Abstract
Background
Bipolar disorder (BD) is a frequent cause of disability, health care costs, and risk of suicide. Pharmacogenetic tests (PGTs) could help clinicians to identify those patients predisposed to the occurrence of adverse events (AEs) improving the understanding of the correlation between genetic variants and drug response.
Materials and methods
The study evaluated 30 patients affected by BD type I or II (according to Diagnostic and Statistical Manual of Mental Disorders, version 5) who underwent the PGT Neurofarmagen® (AB-BIOTICS SA, Barcelona, Spain) between March 2016 and March 2017. The primary aim of this study was to identify if the treatment prescribed by the psychiatrists was consistent with the treatment suggested by the PGT at T0 (corresponding to the test report communication). As a secondary aim, we wanted to assess if clinicians had changed the treatment (in case of discordance) at T1 (3-month follow-up visit) according to the results of the PGT.
Results
At T0, only 4 patients (13%) had an optimal therapy in line with the PGT suggestions. At 3-month follow-up, 13 patients (40%) had received a change of therapy consistent to the test, showing a significant statistical improvement in the Clinical Global Impression item Severity (CGI-S) score over time compared to those not having changes consistent with the test. Regarding AEs, at baseline 9 out of 10 (90%) of the patients who received a therapy modification according to the test presented AEs, and a significant within-group reduction was observed after 3 months (p = 0.031).
Conclusion
Despite the small sample size, the study shows promising data about the usefulness of PGT to support clinicians in reaching a more effective and tolerated treatment in the routine approach of BD.
Keywords: pharmacogenetics testing, bipolar disorder, personalized medicine, pharmacogenetics, adverse events, tolerability, mood disorder
Introduction
Bipolar disorder (BD) is a severe psychiatric disease characterized by mood swings between mania and depression, with a life-time prevalence of approximately 2.4%.1 This is a chronic disease with affective episodes that may produce significant personal distress, social dysfunction and devastating effects on sufferers’ psychological, professional, and social welfare.2,3 Although several effective treatments have been already proposed,4,5 a misdiagnosis of BD may frequently lead to several problems due to drug-resistance, rapid-cycling, and cognitive decline.6,7
It is well established that interpersonal variability in drug response depends on different factors such as diagnostic accuracy, drug–drug interactions, renal and hepatic function, medical and psychiatric comorbidity. Additionally, the drug response can be influenced by genetically determined pharmacokinetic and pharmacodynamic variability,8 and it is known that genetic factors account for 20–40% of differences in individual drug metabolism and response.9,10
Pharmacogenetic tests (PGTs) have been proposed as a method to expedite the process of determining the most efficacious treatment with the lowest side effects, recognizing individual variability in genetics as a key component of drug response.11 Although the Human Genome Project predicted pharmacogenomics would become the approach for predicting drug responsiveness in the standard practice for many disorders and drugs,12 PGTs have been occasionally used in clinical practice up to now and the clinical utility of PGT is an empirical question that has remained largely untested.5
Our hypothesis is that PGTs are a useful tool to offer the most adequate treatment to patients affected by BD and to shorten the time between diagnosis and the finding of the correct therapy.
The primary aim of this study was to identify at T0 (corresponding to the test report communication) if the treatment prescribed by the psychiatrists in patients with a Clinical Global Impression item Severity (CGI-S) ≥ 3 was consistent with the treatment suggested by the PGT Neurofarmagen® (AB-BIOTICS SA, Barcelona, Spain), whose clinical utility in major depression was recently assessed in a multicentric trial.13 As a secondary aim, the study assessed if clinicians changed the treatment (in case of discordance) according to the results of the PGT Neurofarmagen at T1, corresponding to 3-month follow-up visit.
Materials and methods
Study design
This was an observational study with follow-up visits at 3 months, set up in two psychiatric institutes (ASST of Varese, Italy and ASST Santi Paolo and Carlo of Milan, Italy).
Population
Patients affected by BD type I or II (according to Diagnostic and Statistical Manual of Mental Disorders, version 5) who underwent PGT Neurofarmagen between March 2016 and March 2017 were included in the present study; patients had to fulfill the following inclusion criteria:
aged ≥ 18 years old;
written informed consent both for the execution of the test and for the use of the data for research purposes;
a score of CGIs ≥ 3, index of non-clinical stability;
taking the baseline medication for at least 3 months.
Scales
Psychopathological evaluation was carried out using a battery of multiple scales (respectively corresponding to T0, corresponding to the communication of the test report, and T1, corresponding to the 3-month follow-up visit). The scales were:
Clinical Global Impression (CGI), used for patient overall assessment and for monitoring clinical evolution. It takes into consideration three areas: 1, severity of the disease; 2, overall symptom improvement; 3, efficacy of drugs related to their side effects.14 This scale was given at all scheduled appointments and the minimum score of 3 on the severity item served as the patient inclusion criterion.
Hamilton Depression Rating Scale (HDRS): a tool considered as a gold standard for assessing anxiety-depressive symptoms. The most popular version15 is composed of 21 items. Items are differently evaluated: some of them (10) on a 5-point scale (0–4), others (2) on a 4-point scale (0–3) and the remaining 9 on a 3-point scale (0–2). Severity levels are for most well-defined items.16 It was also assessed at different times (T0 and T1).
Young Mania Rating Scale (YMRS): an 11-item scale to assess the severity of manic symptoms. Scoring is obtained on the basis of the subjective symptoms reported by the patient and based on the clinical observation of the patient during the interview. The scale is appropriate both for evaluating maniac symptoms and for evaluating treatment response in patients with Bipolar I and II Diagnosis. As it does not measure depressive symptoms, it has to be co-administered with a scale for depressive symptoms. Four items are rated on a scale of 0–8 points, the remaining 5 items on a scale of 0–4 points. The score obtained must support the clinician in assessing the severity of the symptoms. A score of less than or equal to 12 indicates the remission of symptoms.17 It was also assessed both times (T0 and T1).
Dosage Record and Treatment Emergent Symptom Scale (DOTES): this scale evaluates the onset of side effects in relation to ongoing pharmacological therapy; the fundamental feature of the scale is to investigate not only the presence and severity of the symptoms, during the treatment, but also to determine the likelihood of correlation between symptoms and treatment and to take note of any measures that the appearance of the symptom may have required. The essential element to consider is that the recording of the doses of the treatments is prospective, while that of the undesirable effects is retrospective.18 There is a score for the severity of the symptoms, the judgment on the relationship with treatment and for the overall judgment; the scale used is always 5 points, but the meaning of the score changes: in the case of gravity, 0 is “unrated,” 1 “absent,” 2 “small,” 3 “moderate,” and 4 “severe”; for judgment on the relationship between symptoms and treatment, 0 corresponds to “no relationship,” 1 “remote, <10%,” 2 “possible, 10–50%,” 3 probable, 50–90%,” and 4 “safe, > 90%”; finally, 0 corresponds to “nothing,” 1 “minimum,” 2 “moderate,” 3 “probable,” and 4 “unappreciated.”18 It was also assessed at both times (T0 and T1).
Genetic analysis
Neurofarmagen was used for pharmacogenetic analysis, a PGT for the specific analysis of genetic polymorphisms related to the pharmacokinetics and pharmacodynamics of principles commonly used in neuropsychiatry.
The test evaluates more than 25 different genes (Table S1 for the list of polymorphisms analyzed), putting them in relation to 59 active substances. The test report contains a table where all active substances matched to a color coding: 1) green, expectation of higher likelihood of good response to treatment or a good tolerability profile than average; 2) white, index of a standard response, not different from the general population; 3) yellow, requiring more careful dose monitoring; and 4) red, for high risk of adverse effects or not expected efficacy. The test then allows the clinician to locate the most appropriate dosage for each patient by consulting in advance information on possible side effects of the drug. The genetic polymorphisms analyzed with this genetic test can be grouped into three different categories, depending on the effect they have been associated with:
Drug response: the proteins encoded by these genes are direct or indirect targets of drugs (receptors, signaling pathways, etc.). These genes are crucial for evaluating drug efficacy in the patient.
Risk of unwanted effects: genes that have been associated with adverse effects in subjects receiving the specific psychiatric drugs, and that encode non-metabolic proteins.
Dose (metabolism): genes involved in drug activation, in penetration, and in its elimination rate. Ultimately, the genes controlling the blood levels of the drug.
The administration of the genetic test is carried out on a patient’s saliva sample, collected through a kit; for 30 minutes before the sample collection, the patient should not consume any food, drink, or chewing gum, should not smoke and should have removed any cosmetics from the lips.
After collection, the saliva sample (about 1 ml) remains stable at room temperature for a maximum of 15 days, the time it takes to be sent to the laboratory. The results are available within 10 working days of the sample’s arrival date at the laboratory of AB-BIOTICS S.A., which is in possession of the required authorization to operate as a health laboratory (code E17867643) and to import biological samples.
DNA was extracted from patient saliva samples with the Genomic DNA Isolation Kit (Norgen Biotek Corp. Thorold, ON, Canada). DNA quality was evaluated by 2000 nanodrop microvolume spectrometry. Genotyping of single nucleotide polymorphisms was performed by OpenArray® Technology on the QuantStudio™ 12K Flex Real-Time PCR System (Thermo Fisher Scientific Inc., Waltham, MA, USA) using a custom designed array. CYP2D6 copy number analysis was performed in an Applied Biosystems® 7500 Real-Time PCR System using Hs04083572_cn and Hs04502391_cn TaqMan copy number assays targeting CYP2D6 intron 2 and intron 6, respectively, and RNase P copy number assay as a reference (Thermo Fisher Scientific Inc.).
Conservation of biological material
Saliva samples were tagged with a code associated with the patient and sent to the laboratory of AB-BIOTICS S.A. to extract genomic DNA. The genetic data and the identification code were stored in a key archive at the participating hospitals (Circle Hospital and Macchi Foundation of Varese, Italy and San Carlo Borromeo Hospital in Milan, Italy). This archive containing these correspondences serves to associate each patient’s clinical data with the Neurofarmagen report. At the end of the experiment, the above archive will be destroyed. DNA samples will not be conserved.
All information collected in this study was treated in accordance with the Italian Personal Data Protection Act (D.Lgs. 196/2003). Personal data care was processed electronically with all the criteria to make it confidential and used exclusively for the study.
Genetic data are rendered anonymous in electronic treatment, and after collection it was kept separate from the master data. An encryption system allowed only the person in charge to connect the genetic data to the patients. The collected data was used only for scientific research purposes in aggregate form, thus, anonymously.
Ethics approval
The research protocol was approved by the Ethics Committee of Insubria, Varese on March 1, 2016 (protocol number n 159).
The study was conducted taking into account regulatory requirements and legal requirements (DL n.211, June 24, 2003, and DM December 17, 2004) and in accordance with the ethical principles of the Declaration of Helsinki.
Each recruited patient had seen specific information regarding the request to take part in the study and signed written informed consent, both for the purpose of using the data for study purposes and for the performance of the test.
Statistical analysis
Statistical analysis was performed by Graph-Pad and SAS version 19. Descriptive analysis has been reported as absolute number and percentage (%). The Random Effect Model was used to evaluate the effects of treatment across time in the two subgroups of patients followed up for 3 months. This test allows the estimation of the relationships between two or more variables. It is applicable to a wide range of data types, estimating various types of effects. It is applicable to small comparison groups. The Student’s t-test and the Fisher test were used to assess differences between groups at baseline for continuous and dichotomous variables, respectively, and the McNemar’s test to assess within-group differences across time for dichotomous variables. Significance for all tests was set at p = 0.05, two-tailed.
Results
Demographic and clinical data
The demographic and clinical characteristics of the participants are shown in Table 1. The average age was 54.8 y.o. (SD 15.22), with 52% of males and 48% females. All patients were Caucasian. A total of 52% of patients had a diagnosis of BD type I and 48% of BD type II. At recruitment, 56% of patients suffered from depression, 24% from mania and 20% were in a mix state. The most prescribed mood stabilizers were lithium (28%) and valproate (24%), followed by lamotrigine (8%); among antidepressants, paroxetine (20%) and bupropion (12%) were the most prescribed drugs; among antipsychotics, quetiapine, aripiprazole, and olanzapine were the most prescribed (each one in 16% of the patients). At T0, psychopathological evaluation of the 30 subjects recruited yielded an average CGI-S score of 4.8 (SD 3.7), an average YMRS score of 14.7 (SD 5.6), and an average HDRS score of 18.3 (SD 9).
Table 1.
Demographic characteristics of the patients
| Sociodemographic characteristic | Data | N | % |
|---|---|---|---|
| Gender | Male | 14 | 48 |
| Female | 16 | 52 | |
| Nationality | Italian | 29 | 97 |
| Other | 1 | 3 | |
| Average age | 55 y.o. (SD 15.22) | ||
| Caregiver | 1 | 12 | 40 |
| 2 | 13 | 44 | |
| 3 or more | 5 | 16 | |
| Occupation | Employed | 12 | 40 |
| Unemployed | 5 | 16 | |
| Retired | 11 | 36 | |
| Invalid | 2 | 8 | |
| Years on treatment | 9.5 y (SD 7.2) | ||
| Number of previous treatments | 3.5 (SD 1.3) |
Abbreviations: SD, standard deviation; y.o., years old; y, years.
Primary result
At T0 according to the Neurofarmagen test, 4 patients (13%) received an optimal therapy in line with the test suggestions (i.e. “green color”); for the remaining patients the Neurofarmagen test identified: 8 patients (27%) with a standard therapy (i.e. “white color”), 8 patients (27%) with an idiosyncratic therapy, 7 patients (23%) with an idiosyncratic positive/negative therapy (e.g. a patient can have one genetic variation associated to good response and a second variation in another gene associated to a specific adverse effect), and 3 patients (10%) with a therapy potentially subject to an altered metabolism rate (i.e. “yellow color”).
At the 3-month follow-up evaluation, 13 patients (40%) had received a change of therapy consistent with the Neurofarmagen test; this definition means that the ongoing therapy was according to the test report appropriate for efficacy and/or tolerability, without alteration in metabolic rate or high collateral risk. Overall, 10 patients (32%) maintained a therapy discordant to the test. The other 7 patients (28%) included 4 patients with missing follow-up data, 3 patients who received simultaneously a modification agreeing and a modification not agreeing with the test result at the same time.
Secondary results
A sub analysis of the sample distribution was performed making a comparison between two small subgroups in terms of psychopathology and tolerability. Comparing the subgroup receiving a therapy consistent with the test after the test result (n = 13 patients, 40%) with the subgroup receiving a therapy not consistent with the test after the test result (n = 10 patients, 32%) in term of psychopathology, a significant statistical difference of treatment over time (i.e. treatment × time interaction) in the CGI-S (p < 0.001) emerged: a greater improvement in patients receiving a therapy consistent with the test was observed (Figure 1).
Figure 1.
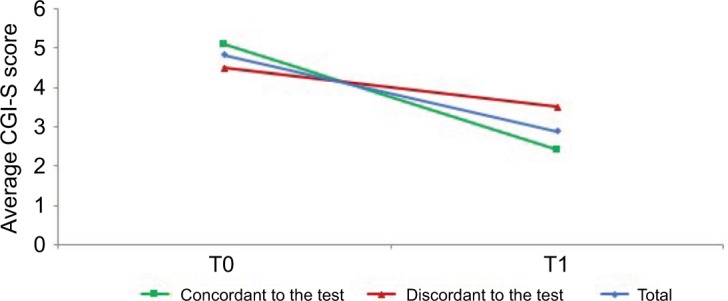
Clinical Global Impression Severity (CGI-S) average scores at baseline (T0) and 3-month follow-up (T1).
Importantly, this effect was still observed when including the baseline HDRS score and baseline AEs as covariates. At the same time, a significant statistical difference over time emerged for HDRS (p = 0.001), with a greater improvement in the subpopulation which received a therapy consistent to the PGT (Figure 2), which was due to this group of patients showing a trend for higher (i.e. worse) HDRS score at baseline (p < 0.1). No significant statistical differences between the two subgroups across time emerged for YMRS (p = 0.9), as shown in Figure 3.
Figure 2.
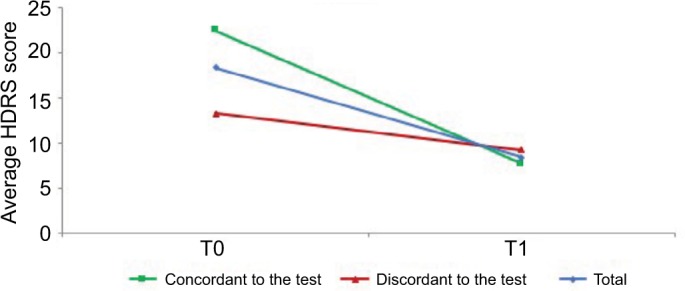
The Hamilton Depression Rating Scale (HDRS) average scores at baseline (T0) and 3-month follow-up (T1).
Figure 3.
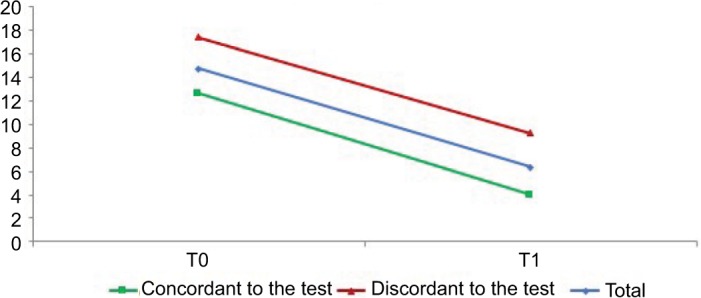
Young Mania Rating Scale (YMRS) average scores at baseline (T0) and 3-month follow-up (T1).
Regarding the adverse events (AEs) recorded through the DOTES scale, an interesting result emerged: at baseline, only 2 out of 10 patients (20%) who did not receive a change in therapy had AEs, while 9 out of 10 (90%) who later received a therapy modification according to the test presented collateral effects (p = 0.013 for the difference at baseline). After 3 months, the incidence of adverse effects in the first subpopulation did not improve, while the second subgroup presented a significant reduction of AEs as shown in Figure 4, with only 3 out of 10 (30%) of patients showing AEs (p = 0.031 for within-group change from baseline).
Figure 4.
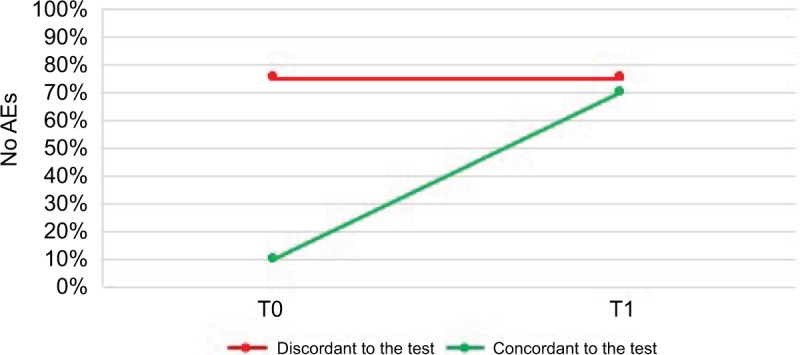
Distribution of the patients according to the percentage of no adverse events (AEs) at baseline (T0) and 3-month follow-up (T1).
Discussion
In psychiatry, it is well known that the discrepancy in response to the same therapies among patients cannot be explained only by physiological, pathological, and environmental factors,19 but frequently arises from multiple factors, not fully understood.20 Being aware of the patient’s pharmacogenetic profile might be helpful to detect the optimal prescription and dosage, and to reduce the trial-and-error-based approach. It is reasonable to suppose that, initially, PGT tests will be performed only in patients whose treatment regimens failed, at least until pharmacogenetics testing become a routine instrument in everyday clinical practice.21 Previous studies on PGT have focused mostly on patients with major depression, although some studies have been performed in different psychiatric populations, sometimes including bipolar patients.21 This study focused on the impact of PGTs exclusively on patients with BD, with a history of disease of an average duration of 9.5 years (SD 7.2).
Our results showed that 4 patients (13%) received an optimal treatment. Despite this data, at 3-month follow-up visit, 13 patients (40%) changed therapy according to the Neurofarmagen test, while 10 patients (32%) maintained a therapy discordant to the test.
Despite some studies showing that psychiatrists’ attitude toward implementing medication use is positive,22,23 in this study some psychiatrists decided to keep the therapy unchanged even if the test suggested a more effective or more tolerated alternative. When occurring, the more frequent reason for changing the initial therapy and following the test results was related to AEs rather than to lack of effectiveness. It is well established that adverse effects represent a huge cost in public health, thus accounting for approximately USD 300 billion on drug prescriptions and USD 136 billion for adverse drug reactions in the US Health Care System in 2014.9 Moreover, adverse effects represent a dramatic issue also in routine clinical practice as previously reported. Clinicians frequently attribute lack of compliance and a consequent high risk of therapeutic failure to AEs.
This result reflects that psychiatrists still have mistrust in following the indications of PGTs. Recent surveys indicate that practicing physicians are aware of PGT and believe that it will represent an important tool in their drug therapy decision-making.24,25 From the comparison sub-analysis between the two subpopulations, it was possible to see how the guided test treatment determined a better outcome in terms of efficacy, particularly on the overall severity of the patient, as already demonstrated in other studies26,27 showing that when PGTs are used to guide the pharmacological treatment of depression, the likelihood of treatment response and remission doubled. Maniac symptoms decreased in both subgroups without differences; this fact may depend on the role of hospitalization and of the passing of time.
In addition, the test proved useful in guiding the clinicians to choose a more tolerable treatment in those patients who complained of collateral effects.
Despite the obvious limitation of this study determined by the small number of the sample, we can affirm that it shows promising data about the usefulness of PGTs in the routine treatment of a complicated pathology such as BD.
Conclusion
The pharmacogenetics approach will evolve the use of medication from a “trial and error” approach to individualized therapy.28 It is necessary to accept the added costs that may be incurred during the transition to genetically guided decisions for choice of drug therapy. In the long run, decreasing the frequency of adverse drug effects and increasing the probability of successful therapy will probably lower health care costs.28 Moving forward, there is a definite need to integrate psychopharmacology with the pharmacogenetics approach to include the development of cost-effective assessment and treatment strategies that have high probability of success in challenging intervention settings.
An interesting future study concerns the evaluation of a wider multidisciplinary patient sample, with a randomized design, and a mirror analysis about the comparison in terms of days of hospitalization and access to emergency services before and after a test-guided therapy approach.
Availability of data and material
The datasets generated and analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Supplementary material
Table S1.
List of genes and polymorphisms analyzed
| Gene symbol | Gene name | Polymorphisms |
|---|---|---|
| ABCB1 | ATP binding cassette subfamily B member 1 | rs2235048, rs11983225 |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | rs1130214 |
| BDNF | Brain-derived neurotrophic factor | rs6265 |
| CACNG2 | Calcium channel, voltage-dependent, gamma subunit 2 | rs2284017 |
| CES1 | Carboxylesterase 1 | rs71647871 |
| COMT | Catechol-O-methyltransferase | rs4680 |
| CRHR1 | Corticotropin releasing hormone receptor 1 | rs4792888 |
| CYP1A2 | Cytochrome P450 family 1 subfamily A member 2 | *1, *1F |
| CYP2B6 | Cytochrome P450 family 2 subfamily B member 6 | *1, *6 |
| CYP2C19 | Cytochrome P450 family 2 subfamily C member 19 | *1, *2, *3, *5, *7, *8, *17, *27 |
| CYP2C9 | Cytochrome P450 family 2 subfamily C member 9 | *1, *2, *3, *6, *8, *27 |
| CYP2D6 | Cytochrome P450 family 2 subfamily D member 6 | *1, *2, *2A, *3, *4, *5, *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, *19, *20, *29, *35, *30, *40, *41, *69, *1xN, *2xN, *35×2 |
| CYP3A4 | Cytochrome P450 family 3 subfamily A member 4 | *1, *22 |
| DDIT4 | DNA damage inducible transcript 4 | rs1053639 |
| DRD3 | Dopamine receptor D3 | rs963468 |
| EPHX1 | Epoxide hydrolase 1, microsomal (xenobiotic) | rs1051740 |
| FCHSD1 | FCH and double SH3 domains 1 | rs456998 |
| GRIK2 | Glutamate receptor, ionotropic, kainate 2 | rs2518224 |
| GRIK4 | Glutamate receptor, ionotropic, kainate 4 | rs1954787 |
| HLA-A | Major histocompatibility complex, class I, A | rs1061235 |
| HTR1A | 5-HTT (serotonin) receptor 1A, G protein-coupled | rs10042486 |
| HTR2A | 5-HTT (serotonin) receptor 2A, G protein-coupled | rs6311, rs6314, rs9316233 |
| HTR2C | 5-HTT (serotonin) receptor 2C, G protein-coupled | rs1414334 |
| LPHN3 | Latrophilin 3 | rs6551665 |
| NEFM | Neurofilament, medium polypeptide | rs1379357, rs1457266 |
| OPRM1 | Opioid receptor, mu 1 | rs1799971 |
| RGS4 | Regulator of G-protein signaling 4 | rs2661319 |
| RPTOR | Regulatory associated protein of MTOR, complex 1 | rs7211818 |
| SLC6A4 | Solute carrier family 6 (neurotransmitter transporter), member 4 | 5-HTTLPR |
| UGT2B15 | UDP glucuronosyltransferase 2 family, polypeptide B15 | rs1902023 |
Footnotes
Author contributions
All authors contributed towards data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
JE and MT are employed by AB-BIOTICS, S.A. Cugat del Vallès (Barcelona). The other authors report no conflicts of interest in this work.
References
- 1.Merikangas KR, Jin R, He JP, et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch Gen Psychiatry. 2011;68(3):241–251. doi: 10.1001/archgenpsychiatry.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips ML, Kupfer DJ. Bipolar disorder diagnosis: challenges and future directions. Lancet. 2013;381(9878):1663–1671. doi: 10.1016/S0140-6736(13)60989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez-Pinto A, Vieta E, Reed C, et al. Effectiveness of olanzapine monotherapy and olanzapine combination treatment in the long term following acute mania – results of a two year observational study in bipolar disorder (EMBLEM) J Affect Disord. 2011;131(1–3):320–329. doi: 10.1016/j.jad.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 4.John M, Eisenberg Center for Clinical Decisions and Communications Science Antipsychotic medicines for treating schizophrenia and bipolar disorder: a review of the research for adults and caregivers. AHRQ. Comparative Effectiveness Reviews. 2005–2013 Apr 10; [PubMed] [Google Scholar]
- 5.Salloum NC, McCarthy MJ, Leckband SG, Kelsoe JR. Towards the clinical implementation of pharmacogenetics in bipolar disorder. BMC Med. 2014;12:90. doi: 10.1186/1741-7015-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nasrallah HA. Consequences of misdiagnosis: inaccurate treatment and poor patient outcomes in bipolar disorder. J Clin Psychiatry. 2015;76(10):e1328. doi: 10.4088/JCP.14016tx2c. [DOI] [PubMed] [Google Scholar]
- 7.Peedicayil J. Epigenetic approaches for bipolar disorder drug discovery. Expert Opin Drug Discov. 2014;9(8):917–930. doi: 10.1517/17460441.2014.922537. [DOI] [PubMed] [Google Scholar]
- 8.Mrazek DA, Biernacka JM, McAlpine DE, et al. Treatment outcomes of depression: the pharmacogenomic research network antidepressant medication pharmacogenomic study. J Clin Psychopharmacol. 2014;34(3):313–317. doi: 10.1097/JCP.0000000000000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saldivar JS, Taylor D, Sugarman EA, et al. Initial assessment of the benefits of implementing pharmacogenetics into the medical management of patients in a long-term care facility. Pharmgenomics Pers Med. 2016;9:1–6. doi: 10.2147/PGPM.S93480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ventola CL. Role of pharmacogenomic biomarkers in predicting and improving drug response: part 1: the clinical significance of pharmacogenetics variants. P.T. 2013;38(9):545–560. [PMC free article] [PubMed] [Google Scholar]
- 11.Knisely MR, Carpenter JS, Von Ah D. Pharmacogenomics in the nursing literature: an integrative review. Nurs Outlook. 2014;62:285–296. doi: 10.1016/j.outlook.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Collins FS, McKusick VA. Implications of the Human Genome Project for medical science. JAMA. 2001;285(5):540–544. doi: 10.1001/jama.285.5.540. [DOI] [PubMed] [Google Scholar]
- 13.Pérez V, Salavert A, Espadaler J, et al. Efficacy of prospective pharmacogenetic testing in the treatment of major depressive disorder: results of a randomized, double-blind clinical trial. BMC Psychiatry. 2017;17(1):250. doi: 10.1186/s12888-017-1412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunlop BW, Gray J, Rapaport MH. Transdiagnostic clinical global impression scoring for routine clinical settings. Behav Sci (Basel) 2017;7(3):E40. doi: 10.3390/bs7030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Pina JA, Sánchez-Meca J, Rosa-Alcázar AI. The Hamilton Rating Scale for Depression: meta-analytic reliability generalization study. Int J Clin Health Psychol. 2009;9(1):143–159. [Google Scholar]
- 16.Giusti E, Pacificio M, Fiume G. Disturbo dell’ umore. Dalla diagnosi DSM-5 al trattamento: Sovera Edizioni. 2014 [Google Scholar]
- 17.Young AH, Ferrier IN, Michalak E. Practical Management of Bipolar Disorder. Cambridge, UK: Cambridge University Press; 2010. [Google Scholar]
- 18.Conti L. Repertorio delle scale di valutazione in psichiatria. Firenze: Società editrice europea di Nicodeno Maggiulli & Co.; 2000. [Google Scholar]
- 19.Wilffert B, Zaal R, Brouwers JR. Pharmacogenetics as a tool in the therapy of schizophrenia. Pharm World Sci. 2005;27(1):20–30. doi: 10.1007/s11096-004-1731-4. [DOI] [PubMed] [Google Scholar]
- 20.Bolla E, Bortolaso P, Ferrari M, et al. Letter to the Editor. Are CYP1A2*1F and *1C associated with clozapine tolerability? A preliminary investigation. Psychiatry Res. 2011;189(3):483. doi: 10.1016/j.psychres.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Espadaler J, Tuson M, Lopez-Ibor JM, Lopez-Ibor F, Lopez-Ibor MI. Pharmacogenetic testing for the guidance of psychiatric treatment: a multicenter retrospective analysis. CNS Spectr. 2017;22(4):315–324. doi: 10.1017/S1092852915000711. [DOI] [PubMed] [Google Scholar]
- 22.Hoop JG, Lapid MI, Paulson RM, Roberts LW. Clinical and ethical considerations in pharmacogenetics testing views of physicians in 3 “early adopting” departments in psychiatry. J Clin Psychiatry. 2010;71(6):745–753. doi: 10.4088/JCP.08m04695whi. [DOI] [PubMed] [Google Scholar]
- 23.Thompson C, Hamilton SP, Hippman C. Psychiatrist attitude towards pharmacogenetics testing, direct-to-consumer genetic testing, and integrating genetic counseling into psychiatric patient care. Psych Res. 2015;226:68–72. doi: 10.1016/j.psychres.2014.11.044. [DOI] [PubMed] [Google Scholar]
- 24.Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet. 2012;82(4):388–394. doi: 10.1111/j.1399-0004.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JA. Pharmacogenetics in clinical practice: how far have we come and where are we going? Pharmacogenomics. 2013;14(7):835–843. doi: 10.2217/pgs.13.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall Flavin DK, Winner JC, Allen JD, et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2013;23(10):535–548. doi: 10.1097/FPC.0b013e3283649b9a. [DOI] [PubMed] [Google Scholar]
- 27.Winner JC, Carhart J, Altar A, Allen J, Dechairo B. A prospective randomized double- blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219–227. [PubMed] [Google Scholar]
- 28.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429(6990):464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
List of genes and polymorphisms analyzed
| Gene symbol | Gene name | Polymorphisms |
|---|---|---|
| ABCB1 | ATP binding cassette subfamily B member 1 | rs2235048, rs11983225 |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | rs1130214 |
| BDNF | Brain-derived neurotrophic factor | rs6265 |
| CACNG2 | Calcium channel, voltage-dependent, gamma subunit 2 | rs2284017 |
| CES1 | Carboxylesterase 1 | rs71647871 |
| COMT | Catechol-O-methyltransferase | rs4680 |
| CRHR1 | Corticotropin releasing hormone receptor 1 | rs4792888 |
| CYP1A2 | Cytochrome P450 family 1 subfamily A member 2 | *1, *1F |
| CYP2B6 | Cytochrome P450 family 2 subfamily B member 6 | *1, *6 |
| CYP2C19 | Cytochrome P450 family 2 subfamily C member 19 | *1, *2, *3, *5, *7, *8, *17, *27 |
| CYP2C9 | Cytochrome P450 family 2 subfamily C member 9 | *1, *2, *3, *6, *8, *27 |
| CYP2D6 | Cytochrome P450 family 2 subfamily D member 6 | *1, *2, *2A, *3, *4, *5, *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, *19, *20, *29, *35, *30, *40, *41, *69, *1xN, *2xN, *35×2 |
| CYP3A4 | Cytochrome P450 family 3 subfamily A member 4 | *1, *22 |
| DDIT4 | DNA damage inducible transcript 4 | rs1053639 |
| DRD3 | Dopamine receptor D3 | rs963468 |
| EPHX1 | Epoxide hydrolase 1, microsomal (xenobiotic) | rs1051740 |
| FCHSD1 | FCH and double SH3 domains 1 | rs456998 |
| GRIK2 | Glutamate receptor, ionotropic, kainate 2 | rs2518224 |
| GRIK4 | Glutamate receptor, ionotropic, kainate 4 | rs1954787 |
| HLA-A | Major histocompatibility complex, class I, A | rs1061235 |
| HTR1A | 5-HTT (serotonin) receptor 1A, G protein-coupled | rs10042486 |
| HTR2A | 5-HTT (serotonin) receptor 2A, G protein-coupled | rs6311, rs6314, rs9316233 |
| HTR2C | 5-HTT (serotonin) receptor 2C, G protein-coupled | rs1414334 |
| LPHN3 | Latrophilin 3 | rs6551665 |
| NEFM | Neurofilament, medium polypeptide | rs1379357, rs1457266 |
| OPRM1 | Opioid receptor, mu 1 | rs1799971 |
| RGS4 | Regulator of G-protein signaling 4 | rs2661319 |
| RPTOR | Regulatory associated protein of MTOR, complex 1 | rs7211818 |
| SLC6A4 | Solute carrier family 6 (neurotransmitter transporter), member 4 | 5-HTTLPR |
| UGT2B15 | UDP glucuronosyltransferase 2 family, polypeptide B15 | rs1902023 |


