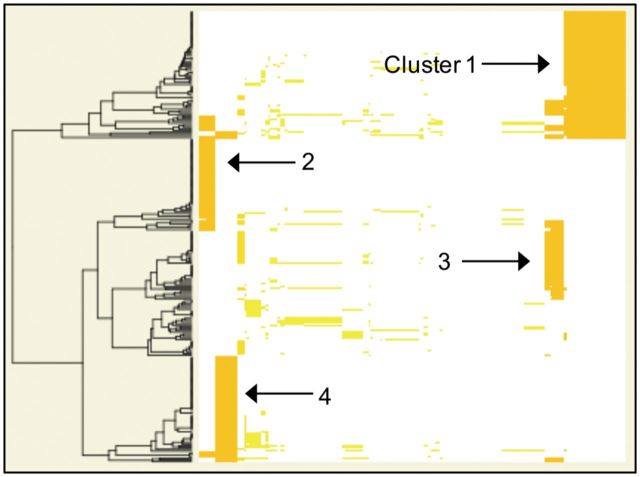
In the table view, many well-known relationships are readily identified at the cellular level at the top of the list, including ‘Endoplasmic Reticulum Stress’, ‘Cell Death’ and ‘Autophagy’, ‘Gene Silencing’, heat-shock response, protein folding and transport, and oxidative phosphorylation. Creating a heatmap of the significantly associated diseases, we saw they fall into four main groups (Fig. 3). Continuing to the interactive heatmap, we saw that the first group corresponded to anemias, deficiencies, toxic poisonings, and a few neurologic diseases that all have in common glutathione, glutamine and several related derivatives. The second group involved blood, bone and heart-related diseases which mainly had calcium-related compounds in common, and the third group consisted mainly of diabetes and nutritional diseases and were related by several sugar compounds, and insulin/velosulin. Finally, the last group of diseases contained many neoplasms having drugs in common, including borozimib, which is known to induce ER stress
Fig. 3.
Screenshot of overview heatmap for diseases associated with Unfolded Protein Response. Each row represents a MeSH disease and each column is a compound. The UPR is associated with four main groups of diseases, defined by the types of overlapping compounds. From here, users may click to proceed to an interactive heatmap view
and lead to apoptosis. Several specific protein-folding related diseases in these groups were Lipoatrophic Diabetes Mellitus, ‘Insulin Resistance’, ‘Fatty Liver’, ‘Neurodegenerative Diseases’, fibrotic diseases (Lenna and Trojanowska, 2012), cadmium poisoning (Gardarin et al., 2010) and lymphomas. We also identified less known relationships with the UPR that are supported by the literature nonetheless. For example, Sturge-Weber syndrome, a rare neurological and skin disease, was identified as significant (q-value = 1.3 × 10−5), and clicking on the number of overlapping compounds shows this relationship includes galactose, hexose and glucose. Recently it was observed that oxidative stress (the UPR is closely linked to OS and is activated upon OS exposure), may play an important role in the pathogenicity of Sturge–Weber syndrome (Kadam et al., 2012).
3.7 Using ConceptMetab to explore relationships between metabolic pathways and diseases
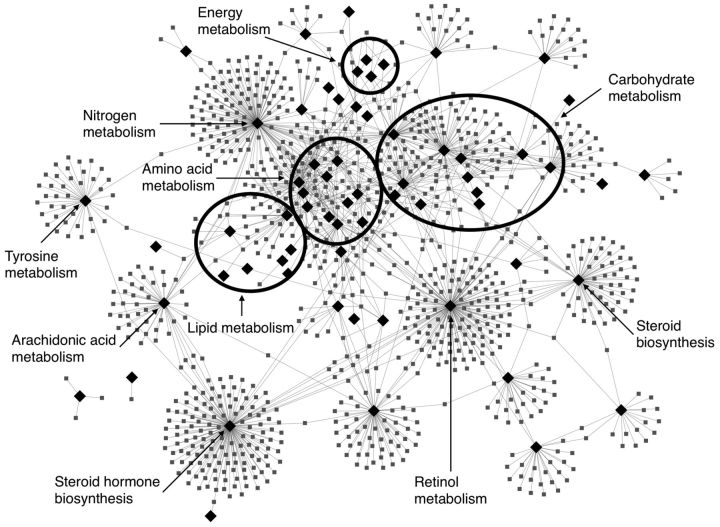
To explore relationships between metabolic pathways and diseases, we took the significant KEGG Pathway-MeSH Disease concept pairs and imported the data into Cytoscape. Figure 4 shows the resulting network. Not surprisingly, there are several network hubs (diamonds) representing a relatively small number of pathways connected to a large number of diseases (squares). Some of the pathway hubs, such as amino acid metabolism, are connected to many well-documented metabolic diseases, e.g. ‘Inborn Errors of Amino Acid Metabolism’ and ‘Ornithine Cabamoyltransferase Deficiency Disease’. Other expected connections include steroid metabolism and hormone dependent neoplasms (e.g. breast and prostate cancer), retinol metabolism and anemia and many others. Interestingly, the central highly interconnected network component includes amino acid (alanine, aspartate arginine, proline, glutamine/glutamate, glycine, serine and threonine, branched chain amino acids), fatty acid and energy metabolism (glycolysis, oxidative phosphorylation) pathways that share many of the same disease connections. Given the central role of these pathways as part of primary metabolism, it is not surprising that their dysregulation has implications for a variety of diseases ranging from brain injuries to cancers to metabolic diseases. Since our pathway – disease network is based on biochemical pathways and literature-derived metabolite concepts, we expect it to be biased towards diseases that are linked to metabolic dysregulations that have been sufficiently described in publications.
Fig. 4.
Bipartite metabolic pathway – disease network identified by ConceptMetab and displayed in Cytoscape. Black diamonds represent pathways; grey squares are diseases. The ovals in the center represent groups of several KEGG pathways, e.g. carbohydrate metabolism includes amino sugar, nucleotide sugar, galactose, fructose and mannose metabolism
3.8 Using ConceptMetab to explore the biological roles of a metabolite
One of the challenges in analyzing metabolomics data is connecting the experimentally observed changes to the associated phenotypes. The usual analysis workflow involves mapping metabolites to known metabolic pathways (Lanza et al., 2010). This helps establish connections between metabolites and a relatively small portion of genes encoding metabolic enzymes, but often neglects broader biological context. Pathway databases have relatively low representation of experimentally detected metabolites, which further limits their utility. ConceptMetab provides an alternative way to explore biological connections of metabolites. To demonstrate the compound analysis capabilities of ConceptMetab, we selected gamma-hydroxybutyrate (GHB, 4-hydroxybutanoate), a compound notoriously known as a date rape drug (Bendinskas et al., 2011) and a club drug (Gahlinger, 2004). GHB also has well-documented medicinal uses (Mamelak et al., 1986), is found naturally at low concentrations in the mammalian brain (Vayer et al., 1987), and accumulates in patients with succinic semialdehyde dehydrogenase (SSADH) deficiency (Pearl et al., 2003).
ConceptMetab shows that GHB is part of 106 concepts that span seven MeSH headings, including Anatomy and Diseases. Predictably, GHB was linked to Central Nervous System (CNS), Alcoholism and Brain Ischemia. Each of these concepts contains hundreds of compounds. We proceeded to select these three concepts and built a complete network (Fig. 5). ConceptMetab provides an easy way to explore the overlap between the concepts displayed in the Complete Network view. Clicking on the edge connecting the concept nodes displays the list of compounds shared between them. The Alcoholism – Brain Ischemia edge and the CNS – Brain Ischemia edge both list taurine, a compound which, like GHB, has neuro-protective properties (Shuaib, 2003). Interestingly, taurine is being tested as a potential treatment in patients with the SSADH deficiency (Pearl et al., 2014). Inspection of the CNS – Alcoholism edge includes baclofen, which is a specific agonist of GABA-B receptors used for alcoholism treatment (Addolorato et al., 2000) but is also known to help with GHB withdrawal (LeTourneau et al., 2008). Thus, ConceptMetab helps find known as well as unexpected useful chemical links between biological concepts.
Fig. 5.
ConceptMetab complete network. Network nodes represent concepts. By clicking on an edge user can obtain the information about compounds that are in common between concepts
4 Discussion
As the ultimate readout of metabolic state, metabolomics has the potential to transform our understanding of mechanisms underlying disease and further enhance knowledge generation through integration with other omics data. As experimental metabolomics matures and the number of measurable metabolites approaches the estimated number of endogenous metabolites, metabolomics together with transcriptomics, proteomics and epigenomics will provide a comprehensive understanding of a biological system as a whole. While gene-based technologies, analysis methods and annotation have well established standards and an abundance of relevant bioinformatics software, the parallel requirements for high throughput metabolomics still lag far behind. As a step towards bridging this gap, we have developed a tool that annotates both endogenous and synthetic small compounds to various types of biological concepts, and that provides interactive exploration of the relationships among these concepts. With the novel MeSH-based annotation source, we have increased the number of annotated metabolites by ∼25-fold and shown that many relevant relationships not identified by genes are identified via metabolites.
The ability to visualize relationships not only between pairs of metabolite sets but also the network structure among many can help bridge the gaps from molecular level to phenotype level to population level biomedical concepts. No other program allows testing for significant enrichment among predefined metabolite sets. The few programs that currently offer enrichment testing of experimental metabolite sets only annotate a small minority of compounds. The next step will be to expand upon ConceptMetab to offer such analysis with greatly expanded annotation.
In ConceptMetab, both KEGG and PubChem IDs were used to maximize annotation, giving us the benefits of both traditional annotation sources such as KEGG and GO, and our MeSH term annotations. We chose KEGG because it is well-established, consistent and cross-referenced with PubChem. We recognize that other databases such as BioCyc (Caspi et al., 2014), Recon2 (Thiele et al., 2013), Reactome (Croft et al., 2014) and SMPDB (Jewison et al., 2014) may provide a complimentary view of metabolic pathways. Overall, ConceptMetab provides a rich resource documenting relationships among different types of metabolite-based concepts, which will aid in understanding the complex and interrelated biological roles of metabolites.
Supplementary Material
Funding
This work was supported by the National Human Genome Research Institute [T32-HG000040; R.C.]; National Institute of Environmental Health Sciences P30 Core Center [P30-ES017885-01A1; M.A.S.]; and Metabolomics Research Core [U24 DK097153; A.K.].
Conflict of Interest: none declared.
References
- Addolorato G. et al. (2000) Ability of baclofen in reducing alcohol craving and intake: II–Preliminary clinical evidence. Alcohol. Clin. Exp. Res., 24, 67–71. [PubMed] [Google Scholar]
- Araki H. et al. (2012) GeneSetDB: A comprehensive meta-database, statistical and visualisation framework for gene set analysis. FEBS Open Bio, 2, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. (2011) Metabolomics: from small molecules to big ideas. Nat. Methods, 8, 117–121. [Google Scholar]
- Bendinskas K. et al. (2011) Enzymatic detection of gamma-hydroxybutyrate using aldo-keto reductase 7A2. J. Forensic Sci., 56, 783–787. [DOI] [PubMed] [Google Scholar]
- Caspi R. et al. (2014) The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res., 42, D459–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagoyen M., Pazos F. (2011) MBRole: enrichment analysis of metabolomic data. Bioinformatics, 27, 730–731. [DOI] [PubMed] [Google Scholar]
- Coletti M.H., Bleich H.L. (2001) Medical subject headings used to search the biomedical literature. J. Am. Med. Inf. Assoc.: JAMIA, 8, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D. et al. (2014) The Reactome pathway knowledgebase. Nucleic Acids Res., 42, D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlinger P.M. (2004) Club drugs: MDMA, gamma-hydroxybutyrate (GHB), Rohypnol, and ketamine. Am. Fam. Phys., 69, 2619–2626. [PubMed] [Google Scholar]
- Gardarin A. et al. (2010) Endoplasmic reticulum is a major target of cadmium toxicity in yeast. Mol. Microbiol., 76, 1034–1048. [DOI] [PubMed] [Google Scholar]
- Harris M.A. et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res., 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W. et al. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Jewison T. et al. (2014) SMPDB 2.0: big improvements to the small molecule pathway database. Nucleic Acids Res., 42, D478–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson P. et al. (2004) A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Anal. Chem., 76, 1738–1745. [DOI] [PubMed] [Google Scholar]
- Kadam S.D. et al. (2012) Cell proliferation and oxidative stress pathways are modified in fibroblasts from Sturge–Weber syndrome patients. Arch. Dermatol. Res., 304, 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res., 40, D109–D114. (Database issue): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky A. et al. (2012) Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics, 28, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P. et al. (2012) Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol., 8, e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza I.R. et al. (2010) Quantitative metabolomics by H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS One, 5, e10538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenna S., Trojanowska M. (2012) The role of endoplasmic reticulum stress and the unfolded protein response in fibrosis. Curr. Opin. Rheumatol., 24, 663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeTourneau J.L. et al. (2008) Baclofen and gamma-hydroxybutyrate withdrawal. Neurocrit. Care, 8, 430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamelak M. et al. (1986) Treatment of narcolepsy with gamma-hydroxybutyrate. A review of clinical and sleep laboratory findings. Sleep, 9, 285–289. [DOI] [PubMed] [Google Scholar]
- Patti G.J. et al. (2012) Innovation: Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol., 13, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl P.L. et al. (2003) Clinical spectrum of succinic semialdehyde dehydrogenase deficiency. Neurology, 60, 1413–1417. [DOI] [PubMed] [Google Scholar]
- Pearl P.L. et al. (2014) Taurine trial in succinic semialdehyde dehydrogenase deficiency and elevated CNS GABA. Neurology, 82, 940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Llamas C., Lopez-Bigas N. (2011) Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One, 6, e19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D.R. et al. (2007) Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia, 9, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R. (1993) The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature, 362, 801–809. [DOI] [PubMed] [Google Scholar]
- Ross R. (1999) Atherosclerosis–an inflammatory disease. N. Engl. J. Med., 340, 115–126. [DOI] [PubMed] [Google Scholar]
- Sartor M.A. et al. (2012) Metab2MeSH: annotating compounds with medical subject headings. Bioinformatics, 28, 1408–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor M.A. et al. (2010) ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics, 26, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas K.M. et al. (2015) Metabolomics and diabetes: analytical and computational approaches. Diabetes, 64, 718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib A. (2003) The role of taurine in cerebral ischemia: studies in transient forebrain ischemia and embolic focal ischemia in rodents. Adv. Exp. Med. Biol., 526, 421–431. [DOI] [PubMed] [Google Scholar]
- Sreekumar A. et al. (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature, 457, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Thiele I. et al. (2013) A community-driven global reconstruction of human metabolism. Nat. Biotechnol., 31, 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urayama S. et al. (2010) Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Commun. Mass Spectrom., 24, 613–620. [DOI] [PubMed] [Google Scholar]
- Vayer P. et al. (1987) Gamma-hydroxybutyrate, a possible neurotransmitter. Life Sci., 41, 1547–1557. [DOI] [PubMed] [Google Scholar]
- Wang T.J. et al. (2011) Metabolite profiles and the risk of developing diabetes. Nat. Med., 17, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2009) PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res., 37, W623–W633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S. (2011) Advances in metabolite identification. Bioanalysis, 3, 1769–1782. [DOI] [PubMed] [Google Scholar]
- Wisloff U. et al. (2005) Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science, 307, 418–420. [DOI] [PubMed] [Google Scholar]
- Xia J. et al. (2012) MetaboAnalyst 2.0 – a comprehensive server for metabolomic data analysis. Nucleic Acids Res., 40, W127–W133. (Web Server issue): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Wishart D.S. (2010) MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res., 38, W71–W77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap I.K. et al. (2010) Metabolome-wide association study identifies multiple biomarkers that discriminate north and south Chinese populations at differing risks of cardiovascular disease: INTERMAP study. J. Proteome Res., 9, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.